The most frequent chromosomal aberrations in B-cell chronic lymphocytic leukemia (B-CLL) are deletions on 13q, 11q, and 17p, and trisomy 12, all of which are of prognostic significance. Conventional cytogenetic analysis and fluorescence in situ hybridization (FISH) are used for their detection, but cytogenetic analysis is hampered by the low mitotic index of B-CLL cells, and FISH depends on accurate information about candidate regions. We used a set of 400 highly informative microsatellite markers covering all chromosomal arms (allelotyping) and automated polymerase chain reaction (PCR) protocols to screen 46 patients with typical B-CLL for chromosomal aberrations. For validation, we compared data with our conventional karyotype results and fine mapping with conventional single-site PCR. All clonal cytogenetic abnormalities potentially detectable by our microsatellite PCR (eg, del13q14 and trisomy 12) were picked up. Allelotyping revealed additional complex aberrations in patients with both normal and abnormal B-CLL karyotypes. Aberrations detectable in the samples with our microsatellite panel were found on almost all chromosomal arms. We detected new aberrant loci in typical B-CLL, such as allelic losses on 1q, 9q, and 22q in up to 25% of our patients, and allelic imbalances mirroring chromosomal duplications, amplifications, or aneuploidies on 2q, 10p, and 22q in up to 27% of our patients. We conclude that allelotyping with our battery of informative microsatellites is suitable for molecular screening of B-CLL. The technique is well suited for analyses in clinical trials, it provides a comprehensive view of genetic alterations, and it may identify new loci with candidate genes relevant in the molecular biology of B-CLL.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL), the most frequent adult leukemia in Western countries, results from slow clonal accumulation of neoplastic B-lymphoid cells with a phenotype of mature B-lymphocytes.1 Despite a number of common clinical features, B-CLL is a heterogeneous disease.1 The cytogenetic abnormalities most often seen in B-CLL differ from those found in other types of leukemias or lymphomas. Chromosomal translocations are rare; chromosomal deletions represent the most common type of abnormality.2 The most frequently deleted loci are on 13q14, 11q22-q23, 17p13, and 6q21. Another fairly distinct cytogenetic feature of B-CLL is trisomy of the chromosome 12.2-4 To date, surprisingly little is known about specific genes involved in the molecular pathology of particular subtypes of B-CLL. Although some chromosomal deletions, such as those involving 13q14, have been mapped quite extensively, no classic tumor suppressor genes specifically involved in the molecular pathology of B-CLL have been reported so far.2,5-7 Nevertheless, several studies have shown that particular clonal chromosomal abnormalities provide specific prognostic information and thus may help to classify B-CLL into clinically relevant distinct entities.4
A number of different diagnostic techniques are available to identify such cytogenetic or molecular “B-CLL” entities. Conventional cytogenetic analysis has been hampered by the low mitotic index of B-CLL leukemic cells in vitro and their relatively poor response to mitogens,8 and clonal chromosome aberrations have been detected in only 40% to 50% of cases.2 Fluorescence in situ hybridization (FISH) has substantially enhanced the sensitivity of cytogenetic analysis in detecting frequent chromosomal aberrations, as the technique is applicable not only to dividing cells but also to interphase nuclei.2 However, the technique is not applicable to a priori screening.9 Comparative genomic hybridization (CGH) may detect many chromosomal losses and gains in B-CLL,10 but this method has so far not been established for routine diagnostic purposes.11
Microsatellites represent repeats of short di-, tri-, or tetranucleotide motifs in human DNA (for example, -CA-CA-CA-). Microsatellites are abundant throughout the genome and represent highly polymorphic, hence highly informative, molecular markers.12 DNA for microsatellite typing can be used from cells irrespective of their cell-cycle stage, and because of the small size of the polymerase chain reaction (PCR) fragments (in the range of a few hundred base pairs), archival material can also be exploited.13 Microsatellite markers represent a valuable tool for detecting loss of heterozygosity (LOH) in regions with candidate tumor suppressor genes,14 and for depicting allelic imbalances representing chromosomal duplications, DNA amplification, or aneuploidy15-18 as well as microsatellite instability (MSI) caused by disrupted DNA mismatch repair genes.19
Previous studies using limited sets of microsatellite markers have demonstrated a considerable number of LOHs in B-CLL patients.20 21 To extend these data in a systematic fashion, we performed a largely automated comprehensive genome-wide allelotype analysis of 46 B-CLL patient samples, using a set of 400 microsatellite markers evenly spaced throughout the human genome. We identified new loci harboring allelic losses and imbalances in B-CLL and demonstrated the applicability of this diagnostic tool in screening for molecular abnormalities in B-CLL.
Patients, materials, and methods
Patient characteristics, samples, and DNA extraction
Peripheral blood samples of unrelated patients with B-CLL were prospectively collected with the patients' informed consent. Approval was obtained from the institutional review board (University Hospital, Bern, Departments of Medical Oncology and Hematology). None of the patients reported any positive family history for leukemia. The diagnosis of B-CLL was assessed according to standard clinical and laboratory criteria,22 including clonal restriction of immunoglobulin light chain expression and a minimal Matutes score of 4.23 To avoid contamination of leukemic DNA with normal DNA, masking possible genetic alterations, we made a leukocyte count of more than 15 × 109/L mandatory. All 46 patients included in the analysis fulfilled the inclusion criteria. Their mean age was 65 years (range, 38-85 years), and the male-to-female ratio was 2:1. According to the modified Rai stage,24 14 (30.4%) of the 46 patients were in the high-risk group, 26 (56.6%) were in the intermediate-risk group, and 6 (13.0%) were in the low-risk group. The mean disease duration was 50.6 months; in 6 patients (13.0%), the diagnosis had been established for less than 1 month prior to sampling. The mean peripheral blood leukocyte count at sampling was 35.8 × 109/L (range, 17-190 × 109/L). Of the 46 patients, 18 (39%) had never received any antineoplastic treatment at the time of sampling. Mononuclear cells were enriched by density gradient centrifugation on Lymphoprep (1.077 g/mL; Nycomed Pharma, Oslo, Norway). A mean purity of 96.5% mononuclear cells was obtained (range, 82.5%-100%), as assessed microscopically. Matched normal samples consisted of polymorphonuclear cells obtained by density centrifugation after lysis of the erythrocytes, with a mean purity of 95% (range, 71%-99.5%, assessed microscopically). DNA was extracted from normal and malignant cell fractions and was quantified fluorometrically with the Pico Green dsDNA Quantitation Kit (Molecular Probes, Leiden, the Netherlands).
Microsatellite markers, multiplex PCR, and electrophoresis
DNA samples were processed at the Centre National de Génotypage (CNG; Evry, France) with a standardized set of 400 microsatellite markers (PRISM Linkage Mapping Set-MD-10, Applied Biosystems, Foster City, CA).12 These markers are organized into 28 panels and cover all chromosomal arms with an average spacing of 10 centimorgan (cM) between 2 markers and an average heterozygosity rate of 75% (http://www.appliedbiosystems.com/apps/). The chromosomal localization of each marker (as shown in Figures 3 and 4) was assessed by combining data from the Généthon genetic map (http://www.cng.fr) and the National Center for Biotechnology Information (NCBI) GeneMap '99 (http://www.ncbi.nlm.nih.gov/genemap/). The GeneMap '99 was also used for gene mapping. Up to 6 markers were combined and amplified in one multiplex PCR (http://www.cng.fr/e/resources). Multiplex PCR amplifications were carried out with 40 ng of DNA with 3.0 mM MgCl2, 1xPCR buffer, 1mM dNTPs, 0.4 units of AmpliTaq Gold, and the appropriate concentration of each primer pair in a total volume of 10 μL in 384-well plates on a PE-9700 VIPER (Applied Biosystems). PCR conditions were 95°C for 12 minutes (to activate AmpliTaq Gold), followed by 30 cycles at 94°C for 30 seconds, 55°C for 15 seconds, and 72°C for 30 seconds, with a final extension step at 72°C for 15 minutes. Initial experiments using quantitative PCR on an ABI 7700 (Applied Biosystems) showed that the majority of PCRs did not reach the plateau phase under these cycling conditions.
To ensure maximal robustness and reproducibility, all liquid handling steps were carried out with a robot (Automation Partnership, Royston, United Kingdom). The amplified products were purified from primers and salts on Sephadex G50 (Amersham Pharmacia Biotech, Uppsala, Sweden). From 10 to 20 markers belonging to a panel were then combined into one well together with capillary loading buffer (Applied Biosystems) and ROX-400 internal size standard, injected, and electrophoresed on an ABI 3700 capillary sequencer (Applied Biosystems) for allele separation. Traces were automatically analyzed with GENESCAN software and genotypes were assigned with GENOTYPER (Applied Biosystems). Genotype analyses were carried out for each marker and the PCR results were evaluated visually and independently by 3 researchers for identification of alleles and calculation of peak sizes. Particular markers that yielded unsatisfactory amplifications were repeated as single PCRs. Of a total of 10′172 genotypes, 10′040 loci (98.7%) were confirmed on repeat analysis.
Selected chromosomal loci where chromosomal aberrations were detected with the screening marker panel were reinvestigated by conventional single-site PCR. Matching microsatellite markers were chosen from the NCBI GeneMap '99; the data bank of the Wellcome Trust Sanger Institute in Cambridge, United Kingdom (http://www.ensembl.org); and the Généthon data bank. Microsatellite conditions were as described above, except that single-site PCRs instead of multiplex reactions were run.
Definition and scoring of results
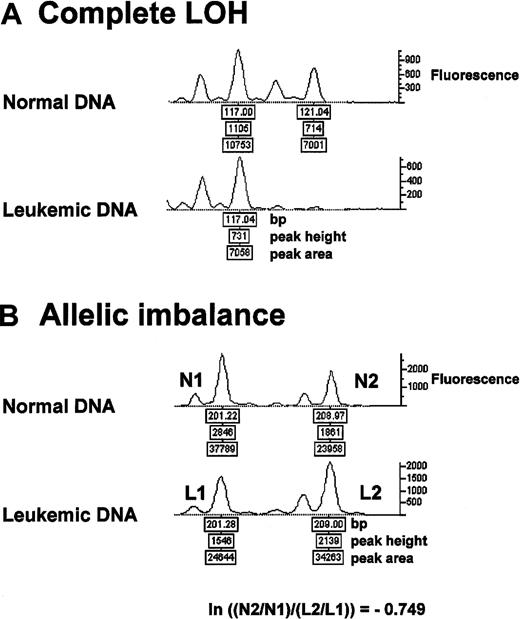
Microsatellite markers showing 2 distinct alleles in the normal DNA were considered informative. Complete absence of 1 allele in leukemic DNA was defined as “complete LOH” (Figure 1A). Loss of both alleles in the leukemic DNA was scored as “biallelic loss,” provided the same result was obtained in 2 different PCR runs from leukemic DNA (data not shown)25 and in the absence of a PCR failure at that locus in the normal DNA. Allelic imbalance was assessed as follows: we compared the allelic ratios of leukemic DNA with those of the matching normal DNA using the formula ln (natural log) [(N2/N1)/(L2/L1)], where N1 and L1 are the peak height values of the shorter allele in normal (N) and leukemic (L) DNA and N2 and L2 are the peak height values of the longer allele in normal and leukemic DNA (Figure 1B). Two-sided conservative cutoff levels were used for the diagnosis of allelic imbalances; all other microsatellite markers were scored as “heterozygous,” that is, normal (Figure 2). The presence of any novel-length allele in leukemia DNA was scored as “microsatellite instability.”26 PCR failure was scored as “no result.”
Complete LOH and allelic imbalance.
Representative electropherogram tracings of microsatellite analysis from the same DNA samples of patient 39. (A) Complete LOH is present at the microsatellite marker D18S70. (B) Allelic imbalance at D17S921 with an allelic ratio of −0.749 (Figure 2). ln = natural log.
Complete LOH and allelic imbalance.
Representative electropherogram tracings of microsatellite analysis from the same DNA samples of patient 39. (A) Complete LOH is present at the microsatellite marker D18S70. (B) Allelic imbalance at D17S921 with an allelic ratio of −0.749 (Figure 2). ln = natural log.
Diagnosis of allelic imbalances.
ln (allelic ratio) of the normal DNA − ln (allelic ratio) of the leukemic DNA was plotted for all patients and all microsatellite markers (except noninformative markers and markers showing complete LOH or biallelic losses). Note that ln (allelic ratio) = ln [(N2/N1)/(L2/L1)] = ln (N2/N1) − ln (L2/L1) and that the values are symmetrically distributed around 0 (representing the case N2/N1 = L2/L1). A total of 879 markers with allelic ratio values above 0.199 [upper cut-off level, calculated as 75% quartile + 3 × (75% quartile − median)] or below −0.207 [lower cut-off level, calculated as 25% quartile − 3 × (median − 25% quartile)] were assessed as harboring allelic imbalances (see Mosteller et al49 for rationale). All other ratios were scored as heterozygous (ie, normal).
Diagnosis of allelic imbalances.
ln (allelic ratio) of the normal DNA − ln (allelic ratio) of the leukemic DNA was plotted for all patients and all microsatellite markers (except noninformative markers and markers showing complete LOH or biallelic losses). Note that ln (allelic ratio) = ln [(N2/N1)/(L2/L1)] = ln (N2/N1) − ln (L2/L1) and that the values are symmetrically distributed around 0 (representing the case N2/N1 = L2/L1). A total of 879 markers with allelic ratio values above 0.199 [upper cut-off level, calculated as 75% quartile + 3 × (75% quartile − median)] or below −0.207 [lower cut-off level, calculated as 25% quartile − 3 × (median − 25% quartile)] were assessed as harboring allelic imbalances (see Mosteller et al49 for rationale). All other ratios were scored as heterozygous (ie, normal).
Cytogenetic analysis
Purified peripheral blood mononuclear cells (16 to 48 × 106) were cultured at 2 × 106/mL for 72 to 96 hours (37°C, 5% CO2) in RPMI 1640 containing glutamine (Seromed/Oxoid, Basel, Switzerland) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin (Life Technologies, Basel, Switzerland), 50 U/mL heparin (Liquemine, Roche, Basel, Switzerland), and 20% human AB serum. Four different culture conditions were used in parallel: (1) basic culture medium without any addition (control), (2) basic medium with 0.02% phytohemagglutinin M (Difco/Brunschwig, Basel, Switzerland), (3) basic medium with 2 μg/mL phorbol-12,13-dibutyrate (Sigma, St Louis, MO), and (4) basic medium with 10% phytohemagglutinin tetradecanoyl phorbol acetate–leukocyte conditioned medium (PT-LCM). PT-LCM was prepared by cultivating peripheral blood mononuclear cells from 10 healthy donors in RPMI 1640 (1 × 106 cells/mL) supplemented with 20% AB serum, 0.04% phytohemagglutinin M (Difco/Brunschwig), and 12-O-tetradecanoyl-phorbol 13-acetate (30 ng/mL; Sigma) for 7 days. Supernatant was harvested, filtered (0.22 μm), and stored at −20°C. Colchicine was added at 0.2 μg/mL for 30 minutes. Hypotonic shock, fixation, and spreading were performed as previously described.27 Chromosomes were stained in G bands. Chromosome defects were defined according to the International System for Human Cytogenetic Nomenclature (ISCN).28 The criteria adopted at the First International Workshop on Chromosomes in Leukemia29 were used for identification of abnormal clones: at least 2 cells with the same trisomy or structural rearrangement or at least 3 cells with the same monosomy.
Results
Technical feasibility of PCR microsatellite allelotyping with the chosen set of 400 markers
Forty-six patients with typical B-CLL were analyzed by means of a panel of 400 microsatellite markers covering all 22 autosomes and the X chromosome with an averaged interval of about 10 cM between 2 adjacent markers. A total of ∼ 47′000 PCR reactions were performed for this study. The overall mean PCR success rate was 84.3% (range, 50.8%-96.3%) per patient. PCR amplification completely failed for 3 markers (D4S1597, D16S515, DXS1226). As expected, overall, 78.2% of the markers were informative in our patient population, providing a mean of 260 informative loci per patient (65.5%; range, 153-308 informative loci; markers on X chromosome included in female patients). Detailed information on informativity rates is available on our Web site (http://www.cx.unibe.ch/dkf5/haemat_onco/Homepage/entry.html).
Data assessment
Figure 1 shows representative output data from the ABI genotyper. To assess allelic imbalances, we adopted the approach of allelic ratios between the leukemic DNA and the matching normal DNA, as proposed by Cawkwell et al,30 but we calculated the ln (natural log; allelic ratio), evaluated different cutoff levels, and then used 2-sided conservative cut-off levels for the diagnosis of allelic imbalances for outliers (Figure2). The symmetrical property and the desirable relation ln (a/b) = −ln (b/a) pushed us to choose this ratio from alternatives including published definitions.18,25,30 In the literature, both allelic peak height25,31 and allelic peak area values32have been used to assess LOH and allelic imbalances. We compared allelic imbalances determined by both approaches in our data set. The Pearson correlation coefficient of the allelic ratio values calculated with peak height and peak area values was 0.8397, and we did not observe a difference in the behavior of these 2 values (detailed data at http://www.cx.unibe.ch/dkf5/haemat_onco/Homepage/entry.html). On the basis of the smaller variance (0.0178 vs 0.028), we decided to use the allelic peak height values.
Allelotyping reveals various regions with high frequency of allelic gains and deletions
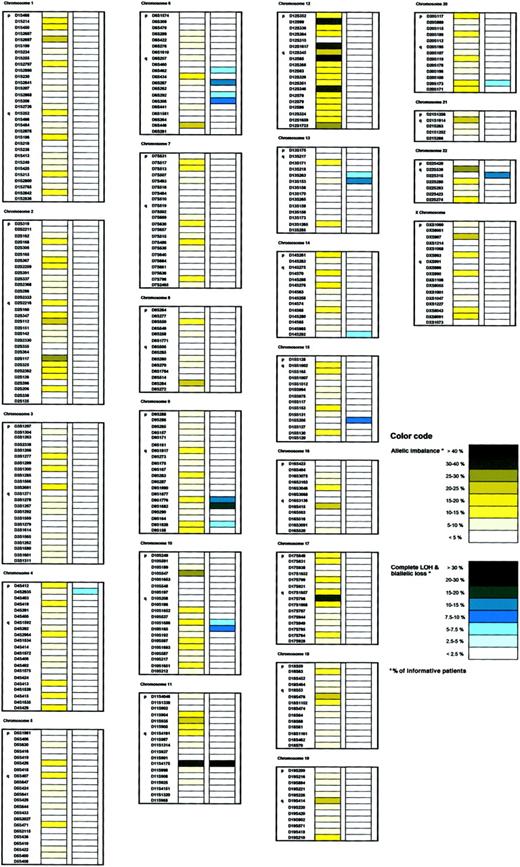
The complete allelotyping results can be downloaded from our Web site (http://www.cx.unibe.ch/dkf5/haemat_onco/Homepage/entry.html). Many patients showed more than one aberration. A total of 879 allelic imbalances were observed (range, 1-63 per patient) at 312 of 397 microsatellite markers (Figure 2) on all chromosomal arms except 13p and 22p, which were represented by only one marker each. Complete LOH was found at 70 microsatellite loci on up to 4 different chromosomal arms in 20 of 46 analyzed patients. Biallelic losses were detected in 8 patients at 11 markers on the long arms of chromosomes 6, 9, and 10. No complete LOH or biallelic losses were found on chromosomes 5, 7, 8, 12, 19, or on the X chromosome. Partial allelic losses due to subclonal deletions and allelic gains such as duplications, DNA amplifications, and aneuploidies (ie, trisomies) present as allelic imbalances in our assay.15-18,33,34While complete LOH and biallelic losses point to clonal monoallelic or biallelic deletions present within the entire population of CLL cells in a sample,30 34 allelic imbalances in regions of clonal deletions most likely reflect subclonal deletions. Total allelic losses comprising complete LOH, biallelic losses, and allelic imbalances were most frequent in 2 distinct regions on chromosome 6q (up to 15% of informative patients) and in one region on chromosomes 9q (up to 25%), 10q (up to 22%), 11q (67%), 13q (up to 22%), 14q (7%), 15q (14%), and 22q (14%) (Figure 3). Additional loci with complete LOH were seen in a subset of patients. Loci with allelic imbalances, but no complete LOH or biallelic deletions in our patient set, were most frequent on chromosome 1p (up to 19% of informative patients), and on chromosomes 2q (up to 25%, with 2 distinct regions identified at this locus), on 4q (19%), 6q (21%), 8q (20%), 10p (27%), 11p (21%), 16q (23%), 17q (up to 35%), 18q (21%), 19q (21%), 21q (22%), 22q (27%), and Xp (22%) (Figure3). Interestingly, allelic imbalances on chromosome 12 were not equally dispersed over the whole chromosome. Four distinct regions on chromosome 12 showed the highest frequency of allelic imbalances: on 12p (up to 33% of informative patients), one around the centromere (up to 44%), and in 2 regions on 12q (up to 31%).
Distribution of allelic imbalances and complete LOH/biallelic losses.
Frequencies of allelic imbalances (yellow) and complete LOH and biallelic losses (blue) per informative patient. See legend in figure for color code. Microsatellite markers per chromosome according to the Généthon map and the NCBI GeneMap '99. Microsatellite markers D4S1597, D16S515, and DXS1226 resulted in PCR failures and are not shown. As allelic imbalances in regions with clonal deletions (complete LOH and biallelic losses) most likely represent subclonal deletions, their frequencies are included in the blue column.
Distribution of allelic imbalances and complete LOH/biallelic losses.
Frequencies of allelic imbalances (yellow) and complete LOH and biallelic losses (blue) per informative patient. See legend in figure for color code. Microsatellite markers per chromosome according to the Généthon map and the NCBI GeneMap '99. Microsatellite markers D4S1597, D16S515, and DXS1226 resulted in PCR failures and are not shown. As allelic imbalances in regions with clonal deletions (complete LOH and biallelic losses) most likely represent subclonal deletions, their frequencies are included in the blue column.
Selected cases with chromosomal aberrations detected by our screening panel of microsatellites were reinvestigated with conventional single-site microsatellite PCR. For example, we were able to confirm the presence of LOH at 1q32-41 in case 9 with additional markers with an average spacing of up to 3.46 cM (data not shown).
Comparison of microsatellite allelotyping and cytogenetic analysis
Conventional cytogenetic analysis was successfully performed on 15 of 16 patient samples on which it was attempted. The number of metaphases fully analyzed varied between 6 and 70, depending on availability of the material and cytogenetic findings such as the presence of nonclonal defects. Chromosome resolution ranged from 350 to 650 bands. Cytogenetic data of the patients and their complete allelotyping results are shown in Figure4. Eight patients (53%) had clonal cytogenetic abnormalities. In 2 of them, the abnormalities consisted of sex chromosome aneuploidies. Interphase FISH analysis performed on the sample from patient 36 revealed 0.6% of cells (1000 nuclei scored) with either 3 (0.4%) or 4 signals for the X chromosome, suggesting either the nonclonal nature of the observed aneuploidy or the existence of a very small population of aneuploid cells (< 1%) whose constitutional character cannot be excluded. In patient 39, Y loss was probably age-related, as he was 67 years old at the time of sampling, and +Y cells could represent a small abnormal constitutional clone. Ten patient samples presented nonclonal aberrations (data not shown).
Comparison of allelotyping results and cytogenetic analysis.
Matched allelotype and cytogenetic data. Allelotyping: patients (numbers indicated on top) in rows, microsatellite markers on the left in lanes per chromosome as in Figure 3. Note color codes in figure legend. Karyotypes according to ISCN.28
Comparison of allelotyping results and cytogenetic analysis.
Matched allelotype and cytogenetic data. Allelotyping: patients (numbers indicated on top) in rows, microsatellite markers on the left in lanes per chromosome as in Figure 3. Note color codes in figure legend. Karyotypes according to ISCN.28
Concordant findings.
All clonal unbalanced anomalies (deletions, trisomy, and duplication) described by conventional cytogenetic analysis were detected by the corresponding microsatellite markers. The trisomy 12 in patient 35, as well as del(6)(q?13q?21) and dup(1)(q21q32) in patient 32 and del(13)(q12q14) in patient 12, detected by cytogenetic analysis presented either as complete LOH or as allelic imbalances on microsatellite analysis.
Additional aberrations detected by allelotyping only.
In addition to the aberrations detected by conventional cyogenetic analysis, 14 loci with complete LOH or biallelic losses and 252 loci with allelic imbalances were detected by microsatellite analysis, for instance, on 6q in patient 22 and on 15q in patient 34, both of which had a normal karyotype. Patient 39 revealed allelic imbalances at several markers on 9q that were not detected by cytogenetic analysis. The loss of one allele on microsatellite analysis despite the presence of 2 cytogenetically normal chromosomes 20 in patient 2 might be caused by a loss of one chromosome 20 followed by duplication of the remaining homolog through a submicroscopic rearrangement (deletion or somatic recombination), all of which occurred relatively early in the evolution of the disease in this patient.
Cytogenetic findings not detected by allelotyping.
As predicted, balanced translocations such as t(2;3)(q?33;q?21), in patient 33, were not detected by microsatellite analysis.
Clinical correlations
In the 6 early-stage patients, the frequency and localization of LOH differed from the overall results for the entire patient population. Genomic alterations detected by our microsatellite panel were particularly frequent on 1q (D1S213, 50%), 4p (D4S2935, 50%), and 1p (D1S255, 17%). In all 6 patients allelic imbalances were detected at D12S85 on 12q/cen, 4 patients had additional allelic imbalances at markers on 12p (D12S99, D12S336, D12S1617) and 12q (D12S346, D12S79), and 3 patients showed allelic imbalance on 8q (D8S1784). In the entire patient population, no single aberrant marker correlated with disease stage. The mean number of microsatellite markers showing allelic imbalances (excluding complete LOH) was slightly higher in pretreated patients (22.4) than in untreated patients (14.0) (P =.029 [2-sided Wilcoxon test], with asymmetric distribution of the data set).
Microsatellite instability in B-CLL is infrequent
In 22 patients (48%), a total of 41 novel alleles (range, 1-11 markers per patient) were seen in the leukemic DNA. In untreated patients, a mean of 0.6 microsatellite markers showed microsatellite instability, compared with 1.1 microsatellite markers in treated patients. This confirms data from prior reports20 35suggesting that microsatellite instability is present in a subset of patients with B-CLL but is infrequent overall (0.003% of all successfully amplified markers in our patients).
Discussion
We would like to highlight a number of conceptual and technical issues arising from this high-resolution, genome-wide allelotype analysis in B-CLL, the first of its kind in the literature. Molecular allelotyping based on a microsatellite PCR system with a large pretested set of markers representing genetic landmarks suitably spaced within the entire human genome has so far been conducted in solid tumors18,25,31,36 and some hematological malignancies.37 We show in this report that the technique is applicable to B-CLL, where highly purified neoplastic cells are available from peripheral blood and neutrophil granulocytes separated by density centrifugation serve as an easily accessible and pure source of normal (constitutional) DNA. For the interpretation of allelotype raw data, we established reproducible criteria, notably a rationale for using allele height rather than peak area values to quantify particular alleles. In addition to complete LOH and biallelic losses reflecting clonal deletions, B-CLL allelotype data should include allelic imbalances, as proposed here. Chromosomal aneuploidies (ie, the frequent trisomy 12 in B-CLL), duplications, and amplifications,15-18 as well as subclonal chromosomal deletions,33 34 typically light up in this pattern. Contamination as a cause of such imbalances was ruled out by careful preparation of both leukemic and normal cell fractions (Figure1).
Up to now, information on genetic alterations in B-CLL was gained by techniques such as conventional cytogenetic analysis, FISH, and CGH, which, despite their advantages, are hampered by a number of logistical or technical limitations. Conventional cytogenetic analysis permits the study of the whole karyotype by a single test, but it may be limited by the low mitotic index of B-CLL leukemic cells.2,8 The FISH technique is sensitive and specifically targets regions of interest, but it depends on the preknowledge of candidate regions, is limited to currently available probes, and cannot readily be automated.9 Likewise, CGH is not ideally suited for routine diagnostic use.11 As a validation of our molecular approach, we compared microsatellite PCR data with conventional karyotypes from a representative subpopulation of our patients. Using an optimized protocol for cell culture, it was possible to obtain cytogenetic data from 15 of 16 patients. All clonal cytogenetic abnormalities found in these patients, except the translocations, were indeed identified by microsatellite analysis. In addition, allelotyping showed several allelic losses and imbalances on different chromosomal arms in patients with both normal and abnormal karyotypes. Cytogenetic analysis detects gross aberrations of 10 to 12 Mb,38 and CGH may detect alterations in the 2 to 10-Mb range.11 FISH will identify even much smaller aberrant regions targeted by the DNA probes (usually 20-50 kilobase [Kb]) with a high sensitivity and in the submicroscopic range.2,9 Our set of microsatellite markers scans the genome with an average of 10 cM (about 6 Mb) between 2 markers. Thus, homozygous deletions, which are often less than 1 Mb,25 may be missed. Microsatellite PCR predictably fails to pick up the clonal chromosomal translocations seen in a subset of CLL cases.3,4 The detection of these abnormalities would still require a complementary cytogenetic approach or the use of specific PCR techniques. With the advent of PCR robots, optimized multiplex PCR, and high-throughput allele detection of fluorescent end-labeled products, allelotyping, as confirmed in this report, can be performed in a semiautomated fashion, providing maximal robustness and reproducibilty.39 This aspect provides a clear advantage over techniques that are labor-intensive and require skilled personnel, such as cytogenetic analysis or CGH. Furthermore, given the small size of the PCR fragments, archival material may be exploited by this technique,13 33 which greatly expands the scope of this methodology to include rapid retrospective diagnostic analyses.
Well-known chromosomal aberrations in B-CLL, such as deletions on 6q21, 11q22-q23, 13q14, and 17p13 or trisomy 12,3,4 were confirmed by our allelotype analysis. An attempt to compare our results with data from the literature is therefore of interest. For example, Döhner et al4 have recently reported deletions at 13q14 in 55% of B-CLL cases, detected by FISH, while our method revealed allelic losses at D13S153 in 22.4% of our cases. However, frequencies of the respective data sets cannot be directly compared, since, in contrast to the FISH probes, our panel of microsatellite markers was not specifically chosen to target the minimally deleted region at 13q14.2 The example of data from chromosome 12 illustrates an additional problem. The incidence of trisomy 12 in the study of Döhner et al4 was 16%, compared with the 43.5% allelic imbalances at D12S85 we found in our patients by PCR analysis. While the FISH approach is based on a single targeted probe for the region of interest,2 we used several microsatellite markers on chromosome 12. Aberrations in the allelotype were found to cluster in 4 distinct regions, some of which would be missed by a single specific FISH probe.
The unbiased and global screening approach presented here clearly extends previous reports on LOH in CLL using small sets of microsatellite markers.20,21 It allowed the detection of several aberrant loci that, to the best of our knowledge, have not been detected in typical B-CLL by any other approach. Novel allelic losses on 9q and 22q were detected in up to 25% of our informative patients (Figure 3). Other new loci harboring clonal deletions were found on 1q, 2q, and 21q in a subset of patients. Allelic imbalances possibly mirroring chromosomal duplications, amplifications, or aneuploidies such as trisomies were detected on various chromosomal arms that have not been reported in B-CLL, including 2q, 10p, and 22q (up to 27% of our informative patients). In distinct contrast to other types of leukemias,37 B-CLL seems to be characterized at the molecular level by widespread allelic alterations of relatively small chromosomal regions. As this finding was seen both in early-stage and untreated patients and in pretreated patients, we believe that such chromosomal alterations are rather unlikely to be linked to possible effects of cytostatic drugs typically used in CLL. On the other hand, ionizing radiation and alkylating agents may produce interstrand DNA crosslinks, which are target areas for DNA double-strand repair mechanisms. This in turn might result in mitotic recombination and slippage, potentially leading to loss of chromosomal material.40-44
It is noteworthy that we did not find a particularly high number of cases with clustered losses at a common chromosomal region. Only a few tumor suppressor genes that have been deposited in the GenBank match loci showing complete LOH or biallelic losses in our population of B-CLL patients. Candidate tumor suppressors include the DBCCR1 gene on 9q32-q33, which was altered in up to 25% of our informative B-CLL patients, including early-stage patients. In 22% of our patients, 10q23.3 was altered. This locus contains the tumor suppressor gene PTEN, shown to be inactivated in some cases of lymphoma and multiple myeloma.45,46 Another candidate tumor suppressor gene is the SHBP2 gene on 4p16, a potential negative regulator of the ABL oncogene.47 Half of our early-stage B-CLL patients showed deletions at this locus. Candidate genes in regions showing allelic imbalances include only MYC on 8q24.12-q24 (20% of our patients showed imbalances with this marker) and CXCR4, involved in B-cell lymphopoiesis and myelopoiesis,48 on 2q21 (24%). The MYC gene has previously been suggested to play a role in B-CLL.2 To date, no specific “B-CLL” genes, including tumor suppressor genes, have been identified; further characterization of such target loci, including a fine mapping with additional microsatellite markers, can now be undertaken.
Allelotyping is a novel diagnostic tool complementing established techniques such as morphology and immunophenotyping. Microsatellite PCR can successfully be performed on samples that are harvested, stored, and shipped under less than ideal conditions.13 33Clearly, the precise prognostic relevance of B-CLL allelotypes will have to be assessed in prospective clinical trials, and we believe that from a technical point of view this method is very well suited for such endeavors. In addition, a great potential of this genome-wide screening method is the identification of new loci harboring candidate genes that are potentially relevant in the molecular biology of B-CLL.
We express our appreciation to Madeleine Oestreicher for excellent technical assistance and to D. Assoulin (Department of Mathematical Statistics and Actuarial Sciences, University of Bern, Switzerland) for statistical support. We are grateful to C. Castagné and all collaborators from the Division of Medical Genetics in Lausanne, Switzerland, for skillful technical assistance. A. Tichelli (Department of Hematology/Central Laboratories, University of Basel, Switzerland) contributed valuable patient material. We thank L. Girard (University of Texas Southwestern Medical Center, Dallas, TX) for the kind gift of the software for color formatting.
Supported by grants from the Swiss National Foundation (3200-053596.98), the Swiss Cancer League (KFS 156-9-1995), the Bernese Foundation for Clinical Cancer Research, the Marlies-Schwegler Foundation for Cancer Research, and the Ursula-Hecht-Stiftung for Leukemia Research.
Some of these data were presented at the 42nd annual meeting of the American Society of Hematology, San Francisco, CA, December 1-5, 2000 (abstract no. 3075).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Martin F. Fey, Institute of Medical Oncology, University of Bern, Inselspital, CH-3010 Bern, Switzerland; e-mail:martin.fey@insel.ch.

![Fig. 2. Diagnosis of allelic imbalances. / ln (allelic ratio) of the normal DNA − ln (allelic ratio) of the leukemic DNA was plotted for all patients and all microsatellite markers (except noninformative markers and markers showing complete LOH or biallelic losses). Note that ln (allelic ratio) = ln [(N2/N1)/(L2/L1)] = ln (N2/N1) − ln (L2/L1) and that the values are symmetrically distributed around 0 (representing the case N2/N1 = L2/L1). A total of 879 markers with allelic ratio values above 0.199 [upper cut-off level, calculated as 75% quartile + 3 × (75% quartile − median)] or below −0.207 [lower cut-off level, calculated as 25% quartile − 3 × (median − 25% quartile)] were assessed as harboring allelic imbalances (see Mosteller et al49 for rationale). All other ratios were scored as heterozygous (ie, normal).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/5/10.1182_blood.v100.5.1787.h81702001787_1787_1794/3/m_h81723059002.jpeg?Expires=1769279536&Signature=zYulxUWXrA0LgQs-uhrGEwR2TBdyk5tnMsZ6t4ik7uR4qb~3euZ2kAEdREWYDgOK0W0dS1UL04eH1o4i02nlXD2Ccwowsws2ebQZTh8tiuDGCQ-KDaytBeve4EGL38oZDs7zyE4rLzozgFOyFcpgtlMizD92uJTbITe2X1VXBOOyA1IiOTotTB95tXSSJHiEeiZXmESnXG6yM~JSZOoer1fmTXz9AtADTSCvEXpsOumFm2hacplGh6i4m9G5vytZ72hJfupj-an-o0jaJ6-sUkhmA5x3P4rCkT6umFIq~a~lH7Olo1DshMW6rOflxT0QlUdMBsuMVRk5wNdiPx~7OA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal