Recent studies on dendritic cell (DC)–associated genes have been performed using monocyte-derived DCs (MoDCs) in different maturation stages. In our approach, to uncover the novel DC-associated genes and their expression profiles among the different DC subsets, we constructed a subtracted DC-cDNA library from CD1a+, CD14+, and CD11c− DCs by subtracting the genes shared with T cells, B cells, and monocytes, and we then screened the libraries with the aid of microarray technique. The genes showing remarkable specificity to DCs in the microarray analysis were selected and confirmed by semiquantitative reverse transcriptase–polymerase chain reaction. Our investigations revealed the following: (1) Genes highly expressed in myeloid DCs are those involved in antigen uptake/processing/presentation, cell metamorphosis, or chemotaxis. (2) Most of the genes previously identified in MoDCs, such as TARC, ferritin L-chain, lysosomal acid lipase, α- and β-tubulin, osteopontin (Eta-1), and others, are not markedly expressed in CD11c− DCs regardless of their maturation status. On the other hand, specific transcription factors and MHC class II molecules, such as interferon regulatory factor-4 (IRF4) and HLA-DR, are similarly expressed in both DC subsets. (3) CD14+ DCs retain unique features of tissue DCs, as evidenced by the gene expression profile of “no CCR7 but more CCR1” and “no TARC but abundant MCP1 and Eta-1.” (4) The genes for immunoglobulin (Ig) superfamily Z39Ig, CD20-like precursor, glycoprotein NMB (GPNMB), transforming growth factorβ (TGF-β)–induced protein (TGFBI), myeloid DAP12-associated lectin (MDL-1), and 6 novel genes are newly identified as being associated with the phenotypic expression of the DC subsets. These identifications provide important molecular information for further functional studies of the DC subsets.
Introduction
Dendritic cells (DCs) are specialized to modulate T-cell immunity, either by priming or tolerizing antigen (Ag)–specific T cells, depending on the exact physiological conditions, such as the nature and amount of Ag and the presence of DC-maturating stress signals.1-4 While constituting less than 1% of the total mononuclear cells in mouse spleen and human peripheral blood, DCs are present ubiquitously in all tissues, even in the human central nervous system.5 Unlike other immune cells, DCs arise, upon different signals, from many different progenitor cells of myeloid or lymphoid origin.6-8 The heterogeneity of the DC population is well demonstrated by the multiple DC subsets in human blood and mouse spleens. Although the ontogeny of each type of DC remains unclear, the presence of multiple distinct DC lineages in both humans and mice has raised the possibility that distinct DC subsets might have unique functions in recruiting distinct types of immune responses.9-12 Intriguingly, even for a given type of DC, there is considerable plasticity in DC functions depending on the maturation stage and the duration of Ag exposure, resulting in different outcomes of DC-mediated immune triggering.13-17
Due to their pivotal role in immune induction and tolerance, DCs have been explored for their usefulness in the control of malignant cancers and autoimmune diseases in mouse models.18,19 However, considering the heterogeneity of naturally occurring DCs, the current DC study in association with human immunotherapy might have been skewed in monocyte-derived DCs (MoDCs). Indeed, many clinical trials using MoDCs are being undertaken by independent workers to elicit tumor-specific immunities.20 21 Increasing pressure from translational research, however, necessitates the study of other human DCs, which might be useful to control harmful immune responses, such as autoimmunity and graft rejection.
In the last few years, advances in methodology have enabled us to access various human DCs of high purity and good quantity.22-25 To have better insight into the unique capacities of distinct DC subsets, attempts have been made to disclose DC-associated genes and their expression patterns using a high-throughput analysis system. Several independent approaches have been made to reveal the genes highly expressed in MoDCs by employing sequential analysis of gene expression (SAGE)26,27 or a cDNA microarray system.28 29 The expression profiles from these studies are generally in good agreement with each other and are sufficient to arrive at a consensus about genes that are highly expressed in MoDCs. However, it remains to be seen whether these genes are also prominent in other types of DCs.
In the present study, we attempted to pool out the “DC-associated genes” from 3 different DC subsets, namely, CD11c− DCs isolated from peripheral blood,23CD1a+ DCs, and CD14+DCs24,25,30 31 generated from CD34+hematopoietic progenitor cells. To identify DC-associated messages, we performed cDNA subtraction and microarray analyses and performed sequencing. We have identified a set of DC-specific genes, including 63 known genes and 6 novel genes. These genes were further examined for their expression profiles in each DC subset and other leukocytes by semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR) and microarray analysis. In this approach, we found DC-associated expression of a group of genes, including the ones that have not been reported in similar studies with MoDCs. We also found that gene expression profiles are markedly different between lymphoid (CD11c−) and myeloid (CD1a+, CD14+, and MoDCs) DCs and are also discernable even among the myeloid DC subsets. These newly identified DC-associated genes and their expressions among the different DC subsets may provide information about the molecular features of each DC subset, which we hope will lead us to envisage future applications of a distinct DC subset in the clinical field.
Materials and methods
Cell and RNA preparations
The generation or isolation procedures of each DC subset are summarized in Figure 1A. CD1a+ DCs or CD14+ DCs were generated from CD34+ progenitor cells isolated from umbilical-cord blood. Mononuclear cells from cord blood were obtained by a standard Ficoll density gradient method (density 1.077 g/mL). CD34+cells were isolated using a MACS Separation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany) and then were seeded for expansion in 12-well culture plates (NUNC, Rochester, NY) at 2 × 105 cells per milliliter. Cultures were established in RPMI 1640 medium (GIBCO, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum, either in the presence of tumor necrosis factor-α (TNF-α) (100 U/mL, Endogen, Rockford, IL) plus interleukin-3 (IL-3) (100 U/mL, Endogen) for CD14+ DCs or in the presence of TNF-α (100 U/mL) plus granulocyte-macrophage colony-stimulating factor (GM-CSF) (500 U/mL, LG Chem, Daejeon, Korea) for CD1a+ DCs. Optimal conditions were maintained by splitting these cultures every 2 days using medium containing fresh factors at a cell density of 2 × 105/mL. Cultures of 8 days and 18 days, respectively, for immature and mature nonadherent DCs were collected, stained with anti-CD1a–phycoerythrin and anti-CD14–fluorescein isothiocyanate (Becton Dickinson, Sunnyvale, CA), and then sorted for single-positive cells using FACSort (Becton Dickinson).
Generation, isolation, and characterization of different DC subsets.
(A) A brief scheme for the preparation of each DC subset. Each subset of DCs was prepared as described in “Materials and methods.” CD1a+ DCs and CD14+ DCs were generated from CD34+ hematopoietic progenitor cells in the presence of TNF-α/GM-CSF and TNF-α/IL-3, respectively. CD1a+ DCs and CD14+ DCs were sorted according to their surface phenotype at day 8 and day 18. CD11c− DCs were either freshly isolated from PBMCs at day 0 or matured for 5 days in the presence of IL-3 and CD40L. MoDCs were derived from monocytes by culturing with IL-4 and GM-CSF for 7 days and then cultured for 2 more days for maturation in monocyte-conditioned medium supplemented with TNF-α. Lin− means TCR−CD14−CD16−CD19−CD56−. (B) Surface phenotype of each DC subset in immature and mature stages. Cells were stained with fluorescence-conjugated monoclonal antibodies (Becton Dickinson) shown above each image. Photographs of the DC subset on the right were taken on day 18 for CD1a+ DCs and CD14+ DCs, day 5 for CD11c− DCs, and day 9 for MoDCs, respectively (magnification × 400).
Generation, isolation, and characterization of different DC subsets.
(A) A brief scheme for the preparation of each DC subset. Each subset of DCs was prepared as described in “Materials and methods.” CD1a+ DCs and CD14+ DCs were generated from CD34+ hematopoietic progenitor cells in the presence of TNF-α/GM-CSF and TNF-α/IL-3, respectively. CD1a+ DCs and CD14+ DCs were sorted according to their surface phenotype at day 8 and day 18. CD11c− DCs were either freshly isolated from PBMCs at day 0 or matured for 5 days in the presence of IL-3 and CD40L. MoDCs were derived from monocytes by culturing with IL-4 and GM-CSF for 7 days and then cultured for 2 more days for maturation in monocyte-conditioned medium supplemented with TNF-α. Lin− means TCR−CD14−CD16−CD19−CD56−. (B) Surface phenotype of each DC subset in immature and mature stages. Cells were stained with fluorescence-conjugated monoclonal antibodies (Becton Dickinson) shown above each image. Photographs of the DC subset on the right were taken on day 18 for CD1a+ DCs and CD14+ DCs, day 5 for CD11c− DCs, and day 9 for MoDCs, respectively (magnification × 400).
CD11c− DCs were immunomagnetically isolated from peripheral blood using a BDCA-4 Cell Isolation Kit (Miltenyi Biotec). For matured DCs, freshly isolated CD11c− DCs were cultured for 5 days in RPMI 1640 medium supplemented with 10% autologous human serum, 200 U/mL IL-3 (Endogen), and 5 μg/mL human recombinant CD40L (expressed, purified, and endotoxin-tested in our laboratory).
T lymphocytes were purified from peripheral blood mononuclear cells (PBMCs) by immunoaffinity depletion using a T-cell isolation kit (Pierce, Rockford, IL). B lymphocytes were obtained from whole blood using RossettSep (StemCell Technologies, Vancouver, BC, Canada). Monocytes were purified from PBMCs based on their tendency to adhere to human γ globulin–coated plates.
MoDCs were generated from adherent mononuclear cells. PBMCs were seeded in 6-well culture plates at a density of 5 × 106/mL, allowed to adhere for 1 hour at 37°C, and nonadherent cells were washed away with prewarmed RPMI 1640. Adherent cells were cultured for 7 days in RPMI 1640 medium supplemented with 10% autologous human serum and 1000 U/mL each of IL-4 (Endogen) and GM-CSF (LG Chem). Media were refreshed on days 3 and 5. On day 7, nonadherent cells were collected as immature MoDCs by moderately vigorous agitation. For matured MoDCs, the nonadherent cells of day 7 were cultured for 2 more days in a monocyte-conditioned medium (final concentration 50%, vol/vol) supplemented with 10 ng/mL TNF-α (Pharmingen, San Diego, CA). The dead cells and contaminating lymphocytes were removed by Nycodenz density gradient centrifugation.32 To get CD1a− MoDCs at day 9, autologous human serum was deliberately used as a culture supplement.22
The total RNA was extracted from each DC subset using Trizol reagent (Life Technologies, Carlsbad, CA), and mRNA was affinity-purified by a polyATtrack system (Promega, Madison, WI).
Generation of the subtracted DC-cDNA library
The cDNAs were synthesized as reported previously33 34 using 200 units of Superscript II (Life Technologies), and 200 ng total RNA was extracted from DC subsets and leukocytes. Subtraction was performed as described in the PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, CA). Dendritic cells (CD1a+, CD14+, and CD11c− DCs) were used as testers, while B cells, monocytes, and T cells (BMTs) were used as drivers. To overcome the limitation of the DC supply, DC-cDNA was preamplified using a SMART PCR cDNA Synthesis Kit (Clontech). Similarly, BMT-cDNAs were preamplified in parallel. Amplified cDNAs were mixed either as testers (DC-cDNAs) or as drivers (BMT-cDNAs) and underwent RsaI digestion and adaptor ligation sequentially. Subtractive hybridization was performed twice in a 30-fold molar excess of driver overtester, to remove cDNAs shared with BMTs, resulting in enrichment of DC-specific cDNAs. The cDNAs were then selectively amplified by the 2 consecutive PCRs: the first one for 27 cycles and the second nested one for 12 cycles. The subtracted cDNAs were inserted into a pGEM-T Easy Vector (Promega) and transformed intoEscherichia coli DH5α to generate the DC-specific cDNA library.
Colony PCR and microarray fabrication
For the DC microarray fabrication, 1920 colonies were randomly selected from the subtracted DC-cDNA library. In addition to the DC-cDNA genes, 124 genes, including many CD (cluster of differentiation) and cytokine genes were purchased from Incyte Genomics Inc. (Palo Alto, CA) and then confirmed by DNA sequencing. Additionally, 57 CD genes that were not available from Incyte were PCR-cloned in this laboratory. Three plant genes, agpL(AF184598), agpS (AF184597), and mt45(AF320905), were included as spike genes. The clones were cultured in 96-well plates for PCR. PCR reactions were performed in 100 μL volume with amidated vector-specific primers (lab1 5′-GTGCTGCAAGGCGATTAAG-3′, lab2 5′-GGAATTGTGAGCGGATAAC-3′) for 30 cycles (30 seconds at 94°C, 30 seconds at 62°C, and 1 minute and 30 seconds at 72°C). Amplified DNAs were dissolved in 3 × SSC and then printed on microarrays with Q-bot (Genetix, New Milton, United Kingdom).
Based on the result of DC/BMT differential analysis on DC chips, another DC microarray (HI380 chip; Creagene, Daejeon, Korea) was fabricated with 71 DC-specific genes of high significance. In addition to the DC-specific genes, it included 307 known genes encoding CD antigens, cytokines, chemokines, cytokine receptors, and chemokine receptors, either purchased from Incyte or cloned in this laboratory. The genes were mounted in duplicate.
The complete list of the genes can be accessed athttp://www.creagene.com/Ncreagene/dnachip/genelist.html.
Microarray analysis
In DC/BMT differential analysis on DC chips, the probes were the forward- and the reverse-subtracted cDNAs. In DC subset-specific analysis on HI380 chips, the probes were the amplified cDNA of each DC subset.
The probes were prepared as follows: 1 μg of the cDNA was mixed with 20 to 100 pg of the plant spike DNAs and then fluorescently labeled with either Cy3 or Cy5 dye by the random priming method using Klenow fragment (NEB, Beverly, MA). The labeled cDNAs were purified through ethanol precipitation at room temperature with 2 volumes of ethanol and resuspended in 40 μL of 4 × SSC, 0.2% sodium dodecyl sulfate (SDS), 0.1 μg/μL poly(dA), 0.1 μg/μL yeast tRNA, and 0.25 μg/μL Cot1 DNA.
Finally, the labeled probes were denatured at 100°C for 5 minutes and applied to the microarray for hybridization at 55°C for 12 to 16 hours and then followed by several washings.
Fluorescent images of hybridized microarrays were obtained using a Scanarray 4000 microarray scanner (GSI Lumonics, Northville, MI), and the images were analyzed with GenePix Pro 3.0 (Axon Instruments, Union City, CA). The photomultiplier tube (PMT) and the laser value for scanning were tuned by equalizing the intensities of Cy3 and Cy5 on a spike gene. Fluorescence ratios were calibrated by applying normalization factors calculated from the mean intensity of spike genes (over 6 spots on each microarray).
Back-hybridization
For the clones randomly pooled from the subtracted DC-cDNA library, the redundancy of each clone was examined by its frequency of identification in the sequencing analysis. The clones of high redundancy were then PCR amplified with primers flanking the T-vector insertion site (sense 5′-TGCTCCCGGCCGCCAT, antisense 5′-CGGCCGCGAATTCACTAG). The amplified clones were collected, labeled with Cy3 or Cy5, and then hybridized with the DC microarray (DC chip, Creagene). The single-stranded vector DNA was prepared by asymmetric PCR with lab1 primer using self-ligated pGEM T-Easy PCR product (lab1- and lab2-primed) as a template. To minimize the background hybridization between vector sequences, the single-stranded vector DNA was included in the hybridization reaction as a blocking DNA. Clones identified with an intensity value of higher than 10 000 were screened out.
Sequence analysis
Following back-hybridization, the nonredundant DC-specific clones were recovered from the cell stock and each insert in the pGEM T-Easy was amplified with M13 forward and reverse primers. The PCR products were then sequenced with the Big Dye Terminator Kit (Perkin-Elmer, Boston, MA) and analyzed with a 377 ABI automated 96-lane sequencer (Perkin-Elmer). Approximately 200 to 700 bp sequences were trimmed for vector sequence with Seqman (DNAstar, Madison, WI) and were analyzed with an Advanced BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/).
Quantitative PCR
The initial cDNA content in each sample was normalized with an amount of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Quantitative PCR reactions were performed in a 50 μL volume using 4 ng of each cDNA on a Perkin-Elmer DNA thermocycler 9600 Prism for 30 cycles (15 seconds at 94°C, 20 seconds at 55°C, and 1 minute at 72°C). To evaluate the specificity of each message semiquantitatively, 10 μL each of the PCR product was withdrawn from 25 cycles and from 30 cycles, respectively, and then run simultaneously on 1.1% agarose gels. PCR primers to each selected clone were designed with PrimerSelect (DNAstar). The expected sizes of the PCR products were 300 to 600 bp, and the optimal annealing temperature ranged from 55°C to 65°C. The sequence of the RT-PCR primers is available from the authors upon request.
Results
Immunophenotypes of purified dendritic cells
The purity of CD1a+ DCs, CD14+ DCs, or CD11c− DCs for the construction of the subtracted DC-cDNA library was 90% ± 4%, and the purities of each DC subset in the additional experiments were more than 98% after cell sorting or isolation (Figure 1A). CD1a+ DCs and CD14+ DCs at day 18 were strikingly distinguished not only by their surface phenotypes but also by their morphologies with and without well-developed dendrites, respectively (Figure 1B). However, these 2 DCs were very similar in their levels of HLA-DR, CD83, and CD86 expression. The expression of DC-Lamp was observed only in CD1a+ DCs but not in CD14+ DCs at day 18. In this specific batch of CD14+ DCs (Figure 1B), the up-regulation of CD83 was observed in parallel with an unusual decrease in the levels of HLA-DR and CD86 during their development between days 8 and 18. While Langerin staining was expected in CD1a+ DCs between days 8 and 12,35 CD1a+ DCs at day 8 were not stained by Langerin monoclonal antibody, probably indicating a kinetic variation between different cultures of CD1a+ DCs. The expression of Langerin at day 18 in CD1a+ DCs was not apparent in their immunostaining, although the microarray analysis on the same DCs revealed the up-regulated Langerin expression at day 18 in CD1a+ DCs (Table 2). Thus, CD1a+ DCs at day 18 of their maturation were considerably nonexpressive of the Langerin-positive phenotype.35 Unlike these cytokine-induced DCs, CD11c− DCs freshly isolated from peripheral blood barely expressed CD86 on their cell surfaces and were relatively small and even in size (Figure 1B). On the other hand, CD11c− DCs at day 5 expressed significant levels of CD86 as well as of DC-Lamp. Interestingly, the absence of CD83 up-regulation in the 5-day maturation period of CD11c− DCs was in good contrast with the maturation-associated expression of CD83 in the other DC subsets. The representative phenotypes of MoDCs were CD1a+/CD83−/DC-Lamp− at day 7 and CD1a−/CD83+/DC-Lamp+ at day 9. However, regarding the level of CD1a expression in MoDCs, there was some degree of difference, depending on the donor.
The subtracted DC-cDNA library was very specific to the DC subset
To gain direct access to DC-specific genes without being hampered by highly abundant messages shared by most leukocytes, we have employed a DC cDNA subtraction strategy followed by microarray analysis (Figure2). A subtracted DC-cDNA library was constructed by subtracting B-cell, monocyte, and T-cell messages concurrently from the combined ones of CD1a+, CD11c−, and CD14+ DCs. In this subtraction, we carried out the modified subtractive hybridization, termed PCR-Select,36 which exploits suppressive PCR to selectively enrich the subtracted genes. To compare the specificity of subtraction, not only forward (DCs subtracted by BMT) but also reverse subtraction (BMT subtracted by DCs) was performed in parallel. The profile of the PCR products from either subtraction revealed a unique pattern of discrete bands on agarose gel that was absent in the nonsubtracted control (data not shown.) To examine the integrity of the subtracted DC-cDNA library, 8 clones were randomly selected and sequenced. Sequence analysis revealed 2 cDNAs that corresponded with the sequences from the MMP12 gene, 3 cDNAs from the mitochondrial genes, and 2 novel expressed sequence tags (ESTs). The high efficacy of subtraction was indicated by the absence of cDNAs corresponding to well-known housekeeping genes among these randomly picked clones. To further assess the integrity of subtraction, the forward- and reverse-subtracted cDNAs were labeled with Cy3 and Cy5, respectively, and then hybridized on a microarray with 2000 known genes (a generous gift from Dr J. H. Park at Korea Research Institute of Bioscience and Bioengineering, Daejeon, Korea) in double-blind approaches. Most of the spots developed single fluorescence of either Cy3 or Cy5, and few developed dual fluorescence of both Cy3 and Cy5. These results suggest that the subtraction was successfully performed for the depletion of the common messages in the 2 cDNA populations, so that the subtracted cDNA was acceptable as a specific probe for each population in the following microarray analysis.
Entire strategy for the identification and characterization of DC-associated genes.
DC denotes an equal mixture of 3 different DC subsets, including CD1a+, CD11c−, and CD14+ DCs. BMT denotes the mixture of B cells, monocytes, and T cells in equal amounts. The cDNA subtractions were performed in either direction. Subtracted DC cDNA clones were immobilized on a microarray and subjected to differential screening. Among the clones selected for DC-preferred transcription, redundant clones were screened out and the remainder were further investigated for sequence identity. For the ones of interest, a DC subset–specific expression profile was elucidated by a newly designed microarray and RT-PCR analysis.
Entire strategy for the identification and characterization of DC-associated genes.
DC denotes an equal mixture of 3 different DC subsets, including CD1a+, CD11c−, and CD14+ DCs. BMT denotes the mixture of B cells, monocytes, and T cells in equal amounts. The cDNA subtractions were performed in either direction. Subtracted DC cDNA clones were immobilized on a microarray and subjected to differential screening. Among the clones selected for DC-preferred transcription, redundant clones were screened out and the remainder were further investigated for sequence identity. For the ones of interest, a DC subset–specific expression profile was elucidated by a newly designed microarray and RT-PCR analysis.
Several DC-associated genes were newly identified by microarray analysis
To identify DC-associated genes, 1920 clones from the subtracted DC library and 181 cDNAs of CD and cytokine genes were immobilized on a glass slide and subjected to differential hybridization using cDNA probes manipulated as follows: The forward-subtracted (DC-specific) and the reverse-subtracted (BMT-specific) cDNAs were labeled differentially with Cy3 or Cy5 and then cohybridized with the cDNAs on the same microarray. To normalize the intrinsic signal differences coming from Cy3 and Cy5 labeling, another hybridization was set up for reverse labeling with Cy3 and Cy5. As expected, quick visual inspection of the hybridization signals revealed most of the spots originating from the subtracted DC library were DC specific, so they were not detected among the dual-labeled ones but strongly hybridized with DC-specific probes. In contrast, most of the known CD genes were not DC specific in the sense that they were barely detected with DC-specific probes, and only a few were strongly labeled by BMT-specific probes.
Of the 1920 clones, 1140 were selected for their propensity to adopt highly DC-specific signals (threshold intensity ratio of DC/BMT > 3). To minimize the number of clones to be analyzed, redundant clones had to be screened out. For this purpose, 74 clones were randomly selected and sequenced. Of the 74 clones sequenced, 31 were unique genes. The following genes were most frequently identified: immunoglobulin (Ig) superfamily Z39Ig, mitochondrial genes (COI and COIII, 12S rRNA, 16S rRNA, and cytochrome b), MHC class II DRα, matrix metalloprotease-12 (MMP12), osteopontin (Eta-1), annexin A2, and α-tubulin. Because the combined redundancy of these clones constituted 62% of the sequenced clones, back-hybridization was performed using a pool of these genes to screen out redundant clones. Thereby, the remaining 300 cDNA clones were sequenced and searched by BLAST for gene identification. Finally, these analyses revealed 69 nonoverlapping genes (Table1). Of these, 63 genes were found to encode for known proteins and 6 genes were novel sequences. The novel genes designated as Crea2, Crea8, Crea11, Crea12, Crea13, and Crea14 were matched either to the ones in the EST database or to the ones recently nominated as encoding sequences for hypothetical proteins. The DC specificity of each clone was designated as the ratio of the DC/BMT signal intensity, which was obtained through differential hybridization. It appeared that some of them were still more frequently identified than others, even after the screening out of the redundant ones. Ig superfamily protein (Z39Ig) was not only highly DC specific but was also apparently abundant among the DC-associated messages. In addition to the genes reported previously in association with MoDCs,26,28 29 our screening revealed new members of DC-associated genes, such as Ig superfamily protein (Z39Ig), CD20-like precursor, glycoprotein NMB (GPNMB), transforming growth factor-β (TGF-β)–induced protein (TGFBI), myeloid DAP12-associated lectin (MDL-1), and the 6 novel genes.
DC-associated genes identified from subtraction, microarray, and sequence analysis
| Gene no. . | Name . | GenBank accession no. . | Redundancy in screening . | Intensity of BMT . | Intensity of DCs . | DC/BMT . |
|---|---|---|---|---|---|---|
| 1 | α-tubulin* | K00558 | 4 | 1 312 | 37 669 | 28.7 |
| 2 | S100 calcium-binding protein, β (S100B) | NM_006272 | 2 | 1 036 | 28 546 | 27.6 |
| 3 | Matrix metalloproteinase-12 (MMP12)* | XM_006272 | 5 | 1 402 | 38 171 | 27.2 |
| 4 | Thymus and activation-regulated chemokine (TARC) | D43767 | 1 | 1 109 | 26 047 | 23.5 |
| 5 | Myosin phosphatase, target subunit 1 (MYPT1) | XM_006578 | 1 | 2 376 | 43 787 | 18.4 |
| 6 | CD1B antigen (CD1B) | XM_002174 | 4 | 1 286 | 20 646 | 16.1 |
| 7 | CD20-like precursor | NM_022349 | 15 | 1 746 | 27 393 | 15.7 |
| 8 | Ig superfamily protein (Z39Ig)* | XM_010265 | 23 | 1 430 | 18 815 | 13.2 |
| 9 | MHC class II HLA-DQα chain | U77589 | 1 | 1 071 | 13 284 | 12.4 |
| 10 | Glycoprotein NMB (GPNMB) | XM_004781 | 3 | 1 170 | 13 914 | 11.9 |
| 11 | Osteopontin* | NM_000582 | 6 | 2 964 | 33 178 | 11.2 |
| 12 | 5-lipoxygenase activating protein (FLAP) | M63262 | 6 | 1 126 | 12 316 | 10.9 |
| 13 | Mannose receptor C, type 1 (MRC1) | NM_002438 | 3 | 1 284 | 13 960 | 10.9 |
| 14 | Cytochrome b-245, β polypeptide (CYBB) | NM_000397 | 3 | 1 050 | 11 226 | 10.7 |
| 15 | Monocyte chemotactic proteins 4 (MCP4; SCYA13) | XM_008411 | 4 | 1 429 | 14 456 | 10.1 |
| 16 | MHC class II HLA-DRα chain* | XM_004209 | 8 | 1 454 | 14 578 | 10.0 |
| 17 | MHC class II HLA-DRβ chain | M26038 | 6 | 1 607 | 14 838 | 9.2 |
| 18 | RNase A family 1 (RNASE1) | XM_012375 | 2 | 2 413 | 21 588 | 8.9 |
| 19 | Dihydropyrimidinase related protein-2 | D78013 | 4 | 1 997 | 17 249 | 8.6 |
| 20 | ATP synthase 6 (MTATP6; mitochondria) | AF347015 | 11 | 3 394 | 26 657 | 7.9 |
| 21 | 16S ribosomal RNA (mitochondria)* | AF347015 | 6 | 2 683 | 21 015 | 7.8 |
| 22 | Lysosomal acid lipase | U08464 | 22 | 1 043 | 7 756 | 7.4 |
| 23 | Cystatin A (CSTA) | NM_005213 | 4 | 1 286 | 9 437 | 7.3 |
| 24 | Antigen CD36 | M98399 | 8 | 1 404 | 9 501 | 6.8 |
| 25 | Immunoglobulin transcription factor-2 (IFT2) | XM_012756 | 1 | 1 719 | 11 613 | 6.8 |
| 26 | Monocyte chemotactic proteins-1 (MCP1; MCAF) | M24545 | 2 | 2 311 | 14 802 | 6.4 |
| 27 | β-actin | BC004251 | 1 | 715 | 4 561 | 6.4 |
| 28 | Transforming growth factor, β-induced, 68 kD (TGFBI) | NM_000358 | 2 | 1 318 | 8 002 | 6.1 |
| 29 | Cytochrome b (MTCYB; mitochondria)* | AF347015 | 2 | 3 660 | 21 907 | 6.0 |
| 30 | Calmodulin | D45887 | 3 | 752 | 4 383 | 5.8 |
| 31 | Myeloid DAP12-associating lectin (MDL-1) | AJ271684 | 1 | 1 345 | 7 736 | 5.8 |
| 32 | Cytochrome C oxidase II (COII; mitochondria) | AF347015 | 19 | 2 555 | 14 572 | 5.7 |
| 33 | β-tubulin | J00314 | 1 | 1 235 | 6 826 | 5.5 |
| 34 | B cell linker protein (BLNK) | NM_013314 | 2 | 1 615 | 8 909 | 5.5 |
| 35 | Activated RNA polymerase II transcription cofactor 4 (PC4) | XM_011218 | 2 | 1 980 | 10 656 | 5.4 |
| 36 | Cytochrome C oxidase I (COI; mitochondria)* | AF347015 | 4 | 2 499 | 13 410 | 5.4 |
| 37 | Enolase 1, α (ENO1) | NM_001428 | 1 | 520 | 2 607 | 5.0 |
| 38 | 90 kD heat shock protein (hsp90) | M16660 | 1 | 2 040 | 10 213 | 5.0 |
| 39 | IL-2Rγ | NM_000206 | 1 | 1 161 | 5 741 | 4.9 |
| 40 | Thymosin β4, X chromosome (TMSB4X) | NM_021109 | 5 | 1 649 | 8 138 | 4.9 |
| 41 | Ferritin, light polypeptide | BC004245 | 3 | 2 527 | 12 327 | 4.9 |
| 42 | NADH dehydrogenase subunit 4 (MTND4; mitochondria) | AF347015 | 1 | 3 738 | 18 023 | 4.8 |
| 43 | Accessory proteins BAP31/BAP29 | NM_005745 | 5 | 966 | 4 598 | 4.8 |
| 44 | Annexin A2 (ANXA2)* | NM_004039 | 8 | 1 221 | 5 506 | 4.5 |
| 45 | Isocitrate dehydrogenase 3 (NAD+) α (IDH3A) | XM_007580 | 1 | 1 754 | 7 766 | 4.4 |
| 46 | Microsomal glutathione S-transferase 2 (MGST2) | XM_003461 | 1 | 1 540 | 6 767 | 4.4 |
| 47 | 12S ribosomal RNA (mitochondria)* | AF347015 | 4 | 1 439 | 6 317 | 4.4 |
| 48 | Cytochrome C oxidase III (COIII; mitochondria)* | AF347015 | 11 | 1 913 | 8 318 | 4.3 |
| 49 | Coagulation factor XIII, A1 polypeptide | AK001685 | 1 | 1 009 | 4 225 | 4.2 |
| 50 | GABA(A) receptor-associated protein (GABARAP) | NM_007278 | 2 | 1 221 | 4 666 | 3.8 |
| 51 | Serine/threonine kinase-15 | BC002499 | 1 | 1 369 | 5 048 | 3.7 |
| 52 | Vesicle-associated membrane protein-8 (VAMP8) | XM_002561 | 2 | 1 976 | 7 267 | 3.7 |
| 53 | Nicastrin | AF240468 | 1 | 1 359 | 4 997 | 3.7 |
| 54 | Purinergic receptor (family A group 5) | XM_007212 | 1 | 1 592 | 5 833 | 3.7 |
| 55 | Rho GDP dissociation inhibitor beta (ARHGDIB) | NM_001175 | 1 | 1 596 | 5 707 | 3.6 |
| 56 | MAD homolog 2 (MADH2) | XM_008795 | 1 | 719 | 2 523 | 3.5 |
| 57 | Cytochrome c, clone MGC: 12367 | BC005299 | 1 | 1 330 | 4 538 | 3.4 |
| 58 | MHC class II HLA-DMα chain | XM_004220 | 1 | 1 403 | 4 552 | 3.2 |
| 59 | High affinity IgE receptor α subunit (FcERI) | X06948 | 1 | 116 | 369 | 3.2 |
| 60 | MLN51 | X80199 | 1 | 1 405 | 4 431 | 3.2 |
| 61 | DORA protein | AJ223183 | 1 | 1 411 | 4 341 | 3.1 |
| 62 | GTP-binding protein (NGB) | XM_005722 | 1 | 877 | 2 674 | 3.0 |
| 63 | Interferon regulatory factor-4 (IRF4) | NM_002460 | 1 | 565 | 1 680 | 3.0 |
| 64 | Crea2 | AL133392 | 2 | 1 437 | 12 646 | 8.8 |
| 65 | Crea8 | BC020516 | 1 | 893 | 3 331 | 3.7 |
| 66 | Crea11 | AA151535 | 1 | 2 639 | 8 446 | 3.2 |
| 67 | Crea12 | AL354813 | 1 | 1 449 | 4 928 | 3.4 |
| 68 | Crea13 | AW295797 | 1 | 1 336 | 9 218 | 6.9 |
| 69 | Crea14 | XM_071675 | 1 | 2 201 | 14 304 | 6.5 |
| Gene no. . | Name . | GenBank accession no. . | Redundancy in screening . | Intensity of BMT . | Intensity of DCs . | DC/BMT . |
|---|---|---|---|---|---|---|
| 1 | α-tubulin* | K00558 | 4 | 1 312 | 37 669 | 28.7 |
| 2 | S100 calcium-binding protein, β (S100B) | NM_006272 | 2 | 1 036 | 28 546 | 27.6 |
| 3 | Matrix metalloproteinase-12 (MMP12)* | XM_006272 | 5 | 1 402 | 38 171 | 27.2 |
| 4 | Thymus and activation-regulated chemokine (TARC) | D43767 | 1 | 1 109 | 26 047 | 23.5 |
| 5 | Myosin phosphatase, target subunit 1 (MYPT1) | XM_006578 | 1 | 2 376 | 43 787 | 18.4 |
| 6 | CD1B antigen (CD1B) | XM_002174 | 4 | 1 286 | 20 646 | 16.1 |
| 7 | CD20-like precursor | NM_022349 | 15 | 1 746 | 27 393 | 15.7 |
| 8 | Ig superfamily protein (Z39Ig)* | XM_010265 | 23 | 1 430 | 18 815 | 13.2 |
| 9 | MHC class II HLA-DQα chain | U77589 | 1 | 1 071 | 13 284 | 12.4 |
| 10 | Glycoprotein NMB (GPNMB) | XM_004781 | 3 | 1 170 | 13 914 | 11.9 |
| 11 | Osteopontin* | NM_000582 | 6 | 2 964 | 33 178 | 11.2 |
| 12 | 5-lipoxygenase activating protein (FLAP) | M63262 | 6 | 1 126 | 12 316 | 10.9 |
| 13 | Mannose receptor C, type 1 (MRC1) | NM_002438 | 3 | 1 284 | 13 960 | 10.9 |
| 14 | Cytochrome b-245, β polypeptide (CYBB) | NM_000397 | 3 | 1 050 | 11 226 | 10.7 |
| 15 | Monocyte chemotactic proteins 4 (MCP4; SCYA13) | XM_008411 | 4 | 1 429 | 14 456 | 10.1 |
| 16 | MHC class II HLA-DRα chain* | XM_004209 | 8 | 1 454 | 14 578 | 10.0 |
| 17 | MHC class II HLA-DRβ chain | M26038 | 6 | 1 607 | 14 838 | 9.2 |
| 18 | RNase A family 1 (RNASE1) | XM_012375 | 2 | 2 413 | 21 588 | 8.9 |
| 19 | Dihydropyrimidinase related protein-2 | D78013 | 4 | 1 997 | 17 249 | 8.6 |
| 20 | ATP synthase 6 (MTATP6; mitochondria) | AF347015 | 11 | 3 394 | 26 657 | 7.9 |
| 21 | 16S ribosomal RNA (mitochondria)* | AF347015 | 6 | 2 683 | 21 015 | 7.8 |
| 22 | Lysosomal acid lipase | U08464 | 22 | 1 043 | 7 756 | 7.4 |
| 23 | Cystatin A (CSTA) | NM_005213 | 4 | 1 286 | 9 437 | 7.3 |
| 24 | Antigen CD36 | M98399 | 8 | 1 404 | 9 501 | 6.8 |
| 25 | Immunoglobulin transcription factor-2 (IFT2) | XM_012756 | 1 | 1 719 | 11 613 | 6.8 |
| 26 | Monocyte chemotactic proteins-1 (MCP1; MCAF) | M24545 | 2 | 2 311 | 14 802 | 6.4 |
| 27 | β-actin | BC004251 | 1 | 715 | 4 561 | 6.4 |
| 28 | Transforming growth factor, β-induced, 68 kD (TGFBI) | NM_000358 | 2 | 1 318 | 8 002 | 6.1 |
| 29 | Cytochrome b (MTCYB; mitochondria)* | AF347015 | 2 | 3 660 | 21 907 | 6.0 |
| 30 | Calmodulin | D45887 | 3 | 752 | 4 383 | 5.8 |
| 31 | Myeloid DAP12-associating lectin (MDL-1) | AJ271684 | 1 | 1 345 | 7 736 | 5.8 |
| 32 | Cytochrome C oxidase II (COII; mitochondria) | AF347015 | 19 | 2 555 | 14 572 | 5.7 |
| 33 | β-tubulin | J00314 | 1 | 1 235 | 6 826 | 5.5 |
| 34 | B cell linker protein (BLNK) | NM_013314 | 2 | 1 615 | 8 909 | 5.5 |
| 35 | Activated RNA polymerase II transcription cofactor 4 (PC4) | XM_011218 | 2 | 1 980 | 10 656 | 5.4 |
| 36 | Cytochrome C oxidase I (COI; mitochondria)* | AF347015 | 4 | 2 499 | 13 410 | 5.4 |
| 37 | Enolase 1, α (ENO1) | NM_001428 | 1 | 520 | 2 607 | 5.0 |
| 38 | 90 kD heat shock protein (hsp90) | M16660 | 1 | 2 040 | 10 213 | 5.0 |
| 39 | IL-2Rγ | NM_000206 | 1 | 1 161 | 5 741 | 4.9 |
| 40 | Thymosin β4, X chromosome (TMSB4X) | NM_021109 | 5 | 1 649 | 8 138 | 4.9 |
| 41 | Ferritin, light polypeptide | BC004245 | 3 | 2 527 | 12 327 | 4.9 |
| 42 | NADH dehydrogenase subunit 4 (MTND4; mitochondria) | AF347015 | 1 | 3 738 | 18 023 | 4.8 |
| 43 | Accessory proteins BAP31/BAP29 | NM_005745 | 5 | 966 | 4 598 | 4.8 |
| 44 | Annexin A2 (ANXA2)* | NM_004039 | 8 | 1 221 | 5 506 | 4.5 |
| 45 | Isocitrate dehydrogenase 3 (NAD+) α (IDH3A) | XM_007580 | 1 | 1 754 | 7 766 | 4.4 |
| 46 | Microsomal glutathione S-transferase 2 (MGST2) | XM_003461 | 1 | 1 540 | 6 767 | 4.4 |
| 47 | 12S ribosomal RNA (mitochondria)* | AF347015 | 4 | 1 439 | 6 317 | 4.4 |
| 48 | Cytochrome C oxidase III (COIII; mitochondria)* | AF347015 | 11 | 1 913 | 8 318 | 4.3 |
| 49 | Coagulation factor XIII, A1 polypeptide | AK001685 | 1 | 1 009 | 4 225 | 4.2 |
| 50 | GABA(A) receptor-associated protein (GABARAP) | NM_007278 | 2 | 1 221 | 4 666 | 3.8 |
| 51 | Serine/threonine kinase-15 | BC002499 | 1 | 1 369 | 5 048 | 3.7 |
| 52 | Vesicle-associated membrane protein-8 (VAMP8) | XM_002561 | 2 | 1 976 | 7 267 | 3.7 |
| 53 | Nicastrin | AF240468 | 1 | 1 359 | 4 997 | 3.7 |
| 54 | Purinergic receptor (family A group 5) | XM_007212 | 1 | 1 592 | 5 833 | 3.7 |
| 55 | Rho GDP dissociation inhibitor beta (ARHGDIB) | NM_001175 | 1 | 1 596 | 5 707 | 3.6 |
| 56 | MAD homolog 2 (MADH2) | XM_008795 | 1 | 719 | 2 523 | 3.5 |
| 57 | Cytochrome c, clone MGC: 12367 | BC005299 | 1 | 1 330 | 4 538 | 3.4 |
| 58 | MHC class II HLA-DMα chain | XM_004220 | 1 | 1 403 | 4 552 | 3.2 |
| 59 | High affinity IgE receptor α subunit (FcERI) | X06948 | 1 | 116 | 369 | 3.2 |
| 60 | MLN51 | X80199 | 1 | 1 405 | 4 431 | 3.2 |
| 61 | DORA protein | AJ223183 | 1 | 1 411 | 4 341 | 3.1 |
| 62 | GTP-binding protein (NGB) | XM_005722 | 1 | 877 | 2 674 | 3.0 |
| 63 | Interferon regulatory factor-4 (IRF4) | NM_002460 | 1 | 565 | 1 680 | 3.0 |
| 64 | Crea2 | AL133392 | 2 | 1 437 | 12 646 | 8.8 |
| 65 | Crea8 | BC020516 | 1 | 893 | 3 331 | 3.7 |
| 66 | Crea11 | AA151535 | 1 | 2 639 | 8 446 | 3.2 |
| 67 | Crea12 | AL354813 | 1 | 1 449 | 4 928 | 3.4 |
| 68 | Crea13 | AW295797 | 1 | 1 336 | 9 218 | 6.9 |
| 69 | Crea14 | XM_071675 | 1 | 2 201 | 14 304 | 6.5 |
DC-associated clones identified by a series of microarray assays are listed together with their GenBank accession numbers. Redundancy represents the number of identical clones among the selected 374 cDNAs. Intensity of BMT and DC means the average signal intensity of each clone at the laser scan of the microarray after normalization and background subtraction. DC/BMT represents the ratio of fluorescence intensity as determined by forward- (DC-BMT) and backward- (BMT-DC) subtracted probes.
DNA clones used for the screening-out experiments to minimize the number of redundant clones.
Each DC subset shows its own expression profile for DC-associated genes
The DC-associated genes identified in the DC/BMT differential microarray analysis were then further examined for their expression profiles in different DC subsets by using another microarray, HI380, (Table 2) and by semiquantitative RT-PCR (Figure 3). Some other genes of special interest, such as CCR1, CCR7, DC-Lamp, E-cadherin, Langerin, and others, were also included in the subset-specific expression analysis. As shown in Table 2 and Figure 3and as summarized in Figure 4, the results from the microarray were in good agreement with the results revealed in the quantitative RT-PCR.
Analysis of DC-associated gene expressions among the DC subsets using microarray
| Gene no. . | Name . | BMT intensity . | Ratio of DCs and BMTs (DCs/BMTs) . | ||||
|---|---|---|---|---|---|---|---|
| CD34+cells . | MoDCs . | CD1a+DCs . | CD11c− DCs . | CD14+DCs . | |||
| Day 9 . | Day 18 . | Day 0 . | Day 18 . | ||||
| 1 | α-tubulin | 13 617 | 0.2 | 2.9 | 2.9 | 0.2 | 1.7 |
| 2 | S100B | 3 416 | 0.7 | 0.9 | 〈28.6〉 | 0.7 | 〈5.6〉 |
| 3 | MMP12 | 1 420 | 0.9 | 0.7 | 〈4.9〉 | 0.5 | 0.3 |
| 4 | TARC | 4 113 | 0.5 | 〈20.0〉 | 〈24.0〉 | 0.6 | 0.8 |
| 5 | MYPT1 | 3 805 | 0.4 | 1.0 | 2.0 | 0.4 | 〈4.9〉 |
| 6 | CD1B | 3 107 | 0.4 | 2.4 | 〈17.6〉 | 0.8 | 1.5 |
| 7 | CD20-like | 3 171 | 0.4 | 0.8 | 〈18.0〉 | 1.7 | 〈20.7〉 |
| 8 | Ig superfamily Z39Ig | 2 016 | 0.4 | 0.9 | 2.9 | 0.6 | 〈16.4〉 |
| 9 | HLA-DQα | 11 817 | 0.2 | 3.2 | 〈9.6〉 | 0.5 | 2.0 |
| 10 | GPNMB | 3 290 | 0.5 | 1.7 | 1.2 | 0.8 | 〈5.0〉 |
| 11 | Eta-1 | 2 409 | 0.3 | 3.2 | 〈6.3〉 | 0.5 | 〈11.8〉 |
| 12 | FLAP | 8 060 | 0.3 | 0.9 | 〈8.1〉 | 3.0 | 〈5.4〉 |
| 13 | MRC1 | 2 097 | 0.5 | 1.6 | 3.8 | 0.7 | 〈6.1〉 |
| 15 | MCP4 | 3 454 | 0.3 | 2.9 | 〈4.5〉 | 0.4 | 〈5.6〉 |
| 16 | HLA-DRα | 22 066 | 0.7 | 2.2 | 3.8 | 2.4 | 1.3 |
| 17 | HLA-DRβ | 20 288 | 0.2 | 0.7 | 2.7 | 1.2 | 1.4 |
| 18 | RNase1 | 3 684 | 0.2 | 1.6 | 1.1 | 0.4 | 〈11.5〉 |
| 19 | DRP-2 | 1 981 | 〈6.1〉 | 〈8.9〉 | 2.4 | 〈5.9〉 | 1.1 |
| 22 | Lysosomal acid lipase | 2 801 | 0.3 | 〈4.2〉 | 〈8.1〉 | 0.3 | 〈6.1〉 |
| 23 | Cystatin A | 3 317 | 0.2 | 0.8 | 〈17.4〉 | 0.3 | 〈4.3〉 |
| 24 | CD36 | 1 651 | 0.4 | 0.8 | 2.9 | 0.8 | 1.2 |
| 25 | ITF2 | 4 145 | 0.4 | 1.1 | 0.8 | 1.2 | 1.3 |
| 26 | MCP1 | 10 899 | 0.1 | 0.7 | 0.5 | 0.2 | 〈9.6〉 |
| 28 | TGFBI | 3 179 | 0.4 | 1.8 | 3.4 | 1.5 | 〈9.2〉 |
| 30 | Calmodulin | 6 950 | 0.9 | 〈5.1〉 | 1.8 | 0.4 | 1.6 |
| 31 | MDL-1 | 3 245 | 0.3 | 1.0 | 1.3 | 0.6 | 1.9 |
| 33 | β-tubulin | 12 754 | 0.3 | 2.8 | 2.8 | 0.3 | 2.7 |
| 34 | BLNK | 6 112 | 0.2 | 0.5 | 0.8 | 0.4 | 0.5 |
| 35 | PC4 | 9 653 | 0.2 | 0.4 | 0.9 | 0.2 | 0.4 |
| 37 | Enolase 1 | 14 759 | 0.1 | 3.8 | 1.4 | 0.1 | 1.5 |
| 38 | HSP90 | 8 933 | 0.2 | 0.6 | 1.1 | 0.3 | 0.8 |
| 39 | IL-2Rγ | 5 573 | 0.3 | 0.5 | 0.7 | 0.2 | 0.2 |
| 40 | Thymosin β4 | 19 186 | 0.3 | 1.9 | 3.4 | 0.1 | 1.6 |
| 41 | Ferritin | 31 033 | 0.1 | 3.7 | 1.9 | 0.0 | 〈5.1〉 |
| 43 | BAP31/BAP29 | 9 919 | 0.6 | 1.0 | 1.1 | 0.4 | 1.3 |
| 44 | Annexin A2 | 12 178 | 0.1 | 3.1 | 〈4.8〉 | 0.1 | 2.2 |
| 45 | IDH3A | 2 298 | 0.4 | 0.6 | 0.6 | 0.9 | 0.5 |
| 46 | MGST2 | 3 060 | 0.3 | 1.0 | 3.5 | 0.5 | 3.5 |
| 49 | Factor XIIIa | 2 110 | 0.3 | 1.0 | 0.7 | 0.5 | 3.0 |
| 50 | GABARAP | 9 150 | 0.1 | 1.4 | 1.3 | 0.2 | 2.0 |
| 51 | Serine/threonine kinase-15 | 2 595 | 0.4 | 1.3 | 1.6 | 0.7 | 1.4 |
| 52 | VAMP8 | 9 344 | 0.1 | 2.8 | 〈5.1〉 | 0.3 | 〈4.2〉 |
| 53 | Nicastrin | 3 652 | 0.3 | 0.9 | 0.7 | 0.6 | 0.7 |
| 54 | Purinergic receptor | 1 901 | 0.4 | 1.0 | 3.1 | 0.6 | 1.9 |
| 55 | ARHGDIB | 23 059 | 0.1 | 0.2 | 1.0 | 0.1 | 2.0 |
| 56 | MAD homolog 2 | 2 167 | 0.5 | 0.9 | 0.8 | 0.6 | 0.7 |
| 58 | MHC-DMα | 8 207 | 0.4 | 1.9 | 〈4.1〉 | 0.6 | 2.8 |
| 59 | FcERI | 2 989 | 0.3 | 0.7 | 1.9 | 0.3 | 0.7 |
| 60 | MLN51 | 15 001 | 2.2 | 1.4 | 0.9 | 1.9 | 1.4 |
| 61 | DORA | 5 405 | 0.3 | 0.9 | 〈4.0〉 | 0.4 | 1.8 |
| 62 | GTP-binding protein | 3 838 | 0.2 | 1.0 | 1.1 | 0.6 | 0.7 |
| 63 | IRF4 | 2 005 | 0.5 | 〈7.9〉 | 2.2 | 〈9.3〉 | 0.4 |
| 64 | Crea2 | 2 318 | 0.4 | 0.8 | 1.3 | 0.6 | 0.5 |
| 65 | Crea8 | 1 985 | 0.8 | 1.1 | 1.3 | 1.6 | 2.1 |
| 66 | Crea11 | 1 805 | 0.3 | 3.3 | 12.9 | 0.6 | 0.7 |
| 67 | Crea12 | 1 199 | 0.7 | 1.4 | 0.7 | 1.9 | 0.5 |
| 68 | Crea13 | 3 469 | 0.4 | 5.0 | 17.1 | 0.5 | 0.8 |
| 69 | Crea14 | 2 892 | 3.4 | 3.0 | 5.0 | 2.1 | 2.5 |
| S1 | CCR1 | 2 883 | 0.2 | 0.8 | 0.8 | 0.3 | 1.6 |
| S2 | CCR7 | 9 952 | 0.1 | 3.1 | 1.5 | 0.1 | 0.1 |
| S3 | DC-Lamp | 3 440 | 0.3 | 〈7.4〉 | 〈6.3〉 | 0.4 | 0.4 |
| S4 | E-cadherin | 3 589 | 0.4 | 1.0 | 1.0 | 0.9 | 0.9 |
| S5 | DEC205 (CD205) | 2 831 | 0.2 | 2.9 | 1.6 | 0.8 | 0.7 |
| S6 | Mannose receptor (CD206) | 2 319 | 0.5 | 1.8 | 〈6.6〉 | 0.4 | 〈8.6〉 |
| S7 | Langerin (CD207) | 3 482 | 0.3 | 1.2 | 〈5.9〉 | 0.8 | 1.0 |
| S8 | DC-sign (CD209) | 2 305 | 0.3 | 1.0 | 0.8 | 0.4 | 0.9 |
| Gene no. . | Name . | BMT intensity . | Ratio of DCs and BMTs (DCs/BMTs) . | ||||
|---|---|---|---|---|---|---|---|
| CD34+cells . | MoDCs . | CD1a+DCs . | CD11c− DCs . | CD14+DCs . | |||
| Day 9 . | Day 18 . | Day 0 . | Day 18 . | ||||
| 1 | α-tubulin | 13 617 | 0.2 | 2.9 | 2.9 | 0.2 | 1.7 |
| 2 | S100B | 3 416 | 0.7 | 0.9 | 〈28.6〉 | 0.7 | 〈5.6〉 |
| 3 | MMP12 | 1 420 | 0.9 | 0.7 | 〈4.9〉 | 0.5 | 0.3 |
| 4 | TARC | 4 113 | 0.5 | 〈20.0〉 | 〈24.0〉 | 0.6 | 0.8 |
| 5 | MYPT1 | 3 805 | 0.4 | 1.0 | 2.0 | 0.4 | 〈4.9〉 |
| 6 | CD1B | 3 107 | 0.4 | 2.4 | 〈17.6〉 | 0.8 | 1.5 |
| 7 | CD20-like | 3 171 | 0.4 | 0.8 | 〈18.0〉 | 1.7 | 〈20.7〉 |
| 8 | Ig superfamily Z39Ig | 2 016 | 0.4 | 0.9 | 2.9 | 0.6 | 〈16.4〉 |
| 9 | HLA-DQα | 11 817 | 0.2 | 3.2 | 〈9.6〉 | 0.5 | 2.0 |
| 10 | GPNMB | 3 290 | 0.5 | 1.7 | 1.2 | 0.8 | 〈5.0〉 |
| 11 | Eta-1 | 2 409 | 0.3 | 3.2 | 〈6.3〉 | 0.5 | 〈11.8〉 |
| 12 | FLAP | 8 060 | 0.3 | 0.9 | 〈8.1〉 | 3.0 | 〈5.4〉 |
| 13 | MRC1 | 2 097 | 0.5 | 1.6 | 3.8 | 0.7 | 〈6.1〉 |
| 15 | MCP4 | 3 454 | 0.3 | 2.9 | 〈4.5〉 | 0.4 | 〈5.6〉 |
| 16 | HLA-DRα | 22 066 | 0.7 | 2.2 | 3.8 | 2.4 | 1.3 |
| 17 | HLA-DRβ | 20 288 | 0.2 | 0.7 | 2.7 | 1.2 | 1.4 |
| 18 | RNase1 | 3 684 | 0.2 | 1.6 | 1.1 | 0.4 | 〈11.5〉 |
| 19 | DRP-2 | 1 981 | 〈6.1〉 | 〈8.9〉 | 2.4 | 〈5.9〉 | 1.1 |
| 22 | Lysosomal acid lipase | 2 801 | 0.3 | 〈4.2〉 | 〈8.1〉 | 0.3 | 〈6.1〉 |
| 23 | Cystatin A | 3 317 | 0.2 | 0.8 | 〈17.4〉 | 0.3 | 〈4.3〉 |
| 24 | CD36 | 1 651 | 0.4 | 0.8 | 2.9 | 0.8 | 1.2 |
| 25 | ITF2 | 4 145 | 0.4 | 1.1 | 0.8 | 1.2 | 1.3 |
| 26 | MCP1 | 10 899 | 0.1 | 0.7 | 0.5 | 0.2 | 〈9.6〉 |
| 28 | TGFBI | 3 179 | 0.4 | 1.8 | 3.4 | 1.5 | 〈9.2〉 |
| 30 | Calmodulin | 6 950 | 0.9 | 〈5.1〉 | 1.8 | 0.4 | 1.6 |
| 31 | MDL-1 | 3 245 | 0.3 | 1.0 | 1.3 | 0.6 | 1.9 |
| 33 | β-tubulin | 12 754 | 0.3 | 2.8 | 2.8 | 0.3 | 2.7 |
| 34 | BLNK | 6 112 | 0.2 | 0.5 | 0.8 | 0.4 | 0.5 |
| 35 | PC4 | 9 653 | 0.2 | 0.4 | 0.9 | 0.2 | 0.4 |
| 37 | Enolase 1 | 14 759 | 0.1 | 3.8 | 1.4 | 0.1 | 1.5 |
| 38 | HSP90 | 8 933 | 0.2 | 0.6 | 1.1 | 0.3 | 0.8 |
| 39 | IL-2Rγ | 5 573 | 0.3 | 0.5 | 0.7 | 0.2 | 0.2 |
| 40 | Thymosin β4 | 19 186 | 0.3 | 1.9 | 3.4 | 0.1 | 1.6 |
| 41 | Ferritin | 31 033 | 0.1 | 3.7 | 1.9 | 0.0 | 〈5.1〉 |
| 43 | BAP31/BAP29 | 9 919 | 0.6 | 1.0 | 1.1 | 0.4 | 1.3 |
| 44 | Annexin A2 | 12 178 | 0.1 | 3.1 | 〈4.8〉 | 0.1 | 2.2 |
| 45 | IDH3A | 2 298 | 0.4 | 0.6 | 0.6 | 0.9 | 0.5 |
| 46 | MGST2 | 3 060 | 0.3 | 1.0 | 3.5 | 0.5 | 3.5 |
| 49 | Factor XIIIa | 2 110 | 0.3 | 1.0 | 0.7 | 0.5 | 3.0 |
| 50 | GABARAP | 9 150 | 0.1 | 1.4 | 1.3 | 0.2 | 2.0 |
| 51 | Serine/threonine kinase-15 | 2 595 | 0.4 | 1.3 | 1.6 | 0.7 | 1.4 |
| 52 | VAMP8 | 9 344 | 0.1 | 2.8 | 〈5.1〉 | 0.3 | 〈4.2〉 |
| 53 | Nicastrin | 3 652 | 0.3 | 0.9 | 0.7 | 0.6 | 0.7 |
| 54 | Purinergic receptor | 1 901 | 0.4 | 1.0 | 3.1 | 0.6 | 1.9 |
| 55 | ARHGDIB | 23 059 | 0.1 | 0.2 | 1.0 | 0.1 | 2.0 |
| 56 | MAD homolog 2 | 2 167 | 0.5 | 0.9 | 0.8 | 0.6 | 0.7 |
| 58 | MHC-DMα | 8 207 | 0.4 | 1.9 | 〈4.1〉 | 0.6 | 2.8 |
| 59 | FcERI | 2 989 | 0.3 | 0.7 | 1.9 | 0.3 | 0.7 |
| 60 | MLN51 | 15 001 | 2.2 | 1.4 | 0.9 | 1.9 | 1.4 |
| 61 | DORA | 5 405 | 0.3 | 0.9 | 〈4.0〉 | 0.4 | 1.8 |
| 62 | GTP-binding protein | 3 838 | 0.2 | 1.0 | 1.1 | 0.6 | 0.7 |
| 63 | IRF4 | 2 005 | 0.5 | 〈7.9〉 | 2.2 | 〈9.3〉 | 0.4 |
| 64 | Crea2 | 2 318 | 0.4 | 0.8 | 1.3 | 0.6 | 0.5 |
| 65 | Crea8 | 1 985 | 0.8 | 1.1 | 1.3 | 1.6 | 2.1 |
| 66 | Crea11 | 1 805 | 0.3 | 3.3 | 12.9 | 0.6 | 0.7 |
| 67 | Crea12 | 1 199 | 0.7 | 1.4 | 0.7 | 1.9 | 0.5 |
| 68 | Crea13 | 3 469 | 0.4 | 5.0 | 17.1 | 0.5 | 0.8 |
| 69 | Crea14 | 2 892 | 3.4 | 3.0 | 5.0 | 2.1 | 2.5 |
| S1 | CCR1 | 2 883 | 0.2 | 0.8 | 0.8 | 0.3 | 1.6 |
| S2 | CCR7 | 9 952 | 0.1 | 3.1 | 1.5 | 0.1 | 0.1 |
| S3 | DC-Lamp | 3 440 | 0.3 | 〈7.4〉 | 〈6.3〉 | 0.4 | 0.4 |
| S4 | E-cadherin | 3 589 | 0.4 | 1.0 | 1.0 | 0.9 | 0.9 |
| S5 | DEC205 (CD205) | 2 831 | 0.2 | 2.9 | 1.6 | 0.8 | 0.7 |
| S6 | Mannose receptor (CD206) | 2 319 | 0.5 | 1.8 | 〈6.6〉 | 0.4 | 〈8.6〉 |
| S7 | Langerin (CD207) | 3 482 | 0.3 | 1.2 | 〈5.9〉 | 0.8 | 1.0 |
| S8 | DC-sign (CD209) | 2 305 | 0.3 | 1.0 | 0.8 | 0.4 | 0.9 |
DC-associated genes listed in Table 1 were further analyzed among the different DC subsets using HI380 microarray (Creagene). Genes are listed in abbreviated form according to the number used in Table 1, and some other genes of interest were added and numbered from S1 to S8. BMT intensity means the average signal intensity of B, monocyte, and T cells in 5 different experimental sets. The microarray was screened with probes prepared from each DC subset (purity more than 98%) and from BMTs without subtraction. The ratio of DC to BMT for each gene was determined from 2 different sets of experiments after background subtraction and normalization. For better contrast, values over 4 are shown between carats.
Expression profile of DCassociated genes in each DC subset.
Semiquantitative RT-PCR was performed on cDNA isolated from MoDCs, CD1a+ DCs, CD11c− DCs, and CD14+DCs, B cells, monocytes, and T cells, respectively, and the PCR products were analyzed at 25 and 30 cycles of PCR. The result of RT-PCR was summarized with differential marking: (+++) for higher expression and (++) for lower expression detectable after 25 cycles of PCR; (+) and (+/−) for expression detectable only after 30 cycles of PCR and marginally detectable even after 30 cycles of PCR, respectively. GAPDH was used for normalization of each cDNA amount. CD19, CD14, and CD28 were used as control genes for B cells, monocytes, and T cells, respectively. The ratio of DC/BMT represents the degree of DC specificity of each clone as determined by microarray analysis and as described in Table 1. Genes not detected by differential screening were indicated by an asterisk. ND denotes not determined in this study.
Expression profile of DCassociated genes in each DC subset.
Semiquantitative RT-PCR was performed on cDNA isolated from MoDCs, CD1a+ DCs, CD11c− DCs, and CD14+DCs, B cells, monocytes, and T cells, respectively, and the PCR products were analyzed at 25 and 30 cycles of PCR. The result of RT-PCR was summarized with differential marking: (+++) for higher expression and (++) for lower expression detectable after 25 cycles of PCR; (+) and (+/−) for expression detectable only after 30 cycles of PCR and marginally detectable even after 30 cycles of PCR, respectively. GAPDH was used for normalization of each cDNA amount. CD19, CD14, and CD28 were used as control genes for B cells, monocytes, and T cells, respectively. The ratio of DC/BMT represents the degree of DC specificity of each clone as determined by microarray analysis and as described in Table 1. Genes not detected by differential screening were indicated by an asterisk. ND denotes not determined in this study.
Schematic summary of genes differentially expressed in different subsets of DCs.
The genes identified in this study were grouped based on the microarray analysis (DC/BMT > 1.0, Table 2) and the RT-PCR (Figure 3) and summarized in a Venn diagram. The nonoverlapping area represents genes specifically expressed in each subset of DCs. The overlapping area represents genes commonly expressed in different subsets of DCs. Shadowed genes represent the ones also highly expressed in blood CD11c− DCs.
Schematic summary of genes differentially expressed in different subsets of DCs.
The genes identified in this study were grouped based on the microarray analysis (DC/BMT > 1.0, Table 2) and the RT-PCR (Figure 3) and summarized in a Venn diagram. The nonoverlapping area represents genes specifically expressed in each subset of DCs. The overlapping area represents genes commonly expressed in different subsets of DCs. Shadowed genes represent the ones also highly expressed in blood CD11c− DCs.
As expected from their lineage differences, the most striking difference was seen between CD11c− lymphoid DCs and the ex vivo–generated myeloid DCs. Most of the DC-associated genes identified from the primary microarray analysis, such as α- and β-tubulin, osteopontin (Eta-1), glycoprotein NMB (GPNMB), MCP4, lysosomal acid lipase, enolase 1, thymosin β4, ferritin L-chain, annexin A2, VAMP8, and GABARAP, were not highly expressed in CD11c− DCs (Table 2 and Figure 3). On the other hand, interferon regulatory factor-4 (IRF4), which is essential for mature T- and B-cell function,37 was highly expressed in CD11c− DCs, which are the main producers of type I interferon in human blood.38-41 Besides IRF4, the genes encoding for dihydropyrimidinase-related protein-2 (DRP-2) and 5-lipoxygenase activating protein (FLAP) were highly expressed in CD11c− DCs.
While the difference was not as remarkable as that shown in CD11c− DCs, there were some differences in the expression profiles of DC-associated genes among the 3 myeloid DCs (CD1a+ DCs, CD14+ DCs, and MoDCs) (Table 2 and Figure 3). For example, TARC, IRF4, CCR7, and DC-Lamp were highly expressed in 2 DCs: MoDCs and CD1a+ DCs. However, in CD14+ DCs, the expression levels of these genes were comparable to those of the non-DC populations. Reciprocally, many genes, such as MCP1, Eta-1, TGFBI, factor XIIIa, mannose receptor (MRC1), Ig superfamily (Z39Ig), and others, were relatively prominent in CD14+ DCs. Lastly, while the expressions of S100B, MMP12, and CD1b were remarkable in CD1a+ DCs, these genes were not seemingly specific for CD1a+ DCs. The relatively lower expression of these genes in MoDCs at day 9 was likely to reflect the differences in the DCs at their acquired degree of maturation, based on the previous findings of their association with the maturity of MoDCs.26-29,37 It is not clear why, but certain genes, such as thymosin β4, BAP31, and GABARAP, showing a relatively lower DC/BMT ratio (< 6.0), failed to reveal DC specificity in the quantitative RT-PCR analysis (Figure 3). Nevertheless, considering the report that DORA, a gene showing the lowest ratio (3.1) of DC/BMT in our experiment (Table 1 and Figure 3), was screened to be DC specific in another independent study,60 the genes in this range of the DC/BMT ratio in our experiment also seemed to merit further consideration. Of the 6 Crea genes identified in this study, the expression of 2 genes, Crea11 and Crea13, were outstanding in CD1a+ DCs and MoDCs, while the expression of the others was likely to be marginally DC biased. These novel genes are now under investigation for their biologic functions in association with human DC subsets.
Expression of DC-associated genes in different maturation stages
To answer the important question of how the differentiation/maturation stages of DCs affect the difference in DC-associated gene expression among the 4 DC subsets, we undertook similar microarray analyses for the immature myeloid DCs and the matured CD11c− DCs (Table3). After maturation by culturing for 5 days under the influence of CD40L and IL-3, CD11c− DCs did not overexpress the messages commonly up-regulated in fully differentiated DCs of myeloid origin. Thus, genes such as α- and β-tubulin, Eta-1, GPNMB, MCP4, lysosomal acid lipase, enolase 1, thymosin β4, ferritin L-chain, annexin A2, VAMP8, and GABARAP were considered to be truly myeloid DC associated. Interestingly, the high expression of FLAP, implicated in allergic inflammations,42 was not restricted to a certain stage of maturation but was relatively common to the DC subclasses, including CD11c− DCs. The exceptional absence of the FLAP overexpression in MoDCs at day 9 was repeatedly observed in the ones derived from a different donor (data not shown). The IRF4 expression in CD1a+ DCs, CD11c− DCs, and CD14+DCs was markedly down-regulated upon DC maturation but was completely reversed in MoDCs (Table 3), suggesting that the control of IRF4 expression might be cell type specific, even among the myeloid DCs. However, in the other set of experiments, the IRF4 expression was considerably higher in CD1a+ DCs at day 18, suggesting that the control of IRF4 expression might not simply be maturation related but might vary depending upon the multiple signals, at least in CD1a+ DCs (data not shown). At a glance, the expression of most DC-associated genes was not remarkable in the case of immature DCs and seemed to be associated with the maturity of the relevant DC subsets. For example, MMP12, Z39Ig, GPNMB, Eta-1, and others showed a DC/BMT ratio lower than 1 in immature DCs. However, there were genes constitutively overexpressed in the immature stages of the relevant DCs (DC/BMT > 1), while the exact level of overexpression often varied substantially following their maturation. These genes included TARC in MoDCs, MHC class II DRα in all of 4 DC subclasses, CD1b in MoDCs and CD1a+ DCs, CD20-like precursors and MRC1 in the 2 CD34+-derived DCs, lysosomal acid lipase and TGFBI in MoDCs, and others. For the 2 DCs derived from the CD34+cells, the DC subset–specific control appeared in the early stages of DC development. Thus, there was differential expression of MCP1 in CD14+ DCs, just as in the case of DC-Lamp in CD1a+ DCs at day 8, indicating their development through distinct pathways from the same precursor.
Analysis of DC-associated gene expression at different maturation stages for each DC subset
| Gene no. . | Name . | Ratio of DC and BMT . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mo DCs . | CD1a+DCs . | CD11c−DCs . | CD14+DCs . | ||||||
| Day 7 . | Day 9 . | Day 8 . | Day 18 . | Day 0 . | Day 5 . | Day 8 . | Day 18 . | ||
| 2 | S100B | 1.4 | 0.9 | 1.9 | 28.6 | 0.7 | 0.9 | 0.9 | 5.6 |
| 3 | MMP12 | 0.6 | 0.7 | 0.4 | 4.9 | 0.5 | 0.7 | 0.5 | 0.3 |
| 4 | TARC | 2.0 | 20.0 | 0.7 | 24.0 | 0.6 | 0.9 | 0.7 | 0.8 |
| 5 | MYPT1 | 0.6 | 1.0 | 0.6 | 2.0 | 0.4 | 0.4 | 1.0 | 4.9 |
| 6 | CD1B | 1.5 | 2.4 | 1.4 | 17.6 | 0.8 | 0.9 | 0.7 | 1.5 |
| 7 | CD20-like | 1.9 | 0.8 | 6.3 | 18.0 | 1.7 | 0.6 | 6.2 | 20.7 |
| 8 | Ig superfamily Z39Ig | 0.4 | 0.9 | 0.3 | 2.9 | 0.6 | 0.8 | 0.5 | 16.4 |
| 9 | HLA-DQα | 1.2 | 3.2 | 0.8 | 9.6 | 0.5 | 0.5 | 1.0 | 2.0 |
| 10 | GPNMB | 0.4 | 1.7 | 0.7 | 1.2 | 0.8 | 0.7 | 1.0 | 5.0 |
| 11 | Eta-1 | 0.8 | 3.2 | 0.4 | 6.3 | 0.5 | 0.5 | 0.7 | 11.8 |
| 12 | FLAP | 2.7 | 0.9 | 6.8 | 8.1 | 3.0 | 4.9 | 7.0 | 5.4 |
| 13 | MRC1 | 0.9 | 1.6 | 2.1 | 3.8 | 0.7 | 1.0 | 1.2 | 6.1 |
| 15 | MCP4 | 1.1 | 2.9 | 0.3 | 4.5 | 0.4 | 0.6 | 1.2 | 5.6 |
| 16 | HLA-DRα | 2.6 | 2.2 | 3.5 | 3.8 | 2.4 | 3.3 | 3.9 | 1.3 |
| 17 | HLA-DRβ | 1.4 | 0.7 | 1.8 | 2.7 | 1.2 | 0.7 | 1.2 | 1.4 |
| 18 | RNase1 | 0.3 | 1.6 | 0.3 | 1.1 | 0.4 | 0.5 | 0.5 | 11.5 |
| 19 | DRP-2 | 0.9 | 8.9 | 6.4 | 2.4 | 5.9 | 5.7 | 7.9 | 1.1 |
| 22 | Lysosomal acid lipase | 3.4 | 4.2 | 0.3 | 8.1 | 0.3 | 0.4 | 0.5 | 6.1 |
| 23 | Cystatin A | 0.5 | 0.8 | 0.3 | 17.4 | 0.3 | 0.4 | 0.4 | 4.3 |
| 24 | CD36 | 0.4 | 0.8 | 0.8 | 2.9 | 0.8 | 0.8 | 0.9 | 1.2 |
| 26 | MCP1 | 0.3 | 0.7 | 0.2 | 0.5 | 0.2 | 0.3 | 3.3 | 9.6 |
| 28 | TGFBI | 1.8 | 1.8 | 0.5 | 3.4 | 1.5 | 0.9 | 0.7 | 9.2 |
| 30 | Calmodulin | 0.8 | 5.1 | 0.5 | 1.8 | 0.4 | 0.7 | 0.6 | 1.6 |
| 31 | MDL-1 | 0.4 | 1.0 | 0.4 | 1.3 | 0.6 | 0.8 | 0.7 | 1.9 |
| 33 | β-tubulin | 1.1 | 2.8 | 0.4 | 2.8 | 0.3 | 0.3 | 0.6 | 2.7 |
| 37 | Enolase 1 | 0.6 | 3.8 | 0.3 | 1.4 | 0.1 | 0.1 | 0.4 | 1.5 |
| 40 | Thymosin β4 | 1.2 | 1.9 | 0.3 | 3.4 | 0.1 | 0.3 | 0.6 | 1.6 |
| 41 | Ferritin | 1.3 | 3.7 | 0.1 | 1.9 | 0.0 | 0.1 | 0.3 | 5.1 |
| 44 | Annexin A2 | 0.4 | 3.1 | 0.1 | 4.8 | 0.1 | 0.1 | 0.3 | 2.2 |
| 46 | MGST2 | 0.6 | 1.0 | 0.6 | 3.5 | 0.5 | 0.4 | 1.1 | 3.5 |
| 49 | Factor XIIIa | 0.3 | 1.0 | 0.4 | 0.7 | 0.5 | 0.7 | 0.8 | 3.0 |
| 52 | VAMP8 | 2.0 | 2.8 | 0.6 | 5.1 | 0.3 | 0.2 | 1.3 | 4.2 |
| 54 | Purinergic receptor | 0.4 | 1.0 | 0.6 | 3.1 | 0.6 | 0.6 | 1.0 | 1.9 |
| 55 | ARHGDIB | 0.1 | 0.2 | 0.1 | 1.0 | 0.1 | 0.1 | 0.3 | 2.0 |
| 58 | MHC-DMα | 0.8 | 1.9 | 1.3 | 4.1 | 0.6 | 0.4 | 2.2 | 2.8 |
| 61 | DORA | 1.7 | 0.9 | 0.5 | 4.0 | 0.4 | 0.6 | 0.7 | 1.8 |
| 63 | IRF4 | 0.3 | 7.9 | 10.0 | 2.2 | 9.3 | 0.9 | 5.8 | 0.4 |
| S4 | DC-Lamp | 0.3 | 7.4 | 1.4 | 6.3 | 0.4 | 0.8 | 0.5 | 0.4 |
| S5 | E-cadherin | 0.5 | 1.0 | 0.6 | 1.0 | 0.9 | 0.9 | 0.5 | 0.9 |
| S6 | DEC205 (CD205) | 0.4 | 2.9 | 0.3 | 1.6 | 0.8 | 0.6 | 0.5 | 0.7 |
| S7 | Mannose receptor (CD206) | 1.1 | 1.8 | 3.1 | 6.6 | 0.4 | 0.5 | 2.0 | 8.6 |
| S8 | Langerin (CD207) | 0.4 | 1.2 | 0.5 | 5.9 | 0.8 | 0.7 | 0.6 | 1.0 |
| Gene no. . | Name . | Ratio of DC and BMT . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mo DCs . | CD1a+DCs . | CD11c−DCs . | CD14+DCs . | ||||||
| Day 7 . | Day 9 . | Day 8 . | Day 18 . | Day 0 . | Day 5 . | Day 8 . | Day 18 . | ||
| 2 | S100B | 1.4 | 0.9 | 1.9 | 28.6 | 0.7 | 0.9 | 0.9 | 5.6 |
| 3 | MMP12 | 0.6 | 0.7 | 0.4 | 4.9 | 0.5 | 0.7 | 0.5 | 0.3 |
| 4 | TARC | 2.0 | 20.0 | 0.7 | 24.0 | 0.6 | 0.9 | 0.7 | 0.8 |
| 5 | MYPT1 | 0.6 | 1.0 | 0.6 | 2.0 | 0.4 | 0.4 | 1.0 | 4.9 |
| 6 | CD1B | 1.5 | 2.4 | 1.4 | 17.6 | 0.8 | 0.9 | 0.7 | 1.5 |
| 7 | CD20-like | 1.9 | 0.8 | 6.3 | 18.0 | 1.7 | 0.6 | 6.2 | 20.7 |
| 8 | Ig superfamily Z39Ig | 0.4 | 0.9 | 0.3 | 2.9 | 0.6 | 0.8 | 0.5 | 16.4 |
| 9 | HLA-DQα | 1.2 | 3.2 | 0.8 | 9.6 | 0.5 | 0.5 | 1.0 | 2.0 |
| 10 | GPNMB | 0.4 | 1.7 | 0.7 | 1.2 | 0.8 | 0.7 | 1.0 | 5.0 |
| 11 | Eta-1 | 0.8 | 3.2 | 0.4 | 6.3 | 0.5 | 0.5 | 0.7 | 11.8 |
| 12 | FLAP | 2.7 | 0.9 | 6.8 | 8.1 | 3.0 | 4.9 | 7.0 | 5.4 |
| 13 | MRC1 | 0.9 | 1.6 | 2.1 | 3.8 | 0.7 | 1.0 | 1.2 | 6.1 |
| 15 | MCP4 | 1.1 | 2.9 | 0.3 | 4.5 | 0.4 | 0.6 | 1.2 | 5.6 |
| 16 | HLA-DRα | 2.6 | 2.2 | 3.5 | 3.8 | 2.4 | 3.3 | 3.9 | 1.3 |
| 17 | HLA-DRβ | 1.4 | 0.7 | 1.8 | 2.7 | 1.2 | 0.7 | 1.2 | 1.4 |
| 18 | RNase1 | 0.3 | 1.6 | 0.3 | 1.1 | 0.4 | 0.5 | 0.5 | 11.5 |
| 19 | DRP-2 | 0.9 | 8.9 | 6.4 | 2.4 | 5.9 | 5.7 | 7.9 | 1.1 |
| 22 | Lysosomal acid lipase | 3.4 | 4.2 | 0.3 | 8.1 | 0.3 | 0.4 | 0.5 | 6.1 |
| 23 | Cystatin A | 0.5 | 0.8 | 0.3 | 17.4 | 0.3 | 0.4 | 0.4 | 4.3 |
| 24 | CD36 | 0.4 | 0.8 | 0.8 | 2.9 | 0.8 | 0.8 | 0.9 | 1.2 |
| 26 | MCP1 | 0.3 | 0.7 | 0.2 | 0.5 | 0.2 | 0.3 | 3.3 | 9.6 |
| 28 | TGFBI | 1.8 | 1.8 | 0.5 | 3.4 | 1.5 | 0.9 | 0.7 | 9.2 |
| 30 | Calmodulin | 0.8 | 5.1 | 0.5 | 1.8 | 0.4 | 0.7 | 0.6 | 1.6 |
| 31 | MDL-1 | 0.4 | 1.0 | 0.4 | 1.3 | 0.6 | 0.8 | 0.7 | 1.9 |
| 33 | β-tubulin | 1.1 | 2.8 | 0.4 | 2.8 | 0.3 | 0.3 | 0.6 | 2.7 |
| 37 | Enolase 1 | 0.6 | 3.8 | 0.3 | 1.4 | 0.1 | 0.1 | 0.4 | 1.5 |
| 40 | Thymosin β4 | 1.2 | 1.9 | 0.3 | 3.4 | 0.1 | 0.3 | 0.6 | 1.6 |
| 41 | Ferritin | 1.3 | 3.7 | 0.1 | 1.9 | 0.0 | 0.1 | 0.3 | 5.1 |
| 44 | Annexin A2 | 0.4 | 3.1 | 0.1 | 4.8 | 0.1 | 0.1 | 0.3 | 2.2 |
| 46 | MGST2 | 0.6 | 1.0 | 0.6 | 3.5 | 0.5 | 0.4 | 1.1 | 3.5 |
| 49 | Factor XIIIa | 0.3 | 1.0 | 0.4 | 0.7 | 0.5 | 0.7 | 0.8 | 3.0 |
| 52 | VAMP8 | 2.0 | 2.8 | 0.6 | 5.1 | 0.3 | 0.2 | 1.3 | 4.2 |
| 54 | Purinergic receptor | 0.4 | 1.0 | 0.6 | 3.1 | 0.6 | 0.6 | 1.0 | 1.9 |
| 55 | ARHGDIB | 0.1 | 0.2 | 0.1 | 1.0 | 0.1 | 0.1 | 0.3 | 2.0 |
| 58 | MHC-DMα | 0.8 | 1.9 | 1.3 | 4.1 | 0.6 | 0.4 | 2.2 | 2.8 |
| 61 | DORA | 1.7 | 0.9 | 0.5 | 4.0 | 0.4 | 0.6 | 0.7 | 1.8 |
| 63 | IRF4 | 0.3 | 7.9 | 10.0 | 2.2 | 9.3 | 0.9 | 5.8 | 0.4 |
| S4 | DC-Lamp | 0.3 | 7.4 | 1.4 | 6.3 | 0.4 | 0.8 | 0.5 | 0.4 |
| S5 | E-cadherin | 0.5 | 1.0 | 0.6 | 1.0 | 0.9 | 0.9 | 0.5 | 0.9 |
| S6 | DEC205 (CD205) | 0.4 | 2.9 | 0.3 | 1.6 | 0.8 | 0.6 | 0.5 | 0.7 |
| S7 | Mannose receptor (CD206) | 1.1 | 1.8 | 3.1 | 6.6 | 0.4 | 0.5 | 2.0 | 8.6 |
| S8 | Langerin (CD207) | 0.4 | 1.2 | 0.5 | 5.9 | 0.8 | 0.7 | 0.6 | 1.0 |
DC-associated genes listed in Table 1 were further analyzed at different maturation stages for each DC subset using an HI380 microarray (Creagene). Genes are listed in abbreviated form according to the number used in Tables 1 and 2. The microarray was screened with probes prepared from each DC subset (purity more than 98%) at different maturation stages and from BMTs without subtraction. The ratio of DCs to BMTs for each gene was determined from 2 different sets of experiments after background subtraction and normalization. Genes showing little difference between their mature and immature stages were omitted from the list shown in Table 2.
Expression of DC-associated genes in different donors
To examine how donor difference affects the results, we prepared another set of 4 DC subclasses from different donors and undertook microarray analyses with cDNA probes freshly derived from the second set of DCs. As shown in Table 4, most of the representative genes generally showed “relative consistency” in their expression profiles for different donors. A profound relative consistency was found in the expression of TARC, Ig superfamily (Z39Ig), MCP1, TGFBI, CCR1, DC-Lamp, E-cadherin, and DEC205. However, the relative consistency of the expression of Eta-1, MRC1, and IRF4 was not as strong as in those mentioned above. Among the DC-associated genes newly identified, MDL-1 was consistently distinguished by its marginal DC association in both donor sets. Briefly, even though the DC/BMT ratios for the selected genes were not exactly conserved, the relative consistency between different donors for their specificity to each DC subset was present for most of the selected genes.
Analysis of the expression profiles of representative DC-associated genes in 2 different donors for each DC subset
| Gene no. . | Name . | Donor set 1 . | Donor set 2 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mo DCs . | CD1a+DCs . | CD11c−DCs 0 . | CD14+DCs . | Mo DCs . | CD1a+DCs . | CD11c−DCs . | CD14+DCs . | ||
| Day 9 . | Day 18 . | Day 0 . | Day 18 . | Day 9 . | Day 18 . | Day 0 . | Day 18 . | ||
| 4 | TARC | 〈20.0〉 | 〈24.0〉 | 0.6 | 0.8 | 〈12.2〉 | 〈9.2〉 | 0.7 | 0.8 |
| 7 | CD20-like | 0.8 | 〈18.0〉 | 1.7 | 〈20.7〉 | 0.8 | 〈9.0〉 | 〈4.2〉 | 〈12.8〉 |
| 8 | Ig superfamily Z39Ig | 0.9 | 2.9 | 0.6 | 〈16.4〉 | 1.0 | 1.6 | 0.4 | 〈7.4〉 |
| 10 | GPNMB | 1.7 | 1.2 | 0.8 | 〈5.0〉 | 1.3 | 1.2 | 0.8 | 〈4.0〉 |
| 11 | Eta-1 | 3.2 | 〈6.3〉 | 0.5 | 〈11.8〉 | 1.1 | 0.9 | 0.6 | 〈8.2〉 |
| 12 | FLAP | 0.9 | 〈8.1〉 | 3.0 | 〈5.4〉 | 0.7 | 3.2 | 4.2 | 〈5.3〉 |
| 13 | MRC1 | 1.6 | 3.8 | 0.7 | 〈6.1〉 | 1.2 | 1.0 | 0.8 | 2.1 |
| 15 | MCP4 | 2.9 | 〈4.5〉 | 0.4 | 〈5.6〉 | 1.0 | 2.6 | 0.5 | 〈5.1〉 |
| 26 | MCP1 | 0.7 | 0.5 | 0.2 | 〈9.6〉 | 0.3 | 0.3 | 0.2 | 〈22.7〉 |
| 28 | TGFBI | 1.8 | 3.4 | 1.5 | 〈9.2〉 | 1.0 | 2.7 | 2.4 | 〈7.2〉 |
| 31 | MDL-1 | 1.0 | 1.3 | 0.6 | 1.9 | 1.0 | 0.9 | 0.6 | 1.5 |
| 63 | IRF4 | 〈7.9〉 | 2.2 | 〈9.3〉 | 0.4 | 〈10.7〉 | 〈8.1〉 | 〈8.4〉 | 0.8 |
| S2 | CCR1 | 0.8 | 0.8 | 0.3 | 1.6 | 0.6 | 0.6 | 0.5 | 1.8 |
| S3 | CCR7 | 3.1 | 1.5 | 0.1 | 0.1 | 2.7 | 0.8 | 0.2 | 0.2 |
| S4 | DC-Lamp | 〈7.4〉 | 〈6.3〉 | 0.4 | 0.4 | 〈14.5〉 | 〈8.9〉 | 0.6 | 0.6 |
| S5 | E-cadherin | 1.0 | 1.0 | 0.9 | 0.9 | 1.2 | 1.2 | 0.9 | 0.6 |
| S6 | DEC205 | 2.9 | 1.6 | 0.8 | 0.7 | 3.1 | 1.9 | 0.5 | 0.5 |
| Gene no. . | Name . | Donor set 1 . | Donor set 2 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mo DCs . | CD1a+DCs . | CD11c−DCs 0 . | CD14+DCs . | Mo DCs . | CD1a+DCs . | CD11c−DCs . | CD14+DCs . | ||
| Day 9 . | Day 18 . | Day 0 . | Day 18 . | Day 9 . | Day 18 . | Day 0 . | Day 18 . | ||
| 4 | TARC | 〈20.0〉 | 〈24.0〉 | 0.6 | 0.8 | 〈12.2〉 | 〈9.2〉 | 0.7 | 0.8 |
| 7 | CD20-like | 0.8 | 〈18.0〉 | 1.7 | 〈20.7〉 | 0.8 | 〈9.0〉 | 〈4.2〉 | 〈12.8〉 |
| 8 | Ig superfamily Z39Ig | 0.9 | 2.9 | 0.6 | 〈16.4〉 | 1.0 | 1.6 | 0.4 | 〈7.4〉 |
| 10 | GPNMB | 1.7 | 1.2 | 0.8 | 〈5.0〉 | 1.3 | 1.2 | 0.8 | 〈4.0〉 |
| 11 | Eta-1 | 3.2 | 〈6.3〉 | 0.5 | 〈11.8〉 | 1.1 | 0.9 | 0.6 | 〈8.2〉 |
| 12 | FLAP | 0.9 | 〈8.1〉 | 3.0 | 〈5.4〉 | 0.7 | 3.2 | 4.2 | 〈5.3〉 |
| 13 | MRC1 | 1.6 | 3.8 | 0.7 | 〈6.1〉 | 1.2 | 1.0 | 0.8 | 2.1 |
| 15 | MCP4 | 2.9 | 〈4.5〉 | 0.4 | 〈5.6〉 | 1.0 | 2.6 | 0.5 | 〈5.1〉 |
| 26 | MCP1 | 0.7 | 0.5 | 0.2 | 〈9.6〉 | 0.3 | 0.3 | 0.2 | 〈22.7〉 |
| 28 | TGFBI | 1.8 | 3.4 | 1.5 | 〈9.2〉 | 1.0 | 2.7 | 2.4 | 〈7.2〉 |
| 31 | MDL-1 | 1.0 | 1.3 | 0.6 | 1.9 | 1.0 | 0.9 | 0.6 | 1.5 |
| 63 | IRF4 | 〈7.9〉 | 2.2 | 〈9.3〉 | 0.4 | 〈10.7〉 | 〈8.1〉 | 〈8.4〉 | 0.8 |
| S2 | CCR1 | 0.8 | 0.8 | 0.3 | 1.6 | 0.6 | 0.6 | 0.5 | 1.8 |
| S3 | CCR7 | 3.1 | 1.5 | 0.1 | 0.1 | 2.7 | 0.8 | 0.2 | 0.2 |
| S4 | DC-Lamp | 〈7.4〉 | 〈6.3〉 | 0.4 | 0.4 | 〈14.5〉 | 〈8.9〉 | 0.6 | 0.6 |
| S5 | E-cadherin | 1.0 | 1.0 | 0.9 | 0.9 | 1.2 | 1.2 | 0.9 | 0.6 |
| S6 | DEC205 | 2.9 | 1.6 | 0.8 | 0.7 | 3.1 | 1.9 | 0.5 | 0.5 |
DC-associated genes listed in Table 1 were further analyzed in different donors using an HI380 microarray (Creagene). Seventeen representative genes are listed in abbreviated form according to the number used in Table 1. The microarray was screened with probes prepared from each high-purity DC subset (purity more than 98%) and from BMTs without subtraction. The ratio of DCs to BMTs for each gene was determined from 2 different sets of experiments after background subtraction and normalization. For better contrast, values over 4 are shown in brackets.
Discussion
The heterogeneity of DCs has been manifested as distinct surface phenotypes, differences in their tissue homing, and at the level of their development. To understand the functional heterogeneity of human DCs, we attempted to explore the differences in the transcriptional profiles of different DC subclasses. Because previous work has revealed specific gene expression in MoDCs, we aimed to delineate the gene expression related to the remainder of the subsets of human DCs, namely CD11c− DCs, CD1a+ DCs, and CD14+ DCs.
In an effort to pool DC-associated genes, we combined the cDNAs derived from the 3 DC subsets (CD1a+ DCs, CD11c− DCs, and CD14+ DCs) and performed a subtraction using combined BMT-cDNAs. The subtracted DC cDNA clones were then examined for their DC specificity by microarray-based differential hybridization using the subtracted cDNA probes of DCs and BMTs. From these combined experiments, we have identified 69 DC-associated genes, including some genes identified previously in MoDCs.26-29 Regardless of the influences of different cytokines, the DC-associated genes commonly acquired during DC development are mostly the ones necessary for typical phenotype or the functioning of the DCs—that is, Ag-uptake/processing/presentation and cell metamorphosis/migration. In addition, throughout the analyses there were frequent appearances of the mitochondrial genes (such as MTATP6, MTCYB, COII) and the genes for Ca++ binding proteins (such as S100B, calmodulin). The up-regulation of these genes seems to be necessary for the 2 typical DC functions requiring a high level of adenosine triphosphate (ATP) and Ca++ modulation. Lastly, the other class of the genes differentially expressed in the DCs was the chemokine genes, such as TARC, MCP1, and MCP4. The identification of such genes was in accordance with the previous findings on MoDCs.26-29 On the other hand, the DC-associated genes revealed in our study did not include the other chemokine genes previously reported with MoDCs, such as MIP-1, DC-CK1, ELC, and MDC.43
The DC-associated genes identified in this study were further examined for their expression profiles among the different DC subsets using an HI380 microarray. Consistent with the result of DC/BMT differential screening, most of the DC-associated genes showed their specificity to one or more of the DC subsets (Table 2). However, in contrast to the results shown in Table 2, 5 genes—BLNK, PC4, IL-2Rγ, nicastrin, and MAD homolog 2—were not clearly overexpressed (DC/BMT ≤ 1) in any of the 4 DC subsets. This could be due to the hypersensitivity of the subtracted cDNA probes used for screening the DC microarray, which resulted in the amplification of the trivial differences between the DCs and the BMTs.
Although this study was quite limited in uncovering the genes selectively expressed in the lymphoid DCs, our results revealing the total absence of the myeloid DC-related messages in CD11c−DCs were likely to reflect the reservation of lymphoid DCs for the varied needs of the host's defense system. In contrast to the lipopolysaccharide (LPS)–responsive MoDCs, this DC subset is able to respond to CpG but not to LPS among the various formats of bacterial stimuli.39 The best known role of CD11c− DCs is its function as the main type I interferon producer at the interphase between innate and adaptive immunity.44,45 While the other functions of CD11c− DCs in vivo is still vague, our finding of IRF4 as one of the main transcripts in CD11c− DCs is likely suggestive of the potential importance of the lymphoid DCs in the homeostasis of both mature T and B cells. As a DC subset with a unique location in the lymphoid organs, such a high responsiveness to interferon may allow this resident DC to control hyperimmunity through the generation of IL-10–producing regulatory T cells.46
Human myeloid DCs can be developed from various sources, including blood monocytes and CD34+ hematopoietic stem cells. Likely reflecting the heterogeneous DCs developed from different precursors under different tissue-cytokine microenvironments, the 3 DC subclasses of myeloid origin (CD1a+ DCs, CD14+ DCs, and MoDCs) appeared to be somewhat different in their gene regulations. According to our data shown in Table 2, the apparent differences were in the range of “from the subtle to the profound,” indicating the presence of shared as well as unique functions. The most outstanding difference was revealed in their expression levels of chemokine and chemokine receptors, implying their remarkable differences in trafficking properties. Based on the previous findings,47MoDCs and CD1a+ DCs, which showed the unique up-regulation of TARC and CCR7, fitted into the authentic migratory DC pattern of traveling from tissue to the draining lymph nodes to convey Ag to T cells. On the other hand, the results from the subset-specific expression analysis indicated that CD14+ DCs were more likely to be tissue resident DCs with a transcription profile of “no CCR7 but more CCR1” and “no TARC but more MCP1 and Eta-1.” The preferential expression of Eta-1 (osteopontin)48 and MCP1 in CD14+ DCs tends to suggest that CD14+ DCs play multiple roles in the regulation of inflammation and tissue remodeling. The outstanding expression of factor XIIIa in CD14+ DCs further supports the likely connection of CD14+ DCs with dermal DCs. Our finding of Eta-1 overexpression by CD14+ DCs certainly provides the missing link between factor XIIIa+ dermal DCs and wound healing, angiogenic, and fibrogenic processes.49-51
The absence or the lower level of DC-Lamp expression has been described in the immature forms of MoDCs,52 CD1a+DCs,53 and CD11c− DCs.38Interestingly, the results of our microarray analyses indicated that DC-Lamp expression was absent in CD14+ DCs at any stage of their development in vitro. In good contrast to this, CD1a+DCs appeared to up-regulate the DC-Lamp as early as day 8 of the culture (Figure 1B, Table 3). However, comparing the surface phenotypes of CD14+ DCs (day 18) with those of CD1a+ DCs (day 8), it seems inappropriate to consider CD14+ DCs (day 18) as less mature than CD1a+ DCs (day 8). In this regard, our data support the consideration of CD14+ DCs as distinct DCs derived from a common myeloid precursor.
TGF-β dependency of Langerhans cell (LC) development is well documented both in vivo54 and in vitro.55,56 It was possible to develop CD1a+DCs, not only from the myeloid precursor but also from other types of myeloid DCs, including blood CD11c+ DCs,56CD14+ DCs,57 and MoDCs,58 with the provision of TGF-β in the culture system. Certainly, the plasticity of the DCs seemed to be far beyond our imagination. Although this has to be further confirmed, our finding of TGF-β–induced protein (TGFBI) among the DC-associated genes seems to insinuate the endogenous production of TGF-β, thus supporting to a certain degree the development of CD1a+ DCs in almost any culture of myeloid DCs. Actually, it is very common to encounter CD1a+MoDCs in the course of a routine MoDC generation, while further exposure to a cytokine cocktail usually sweeps the process off (Figure1). Not much has been described about the actual function of TGFBI except that it is a secreted protein having an RGD motif and that it inhibits the attachment of adhesive cells to plastic.59 In addition to TGFBI, this study identified new members of DC-associated genes. These included Ig superfamily protein (Z39IG),60glycoprotein NMB (GPNMB),61 CD20-like precursor,62 and myeloid DAP12-associated lectin (MDL-1).63 While these genes are seemingly important in DC biology for encoding membrane proteins at the cell surface and as secretory proteins, it is still uncertain where to put them in the context of their DC functions.
The up-regulation of some DC-associated genes is likely to explain the connection of DC subtypes with certain human diseases. These genes include high-affinity IgE receptor α (FcERI) and CD36 of CD1a+ DCs in atopic dermatitis,64 FLAP of CD11c− DCs in human nasal allergies,65 and Eta-1 of CD14+ DCs in erythema elevatum diutinum.49
These findings provide a great deal of important molecular information about each DC subset, which will be useful in further functional studies of DC subsets. Although the biologic significance of these findings is still unclear, further studies in vitro and in vivo may elucidate the functional milieu of the genes.
We are very grateful to Drs Y. I. Yeom, J. H. Park, K. A. Yoon, and S. J. Yang of The Center for Functional Analysis of the Human Genome at the Korea Research Institute of Bioscience and Bioengineering for their kind help in microarray fabrication and analysis. We thank Drs S. Y. Kim and S. R. Nam of the Faculty of Medicine, Chungnam National University, Dr K. J. Baek of the Motae Obstetrician's Office, Dr J. I. Lee of the Shina Obstetrician's Office, and Dr I. S. Kim of the Sungae Hospital for their generous supplies of cord blood and PBMC. We are also grateful to L. Waldron for her generous editorial help.
Supported by the Korea Ministry of Health and Welfare grant 01-PJ1-PG4-01PT02-0003 and in part by the Korea Science and Engineering Foundation grant 2000-1-20200-002-3.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yong Soo Bae, Department of Microbiology, Hannam University, 133 Ojung-dong-, Daedeok-gu, Daejeon 306-791, South Korea; e-mail: ysbae@mail.hannam.ac.kr.

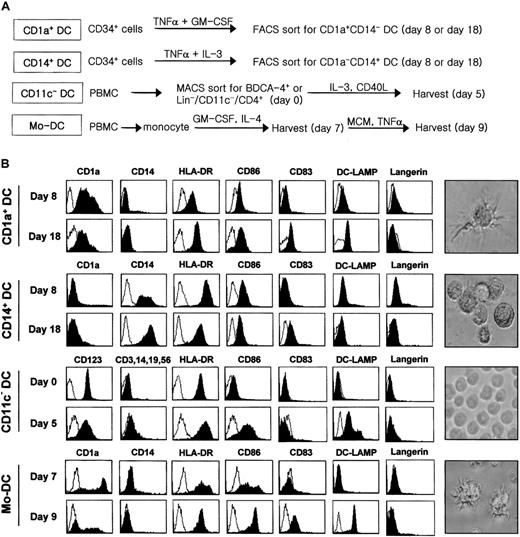
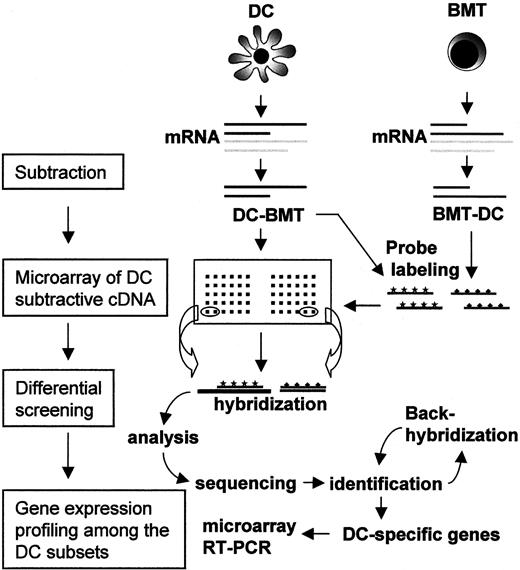

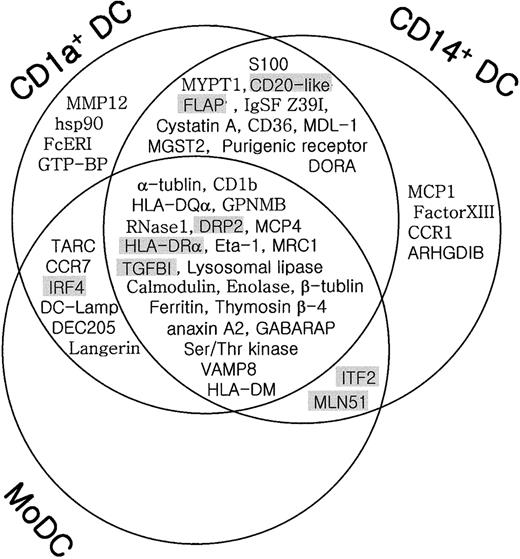
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal