We present data on a patient of South Asian origin with recessive hereditary spherocytosis (HS) due to absence of protein 4.2 [4.2 (−) HS]. Protein 4.2 cDNA sequence analysis showed the presence of a novel 41-bp frameshift deletion that predicts a truncated peptide designated protein 4.2 Hammersmith. Quantitative reverse transcription-polymerase chain reaction indicated that the mutant mRNA was unstable. Sequencing of protein 4.2 genomic DNA revealed that the deletion stems from aberrant splicing. The proband was homozygous for a G>T substitution at position 1747 (cDNA numbering) that activates a cryptic acceptor splice site within exon 11 of the protein 4.2 gene (EPB42). The proband's mother was found to be heterozygous for this substitution. Unlike protein 4.2 null mice, the proband's red cells showed no evidence for abnormal cation permeability. Quantitation of red cell membrane proteins was carried out by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), Western blotting, and flow cytometric measurement. CD47, a protein associated with the Rh complex, was markedly reduced to about 1% (in the proband) and 65% (in the mother) that found in healthy controls. The Rh-associated glycoprotein migrated with a higher than normal apparent molecular weight on SDS-PAGE. There was no obvious reduction in Rh polypeptides. These observations indicate that protein 4.2 and CD47 interact in the human red cell membrane. They provide further evidence for an association between the band 3 complex (band 3, ankyrin, protein 4.2, glycophorin A) and the Rh complex (Rh-associated glycoprotein, Rh polypeptides, glycophorin B, CD47, LW) and define a point of attachment between the Rh complex and the red cell cytoskeleton.
Introduction
Protein 4.2 (pallidin) is a major protein of the red cell membrane.1,2 It is myristoylated at the N-terminal glycine residue3 and is also palmitoylated.4 The protein 4.2 gene, EPB42, encompasses about 20 kb and contains 13 exons.5,6 Protein 4.2 binds, through its N-terminal region, to the N-terminal cytoplasmic domain of the band 3 anion exchanger (AE1). This domain of band 3 links the membrane to the underlying cytoskeleton via ankyrin. Band 3 is known to associate with many other proteins, glycophorin A (GPA), carbonic anhydrase, protein 4.1, various glycolytic enzymes, and hemoglobin (for a recent review, see Tanner7), described here as the band 3 complex. The presence of protein 4.2 in the red cell membrane is completely dependent on the presence of band 3 and reduction in the amount of band 3 causes a corresponding reduction in protein 4.2.8 Very recently, protein 4.2 has also been reported to interact with protein p55, which forms part of the glycophorin C (GPC) complex (GPC, protein 4.1, p55).9
Total or almost total lack of protein 4.2 results in an atypical form of hereditary spherocytosis [4.2 (−) HS] in the homozygous (or the compound heterozygous) state. Seven mutations have been identified in the EPB42 gene, of which half are amino acid substitutions.10 In human 4.2 (−) HS erythrocyte band 3 is not detectably reduced. In contrast, protein 4.2 null mice have reduced amounts of band 3 as well as mild spherocytic anemia, altered red cell cation content, and abnormal regulation of cation transport.11
The existence of the Rh protein complex (Rh-associated glycoprotein, Rh polypeptides, glycophorin B [GPB], CD47, LW) in red cell membranes was suggested by the absence or deficiency of these proteins in human red cells with the Rhnull phenotype (for a review, see Cartron12). The Rh-associated glycoprotein (RhAG, 45-75 kDa) is sequence-related to the Rh polypeptides (Rh, 30 kDa), but is N-glycosylated.13 14
CD47 (47-52 kDa, also referred to as integrin-associated protein [IAP]),15-17 is a multispanning membrane protein with broad tissue distribution (for a review, see Brown and Frazier18), which is much reduced in Rhnullred cells.19 CD47 associates with integrins in many cell types, where it has a role in cell signaling and activation. However, CD47 does not act by the same mechanism in mature red cells because red cells lack integrins. Evidence indicates that CD47 may also have integrin-independent functions in lymphocytes.20Thrombospondin 1 is a major extracellular soluble ligand for CD47 and thrombospondin increases the adhesiveness of sickle red cells under shear stress conditions by a large G protein– and tyrosine kinase–mediated mechanism.21 CD47 null mouse red cells have a normal phenotype22 and contain normal amounts of murine RhAG and Rh polypeptides.23 Recent work indicates that CD47 also acts as a marker of self on murine red cells because CD47 null mouse red cells are rapidly cleared from the circulation in normal mice by splenic red pulp macrophages, whereas CD47 on normal mouse red cells prevents this clearance by interacting with the inhibitory signal regulatory protein α (SIRPα) on macrophages.24
Individuals with Rhnull syndrome have hemolytic anemia, stomatocytosis, and spherocytosis,12 indicating the presence of a cytoskeleton-associated defect in the red cells. Biochemical evidence for an interaction of the Rh proteins with the red cell cytoskeleton25,26 is consistent with this view, which is further supported by recent micropipette aspiration studies.27 28 However, the site(s) of linkage between the cytoskeleton and the Rh complex has not been established.
In this paper we describe a study of the red cells of a patient with protein 4.2 (−) HS (protein 4.2 Hammersmith). Red cell protein 4.2 was entirely absent due to the homozygous presence of a mutation that causes missplicing and instability of the mRNA. Although no mutations were found in the CD47 and band 3 cDNAs, the proband's red cells contained very little CD47 protein. These observations indicate that protein 4.2 and CD47 interact in the human red cell membrane. This interaction forms a point of contact between the band 3 complex (band 3, ankyrin, protein 4.2, and GPA) and the Rh complex (RhAG, Rh, GPB, CD47, and LW) and also acts as a link between the Rh complex and red cell cytoskeleton.
Patient, materials, and methods
Case history
A 20-year-old man whose family originated from South Asia (Pakistan) was referred for investigation of lifelong hemolytic anemia. Jaundice accompanied by anemia and splenomegaly had been apparent since early life. Fluctuation of icterus, being more marked during infections or after fasting and less pronounced following exposure to sunlight, was conspicuous. On investigation he was found to be anemic (hemoglobin, 11.8 g/dL; mean corpuscular volume [MCV], 85.5 fL; mean corpuscular hemoglobin [MCH], 29.1 pg; mean corpuscular hemoglobin concentration [MCHC], 34 g/dL) with reticulocytosis (5.44%, 214.9 × 109/L) and hyperbilirubinemia predominantly unconjugated (total 193 conjugated, 26 μM) in type (Table 1).
Red blood cell indices
| Sample . | Units . | Proband . | Mother . | Normal range . |
|---|---|---|---|---|
| Red blood cell | × 1012/L | 4.06* | 5.00 | 4.30 -5.70 |
| Hemoglobin | g/dL | 11.8* | 13.8 | 13.0 -16.8 |
| Hematocrit | Ratio | 0.347* | 0.414 | 0.39 -0.50 |
| MCV | fL | 85.5 | 82.8 | 82.0 -98.0 |
| MCH | pg | 29.1 | 27.6 | 26.7 -33.0 |
| MCHC | g/dL | 34.0 | 33.3 | 31.4 -35.0 |
| Reticulocytes | × 109/L | 214.9* | 39.80 | 27.7 -86 |
| Reticulocytes | % | 5.44* | 0.88 | 0.60 -1.80 |
| RDWCV | % | 16.0* | 12.7 | 11.6 -15.5 |
| RDW-SD | fL | 50.1 | 38.3 | 38.1 -51.3 |
| Sample . | Units . | Proband . | Mother . | Normal range . |
|---|---|---|---|---|
| Red blood cell | × 1012/L | 4.06* | 5.00 | 4.30 -5.70 |
| Hemoglobin | g/dL | 11.8* | 13.8 | 13.0 -16.8 |
| Hematocrit | Ratio | 0.347* | 0.414 | 0.39 -0.50 |
| MCV | fL | 85.5 | 82.8 | 82.0 -98.0 |
| MCH | pg | 29.1 | 27.6 | 26.7 -33.0 |
| MCHC | g/dL | 34.0 | 33.3 | 31.4 -35.0 |
| Reticulocytes | × 109/L | 214.9* | 39.80 | 27.7 -86 |
| Reticulocytes | % | 5.44* | 0.88 | 0.60 -1.80 |
| RDWCV | % | 16.0* | 12.7 | 11.6 -15.5 |
| RDW-SD | fL | 50.1 | 38.3 | 38.1 -51.3 |
RDWCV indicates red cell distribution width coefficient variation; RDW-SD, RDW–standard deviation.
Outside normal range.
Morphologic examination of peripheral blood revealed atypia that included ovalospherocytic and pincered red cells. Mean channel fluorescence intensity of red cells determined by flow cytometry after labeling with eosin-5-maleimide29 was 41.6 U (reference range 53.9 ± 3.2 U), confirming the presence of HS.29Additional studies including red cell adenosine triphosphate, glycolytic intermediates, reduced glutathione and nucleotides, as well as globin mass spectrometry and expression of glycosylphosphatidylinositol-linked proteins proved normal. Red cell glucose-6-phosphate dehydrogenase activity was elevated (16.3 IU/g hemoglobin) consistent with reticulocytosis. In the face of hyperbilirubinemia disproportionate to the degree of hemolysis, the uridine diphosphate-glucuronosyltransferase 1A1 promoter genotype was determined. The propositus was found to be homozygous for the (TA)7 allele linked to Gilbert syndrome and increased risk of cholelithiasis in HS.30 The appearance of the gallbladder and biliary tract was normal on ultrasonography. Of the parents, who are consanguineous, the mother exhibited no hematologic abnormality. The father was not available for study. Informed consent was sought and given in accordance with the Declaration of Helsinki for the studies undertaken.
Red cell membrane protein analysis
Preparation of red cell membranes, peptide N-glycosidase F (PNGase F) treatment, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), Coomassie blue staining, and Western blotting of membrane proteins were carried out as previously described.31,32 Quantitation of proteins was done by scanning densitometry. Monoclonal antibodies are listed in Tables2 and 3and were used as previously described.33,34 Rabbit polyclonal anti-Rh,35 anti-p55, and anti-4.136 were used. Rabbit anti-CD47 was raised against the synthetic peptide ASNQKTIQPPRNN (with an additional N-terminal cysteine for coupling [single-letter amino acid codes]), which represents the C-terminal sequence. Polyclonal antibodies against protein 4.2 were used as before.37 For flow cytometric measurement, red cells were fixed with dimethyl suberimidate (Sigma-Aldrich, Poole, United Kingdom)38 prior to incubation with appropriate monoclonal antibody, followed by fluorescein isothiocyanate–conjugated antimouse (Fab′)2antibody and analyzed in a gated red cell region on FL-1 of the Coulter EPICS-XL counter (Beckman Coulter, High Wycombe, United Kingdom).
Quantitation of immunoblotting results on the proband and mother
| Protein . | Antibody . | Proband % of controls . | Mother % of controls . | n . |
|---|---|---|---|---|
| Protein 4.2 | Polyclonal | 0 | 97 (± 2) | 4 |
| Band 3 | Coomassie | 102 (± 12) | 103 (± 10) | 4 |
| GPA (dimer) | BRIC 163 | 92 (± 5) | 109 (± 3) | 3 |
| GPA (monomer) | BRIC 163 | 99, 93 | 96, 106 | 2 |
| Rh30 | Polyclonal | 101 (± 12) | 99 (± 8) | 7 |
| RhAG | LA1818 | 99 (± 5) | 95 (± 9) | 6 |
| CD47 | C-terminal polyclonal | 0.8 (± 0.3) | 68 (± 4) | 3 |
| CD47 | BRIC 125 | 1.4 (± 1.1) | 58 (± 10) | 4 |
| LW | BS56 | 211 (± 38) | 204 (± 28) | 3 |
| GPB (dimer) | R1.3 | 71 (± 5) | 95 (± 4) | 4 |
| GPB (monomer) | R1.3 | 91 (± 5) | 90 (± 8) | 4 |
| Protein 4.1 | Polyclonal | 116 (± 16) | 108 (± 13) | 4 |
| GPC | BRIC 4 | 100 | NT | 1 |
| p55 | Polyclonal | 109 (± 13) | NT | 3 |
| Protein . | Antibody . | Proband % of controls . | Mother % of controls . | n . |
|---|---|---|---|---|
| Protein 4.2 | Polyclonal | 0 | 97 (± 2) | 4 |
| Band 3 | Coomassie | 102 (± 12) | 103 (± 10) | 4 |
| GPA (dimer) | BRIC 163 | 92 (± 5) | 109 (± 3) | 3 |
| GPA (monomer) | BRIC 163 | 99, 93 | 96, 106 | 2 |
| Rh30 | Polyclonal | 101 (± 12) | 99 (± 8) | 7 |
| RhAG | LA1818 | 99 (± 5) | 95 (± 9) | 6 |
| CD47 | C-terminal polyclonal | 0.8 (± 0.3) | 68 (± 4) | 3 |
| CD47 | BRIC 125 | 1.4 (± 1.1) | 58 (± 10) | 4 |
| LW | BS56 | 211 (± 38) | 204 (± 28) | 3 |
| GPB (dimer) | R1.3 | 71 (± 5) | 95 (± 4) | 4 |
| GPB (monomer) | R1.3 | 91 (± 5) | 90 (± 8) | 4 |
| Protein 4.1 | Polyclonal | 116 (± 16) | 108 (± 13) | 4 |
| GPC | BRIC 4 | 100 | NT | 1 |
| p55 | Polyclonal | 109 (± 13) | NT | 3 |
NT indicates not tested.
Flow cytometric analysis of monoclonal antibody binding to red cells
| Protein . | Monoclonal antibody . | Controls (n = 3)3-150 . | Proband3-150 . | Mother3-150 . |
|---|---|---|---|---|
| Band 3 | BRIC 6 | 246 (± 15) | 269 | 244 |
| GPA | R10 | 112 (± 6) | 105 | 108 |
| CD47 | BRIC 126 | 102 (± 8) | 25 | 86 |
| CD47 | BRIC 32 | 80 (± 4) | 20 | 68 |
| RhAG | LA1818 | 111 (± 17) | 132 | 74 |
| Rh | BRIC 69 | 184 (± 15) | 120 | 159 |
| GPC | BRIC 10 | 163 (± 2) | 165 | 156 |
| Protein . | Monoclonal antibody . | Controls (n = 3)3-150 . | Proband3-150 . | Mother3-150 . |
|---|---|---|---|---|
| Band 3 | BRIC 6 | 246 (± 15) | 269 | 244 |
| GPA | R10 | 112 (± 6) | 105 | 108 |
| CD47 | BRIC 126 | 102 (± 8) | 25 | 86 |
| CD47 | BRIC 32 | 80 (± 4) | 20 | 68 |
| RhAG | LA1818 | 111 (± 17) | 132 | 74 |
| Rh | BRIC 69 | 184 (± 15) | 120 | 159 |
| GPC | BRIC 10 | 163 (± 2) | 165 | 156 |
Mean channel fluorescence units.
Analysis of genomic DNA
Genomic DNA was isolated from blood samples by using Isocode Stix (Schleicher and Schull, Dassel, Germany) or buffy coats. The coding regions, exons 2-20 of band 3 and 1-11 of CD47, were analyzed for single-stranded conformational polymorphisms (SSCPs) using exon-specific primers to band 339 and CD47 (cDNA) sequence15 compared to chromosome 3 (Human Genome Draft Sequence, National Center for Biotechnology Information). Exon 11 of protein 4.2 was sequenced in the ABI PRISM 310 Genetic Analyser Automatic Sequencer (Applied Biosystems, Warrington, United Kingdom) using exon-specific primers; forward primer: CCTGAGTCCTTTGTATTGTGT and reverse primer: GGGCCTGGATTCCTTCTGA.
Analysis of reticulocyte cDNA
Preparation of reticulocyte mRNA from the proband, mother, and healthy control was carried out as described.40First-strand cDNA was prepared using the RETROscript kit (Ambion, Austin, TX). The entire coding regions of protein 4.2, band 3, and CD47 cDNA were sequenced, in both directions, either manually as described previously32 or automatically (as above) following reverse transcription–polymerase chain reaction (RT-PCR) from the mRNA. The deletion in the protein 4.2 cDNA was found through sequencing the RT-PCR product obtained using the forward and reverse primers GGATGCCCAGATCTCAGTGA and TGTAGCTCCTCTCTCTGTGA, respectively.
Quantitative RT-PCR
Quantitative (real-time) RT-PCR analysis was carried out using the SYBR Green kit and an ABI Prism 7700 Sequence Detector (Applied Biosystems). First-strand cDNA, (2.5% of a preparation from 4 μg total RNA) was used in a 40-cycle 2-step PCR. PCR cycle parameters were 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 58°C for 1 minute. The forward and reverse primers for the band 3 amplicon were ACGCTTCCTCTTTGTGTTGCT and TGTAGGCATCTATGCGGAACA, respectively, and for the protein 4.2 amplicon were TGACTGCATCCAGGCAGAGT and TCCACTTCTCTACCTGCTTGT, respectively. The number of target copies of each gene was interpolated from its detection threshold (CT) value using a control cDNA standard curve included on each plate. Each transcript was assayed 3 times and the median CT values were used for analysis.
Cation transport studies
Intracellular Na+ and K+ contents of red cells were measured within 1 hour of taking the blood, which was kept at 20°C. K+ influx was measured41 using86Rb tracer in a solution containing (mM): Na, 145; K, 5; Cl, 150; MOPS (3[N-Morpholino]propanesulphonic acid; pH 7.4 at room temperature), 15; glucose, 5; and ouabain or bumetanide, 0.1, where appropriate.
Results
Protein 4.2 and the band 3 complex in the proband red cells
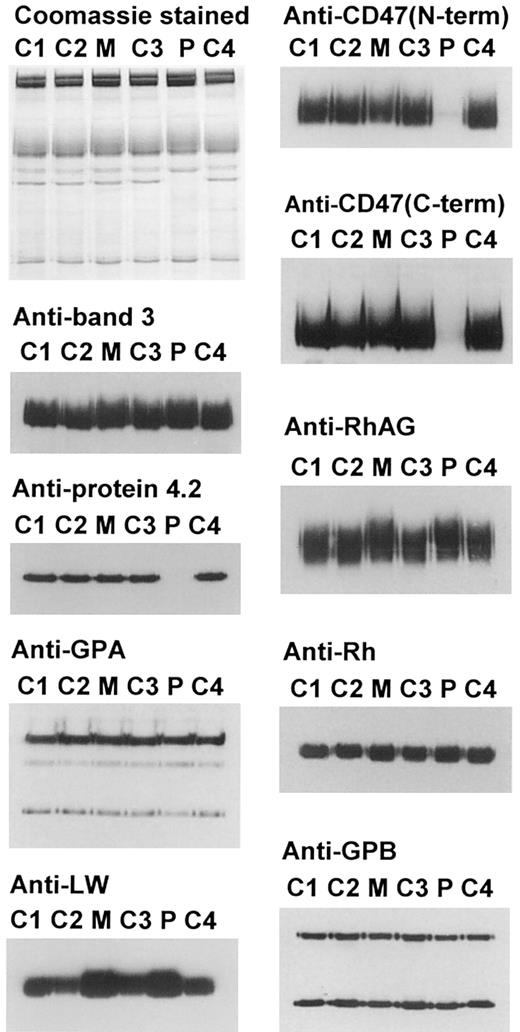
Western blotting and SDS-PAGE of the red cell membrane proteins of the proband showed that protein 4.2 was completely missing but the membranes contained normal amounts of band 3 and GPA, 2 other members of the band 3 complex (Table 2 and Figure1). The trailing edge of the band 3 in the proband and mother migrated slightly more slowly than in the controls, suggesting it may be more highly glycosylated. Flow cytometric analysis of the red cells of the proband and mother (Table3) showed normal binding of antiband 3 antibody (BRIC 6) and anti-GPA antibody (R10).
Electrophoretic and immunostaining of the proteins of the band 3 complex and of the Rh complex.
Erythrocyte ghost membranes were separated on 8% or 10% Laemmli gels. Loading: C1, C2, C3, C4, controls 1-4; M, mother; P, proband. Immunoblotting used polyclonal antibodies against protein 4.2, C-terminal region of CD47 and Rh polypeptides, and monoclonal antibodies: BRIC 170 (band 3), BRIC 163 (GPA), BRIC 125 (N-terminal region of CD47), LA1818 (RhAG), BS56 (LW), and R1.3 (GPB).
Electrophoretic and immunostaining of the proteins of the band 3 complex and of the Rh complex.
Erythrocyte ghost membranes were separated on 8% or 10% Laemmli gels. Loading: C1, C2, C3, C4, controls 1-4; M, mother; P, proband. Immunoblotting used polyclonal antibodies against protein 4.2, C-terminal region of CD47 and Rh polypeptides, and monoclonal antibodies: BRIC 170 (band 3), BRIC 163 (GPA), BRIC 125 (N-terminal region of CD47), LA1818 (RhAG), BS56 (LW), and R1.3 (GPB).
Mutations in band 3 could disrupt the protein 4.2 binding site and cause protein 4.2 to be absent. We therefore sequenced the entire coding region of the band 3 and protein 4.2 cDNAs from the reticulocyte cDNA of the proband. No alterations were found in the band 3 cDNA, but sequencing showed that the proband was homozygous for a deletion in the protein 4.2 cDNA. We named this mutant, protein 4.2 Hammersmith. This 41-nucleotide (nt) frameshift deletion (nt 1709-1749; numbering according to Sung et al2) removes the 5′ region of exon 11 (Figure 2C) and introduces a stop codon (TAG) as the ninth full codon of the frameshifted region following the deletion (this overlaps codon 593 [AGA→Arg593] of the normal cDNA), which results in a truncated protein containing 578 residues compared to the 721 residues of normal protein 4.2.
The 41-bp frameshift deletion (nucleotides 1709-1749) in the 5′ end of exon 11 protein 4.2 cDNA.
(A) Normal protein 4.2 cDNA sequence shown between nucleotides 1699-1779 (numbered according to Sung et al2). (B) The equivalent sequence in protein 4.2 Hammersmith, correctly spliced and containing the 1747G>T mutation (revealed by genomic DNA sequencing, as shown in Figure 4). The 1747G>T mutation introduces a Glu583→Stop substitution. (C) The equivalent misspliced sequence in protein 4.2 Hammersmith is shortened by the 41-bp frameshift deletion (bold and underlined characters in the control), which alters the reading frame and introduces a stop codon (TAG) as the ninth full codon of the frameshifted region following the deletion.
The 41-bp frameshift deletion (nucleotides 1709-1749) in the 5′ end of exon 11 protein 4.2 cDNA.
(A) Normal protein 4.2 cDNA sequence shown between nucleotides 1699-1779 (numbered according to Sung et al2). (B) The equivalent sequence in protein 4.2 Hammersmith, correctly spliced and containing the 1747G>T mutation (revealed by genomic DNA sequencing, as shown in Figure 4). The 1747G>T mutation introduces a Glu583→Stop substitution. (C) The equivalent misspliced sequence in protein 4.2 Hammersmith is shortened by the 41-bp frameshift deletion (bold and underlined characters in the control), which alters the reading frame and introduces a stop codon (TAG) as the ninth full codon of the frameshifted region following the deletion.
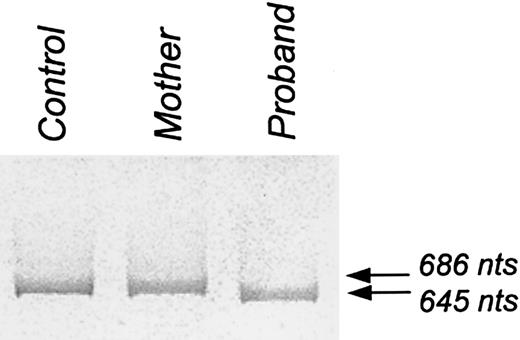
The PCR of the region of protein 4.2 cDNA encompassing exon 11 (nt 1111-1797, numbering as above) produced a single band of 645 nucleotides from the proband and a single band of 686 nucleotides from the control sample (Figure3). Unexpectedly, the mother gave only the normal 686 nucleotide band. Sequence analysis of this region of protein 4.2 cDNA from the mother was normal, indicating that the mother had at least one normal protein 4.2 allele. Quantitative RT-PCR analysis of reticulocyte RNA showed that the level of mutant protein 4.2 mRNA in the proband and the mother was 11% and 62%, respectively, relative to the healthy control. However, the amount of band 3 mRNA in the proband and the mother was found to be 991% and 239%, respectively, of the control, reflecting the hemolytic anemia and reticulocytosis in the proband. When the amount of mutant protein 4.2 mRNA was normalized to the amount of band 3 mRNA, the mutant protein 4.2 mRNA in the proband was calculated to be at a level of only 1.1% of the control, demonstrating that the deletion induced instability in the mutant protein 4.2 mRNA. Consistent with this conclusion, the total amount of protein 4.2 mRNA (normal plus mutant) was also substantially reduced in the mother. The instability of the mutant mRNA accounts for our inability to detect the PCR product corresponding to it in the mother's DNA.
PCR product of a region of protein 4.2 cDNA encompassing exon 11.
PCR of a region of protein 4.2 cNDA encompassing exon 11 (nucleotides 1111-1797; numbered according to Sung et al2) produced a single band of 686 nucleotides from the control and a single band of 645 nucleotides from the proband. Unexpectedly, PCR of this region from the mother showed only a single band of 686 nucleotides.
PCR product of a region of protein 4.2 cDNA encompassing exon 11.
PCR of a region of protein 4.2 cNDA encompassing exon 11 (nucleotides 1111-1797; numbered according to Sung et al2) produced a single band of 686 nucleotides from the control and a single band of 645 nucleotides from the proband. Unexpectedly, PCR of this region from the mother showed only a single band of 686 nucleotides.
To ascertain the underlying cause of the protein 4.2 Hammersmith cDNA deletion we sequenced the genomic DNA from the proband and the mother using exon 11 specific primers (see “Patient, materials, and methods”). We found that the proband was homozygous, and the mother heterozygous, for a 1747G>T substitution (cDNA numbering according to Sung et al2; Figure 4). This mutation activates a cryptic acceptor splice site within exon 11, which leads to the deletion product (Figure 4). The PCR experiment in Figure3 showed that only the mRNA containing the deletion, resulting from abnormal splicing, was detected in the proband. We were unable to amplify any normally spliced mRNA product containing the 1747G>T substitution. If any of the mutant mRNA was correctly spliced the 1747G>T substitution would introduce a different premature stop codon (GAG>TAG, Glu583→Stop; Figure 2B) and we assume that this transcript is so unstable that it is not detected in the PCR reaction.
Genomic mutation (1747G>T) in the proband and the cryptic acceptor splice site.
Normal protein 4.2 genomic DNA sequence is shown between nucleotides 1699-1779 (using cDNA numbered according to Sung et al2), also shown are the “gt” donor and “ag” acceptor sites of intron 10. The equivalent sequence in protein 4.2 Hammersmith shows the cryptic acceptor splice site (nucleotides 1748-1749) activated by the mutation. Wild-type (G) and mutated (t) position 1747 are marked with an arrow. Note that in protein 4.2 Hammersmith the normal “ag” acceptor site of intron 10 is present.
Genomic mutation (1747G>T) in the proband and the cryptic acceptor splice site.
Normal protein 4.2 genomic DNA sequence is shown between nucleotides 1699-1779 (using cDNA numbered according to Sung et al2), also shown are the “gt” donor and “ag” acceptor sites of intron 10. The equivalent sequence in protein 4.2 Hammersmith shows the cryptic acceptor splice site (nucleotides 1748-1749) activated by the mutation. Wild-type (G) and mutated (t) position 1747 are marked with an arrow. Note that in protein 4.2 Hammersmith the normal “ag” acceptor site of intron 10 is present.
CD47 and the Rh complex
Western blotting and SDS-PAGE analysis of the red cell membrane proteins showed that the proband had CD47 content massively reduced to only 1% of healthy controls, whereas the mother had 65% of healthy controls (Table 2 and Figure 1). Similar Western blotting results were obtained with 2 anti-CD47 antibodies reactive with different regions of the protein: a monoclonal antibody reactive with the N-terminal extracellular region and a polyclonal antipeptide antibody directed at the C-terminal sequence of CD47. The absence of CD47 from these 4.2 (−) HS red cells prompted us to examine the CD47 gene in the proband. SSCP of the 11 CD47 exons, using genomic DNA, and sequencing of the coding region of CD47 cDNA showed no mutations were present in the CD47 of the proband.
Protein analysis by immunoblotting of the red cell membranes of the proband and mother showed that the RhAG and Rh polypeptides were present in normal amounts (Table 2 and Figure 1). However, RhAG in the membranes of both the proband and mother migrated slightly slower than normal on SDS-PAGE, and this slower migration was much more pronounced at the trailing edges of the RhAG bands (Figure 1). The RhAG bands of both proband and mother showed normal mobility after treatment with PNGase F, showing that the reduced mobility was due to an increase in size of the RhAG N-glycan in the proband and mother's red cells (data not shown).
Immunoblotting also showed that the proband and mother had increased levels of LW antigens in comparison with the random red cell samples used as controls (Table 2 and Figure 1). The total amount of GPB was also reduced in the proband's red cells and this was reflected in the reduced dimer-to-monomer ratio in these cells (Table 2).
Flow cytometric analysis with 2 different monoclonal anti-CD47 antibodies showed very much reduced reactivity with the proband's red cells and a less marked reduction in reactivity with the mother's red cells (Table 3). The proband's red cells also showed a reduction in reactivity with BRIC 69, a monoclonal antibody reactive with the Rh polypeptides, even though the immunoblotting studies suggest there are normal levels of Rh polypeptides in the membranes. A less significant reduction was observed in the mother's red cells. This probably results from the altered presentation or accessibility of the Rh epitope at the changed cell surface of the proband's and mother's red cells, most likely as a result of the altered glycosylation of RhAG. This altered glycosylation may also affect the binding of anti-RhAG antibody to the red cells of the mother (Table 3). These effects, together with the intrinsic nonlinearity of the fluorescence signals, interfere with quantitative analysis by flow cytometry, and the immunoblotting data (Table 2) more accurately reflect the amount of the proteins present in the red cell membranes.
Protein p55 and the GPC complex
Kuchay et al9 recently reported that an association exists between protein p55 and the C-terminal region of protein 4.2. We therefore investigated protein p55, GPC, and protein 4.1, which are all part of the GPC complex,42 by Western blotting of the mutant red cells. These 3 proteins were present at normal levels in the proband's red cells (Table 2).
Cation transport studies
Because protein 4.2 null mouse red cells show altered cation content and cation fluxes,11 we measured the cation content and potassium fluxes in the freshly drawn red cells of the proband and mother. Table 4 shows that these were all in the normal range, and, in particular, did not show the large changes in bumetanide-sensitive Na+-K+-2Cl− cotransport observed in the mouse protein 4.2 null red cells. The proband and mother did not show any abnormality in the temperature dependence of ouabain plus bumetanide resistant K+ influx (data not shown).
Cation transport studies
| Donor . | Storage time before analysis . | Intracellular Na and K . | K influx at 5 mM external K 86Rb tracer . | |||
|---|---|---|---|---|---|---|
| [Na] . | [K] . | NaK pump . | NaK2Cl cotransport . | Leak . | ||
| mM cells . | Ouabain sensitive . | Bumetanide sensitive . | Ouabain + bumetanide insensitive . | |||
| h/°C . | mM cells/h . | |||||
| Proband | 1/20 | 7.4 | 97.3 | 2.225 | 0.497 | 0.058 |
| Mother | 1/20 | 9.5 | 102.0 | 2.098 | 0.240 | 0.071 |
| Control | 1/20 | 12.1 | 100.6 | 1.851 | 0.846 | 0.096 |
| Normal range | 5-11 | 85-105 | 1-2 | 0-1 | 0.05-0.10 | |
| Donor . | Storage time before analysis . | Intracellular Na and K . | K influx at 5 mM external K 86Rb tracer . | |||
|---|---|---|---|---|---|---|
| [Na] . | [K] . | NaK pump . | NaK2Cl cotransport . | Leak . | ||
| mM cells . | Ouabain sensitive . | Bumetanide sensitive . | Ouabain + bumetanide insensitive . | |||
| h/°C . | mM cells/h . | |||||
| Proband | 1/20 | 7.4 | 97.3 | 2.225 | 0.497 | 0.058 |
| Mother | 1/20 | 9.5 | 102.0 | 2.098 | 0.240 | 0.071 |
| Control | 1/20 | 12.1 | 100.6 | 1.851 | 0.846 | 0.096 |
| Normal range | 5-11 | 85-105 | 1-2 | 0-1 | 0.05-0.10 | |
Discussion
This new case of 4.2 (−) HS has a mild clinical expression, with a well-compensated anemia and a recessive inheritance pattern, in keeping with all cases reported so far. The majority of the previously reported cases are due to missense mutations, which probably alter the binding site of protein 4.2 for band 3.43 However, the absence of protein 4.2 in this case is caused by instability of the mRNA resulting from a premature stop codon, a well-recognized phenomenon,44 with a complex underlying mechanism.45
The marked reduction of CD47 polypeptide in protein 4.2 Hammersmith red cell membranes suggests that, in human red cells, the presence of CD47 is dependent on its interaction with protein 4.2 in the membrane. No mutations were found in the coding region of the CD47 mRNA and no SSCP abnormality was found in the exons of the CD47 genomic DNA, so that it is reasonable to assume that the primary absence of protein 4.2 leads to the nearly total disappearance of CD47 in the proband's red cells. This assumption was strengthened by the very low level of CD47 found in 2 other cases of missing protein 4.246 due to different protein 4.2 mutations, each in the homozygous state and inducing premature stop codons.10,47 It is well known that the primary loss of a component of a complex can lead to the secondary lack, in varying degree, of one or more components of the complex. The relative abundance of the component(s) and the strength of their association with other sites are important determinants for their secondary disappearance. For example, in human red cell membranes the primary absence of protein 4.2 has no influence on the amount of band 3 present, whereas the primary absence of band 3 results in the complete loss of protein 4.2 and the reduction of other proteins.8Our results also show that, although the C-terminal region of protein 4.2 is reported to interact with protein p55,9 the absence of protein 4.2 had no impact on the levels of p55, protein 4.1, or GPC. This suggests that the interaction between protein 4.2 and p55 may not be physiologically relevant in the red cell membrane and contrasts with the situation when GPC or protein 4.1 are missing, because protein p55 is then completely absent.42 Although the primary absence of protein 4.2 affects the level of red cell CD47, the primary absence of CD47 (which has not been described in humans) would be expected to have little effect on the level of protein 4.2. Consistent with this, CD47 null mouse red cell membranes retain normal levels of protein 4.2 (data not shown). Immunoblotting studies also showed that CD47 null mouse red cells have unaltered RhAG and Rh polypeptides (data not shown) confirming a previous abstract that this is the case.23
It is interesting that the protein 4.2 null condition has rather different effects on human and mouse red cells. Although the red cells of protein 4.2 null mice, like the human analogue, exhibit spherocytosis, the mouse red cells have band 3 content reduced to 70% of normal and altered cation content and activity of cation transporters and channels.11 Most striking is the recent report that protein 4.2 null mouse red cells have normal CD47 levels.46 In contrast the human protein 4.2 null red cells examined in this study have normal band 3, cation content, Na+-K+-2Cl− cotransport activity and K+ leak permeability. They show no cation leak even below 37°C. Murine red cells differ from primate red cells in terms of Na/K transport, in that they have more active KCl cotransport.48 In man, protein 4.2 mutations seem unlikely to be a cause of leaky red cell conditions of the hereditary stomatocytosis class.49 The dissimilar consequences of the absence of protein 4.2 in mouse and man could arise either because there are different strengths of association of protein 4.2 with its several binding partners in the 2 species, or because of differences in the structure and organization of the protein 4.2–associated protein complexes.
The other changes observed in the human protein 4.2 null cells are in proteins associated with the Rh complex. The RhAG in the protein 4.2 null red cells had a lower mobility on SDS-PAGE as a result of an increase in size of the N-glycan chain. RhAG carries N-glycan chains of the erythroglycan type containing repeating N-acetyllactosamine units.14 A similar reduction in RhAG mobility is observed in human GPB null red cells and reflects an increase in the size of the N-glycan chain because of a greater number of N-acetyllactosamine repeats on the variant RhAG protein.14 This was attributed to the absence of GPB causing a longer residence time of RhAG within the late Golgi system during the biosynthesis of the Rh complex. The lower mobility of RhAG on SDS-PAGE in the protein 4.2 null red cells also reflects an increase in the size of the N-glycan chain arising from perturbations in the biosynthesis of the Rh complex. The protein 4.2 null membranes also contain reduced amounts of GPB. Both these effects most probably stem from the CD47 deficiency rather than the absence of protein 4.2, because immunoblots of GPB null (S−s− phenotype) and RhAG null (Rhnull U− phenotype) red cell membranes show no reduction in protein 4.2 (data not shown). The reciprocal effects of primary changes in different components of the Rh complex emphasize the intimate structural and biosynthetic relationships between them. Our immunoblotting studies indicate that the proband and mother both had more LW antigens than the random controls used. However, this increase is unlikely to be significant, because both the proband and mother are Rh D+, and the abundance of red cell LW antigens is dependent on the Rh D type (4400 copies in D red cells but 2800 copies in d cells50).
Our evidence suggests that protein 4.2 associates with both the band 3 protein complex and the Rh complex, and comparison of the stoichiometry of these complexes in red cell membranes is of interest. Protein 4.2 is thought to be part of the band 3–cytoskeleton linkage,51,52 and associates with both band 3 and ankyrin.53 Although more than 106 copies of band 3 are present in red cells, only a fraction of these (4 × 105) are present in the ankyrin-associated band 3 tetramers that are linked to the cytoskeleton. The 105 band 3 tetramers complex with 105 ankyrin molecules and 105 protein 4.2 dimers and probably 2 × 105GPA dimers.54 There are also 105 copies of the Rh complex per red cell, which are comprised of heterotetramers of RhAG and the Rh polypeptides associated with GPB dimers. All the Rh complexes could associate with CD47 (2-5 × 104copies/cell) if each CD47 associated with 2 RhAG/Rh core heterotetramers, or up to half of the Rh complexes could be associated with CD47 if the RhAG/Rh heterotetramers bound CD47 with a 1:1 stoichiometry. A small fraction (< 5%) of the Rh complexes associate with the LW glycoprotein (2.8-4.4 × 103 copies/cell). Other studies have suggested that the Rh and band 3 complexes interact directly through a band 3-RhAG/Rh interaction. Coexpression of band 3 enhances the expression of Rh antigens at the surface of K562 erythroleukemic cells.33 34 The relatively minor effect of the absence of protein 4.2 on RhAG and the Rh polypeptides, in contrast to its major effect on CD47, is also consistent with alternative interacting sites for RhAG/Rh. It is well known that GPA and GPB, associated with the band 3 and Rh complexes, respectively, form heterodimers on SDS-PAGE of erythrocyte membranes. Although it is not proven that these glycophorins also heterodimerize in the intact red cell membrane, interactions between these 2 proteins form an additional potential contribution to associations between the 2 protein complexes. From these observations we draw the tentative conclusion that in the membrane the Rh complexes are associated with the subset of band 3 complexes that are tetrameric and associated with ankyrin and the cytoskeleton, and these all form a macrocomplex (shown schematically in Figure 5.) The interaction between the 2 complexes is maintained by protein 4.2–CD47 interactions, direct band 3–RhAG/Rh interactions and very likely also GPA-GPB interactions. The present results explain earlier findings that indicate an association of the Rh antigen complex with the red cell skeleton, and suggest this association occurs by the multiple interactions between the Rh complex and the tetrameric band 3 complexes, ultimately mediated by the band 3-ankyrin–protein 4.2 attachment to the spectrin cytoskeleton.
Schematic representation of the link between the band 3 complex and the Rh complex.
The band 3 complex (band 3, ankyrin, protein 4.2, and GPA) and the Rh complex (Rh polypeptides, RhAG, CD47, and GPB) pack closely together, the association of protein 4.2 with CD47 forming one point of contact. For clarity GPA, GPB, LW, and other peripheral ligands of band 3 are not shown.
Schematic representation of the link between the band 3 complex and the Rh complex.
The band 3 complex (band 3, ankyrin, protein 4.2, and GPA) and the Rh complex (Rh polypeptides, RhAG, CD47, and GPB) pack closely together, the association of protein 4.2 with CD47 forming one point of contact. For clarity GPA, GPB, LW, and other peripheral ligands of band 3 are not shown.
CD47 is present on early hematopoietic cells and remains expressed on cells throughout human erythropoeisis.19,55 RhAG and Rh are expressed before or around the time of band 3 expression, respectively.55 In contrast, protein 4.2 is expressed very late in erythroid maturation, after band 3 and protein 4.1, at a point suggested to be after the assembly of the cytoskeleton on the membrane.56 Our results show that normal protein 4.2 mRNA levels are much reduced in the reticulocytes of the proband's mother. Surprisingly, although the red cells of the proband's mother have a normal complement of protein 4.2, these cells have CD47 content reduced to about half normal and also show altered glycosylation of RhAG. Thus, there are protein 4.2 gene dosage effects on the amount of CD47 and the altered glycosylation of RhAG, but not on the level of protein 4.2 expressed in the mother's red cells. This can be explained if there was a short window during protein 4.2 mRNA expression and translation in which CD47-protein 4.2 associated with Rh complexes can be incorporated into the membrane, and the amount of incorporation during this time is rate-limited by the amount of protein 4.2 synthesized by the cells. This temporal window could be initiated by the start of protein 4.2 mRNA expression and terminated by the cessation of expression of either CD47 itself or any of the major components of the Rh complex (RhAG, Rh, or GPB). Continued expression of protein 4.2 mRNA after this period would allow protein 4.2 from the transcripts of just one protein 4.2 allele to accumulate further and saturate its binding sites on band 3.
Murine CD47 null red cells, when transfused into normal mice, are rapidly cleared from the circulation by splenic red pulp macrophages due to the absence of inhibitory CD47-SIRPα signaling.24 Although the protein 4.2 null mice (which appear to have normal CD47) have a mild HS, it is possible that the almost complete absence of CD47 on the human protein 4.2 null red cells leads to reduced inhibitory CD47-SIRPα signaling in macrophages and contributes to the hemolytic anemia seen in the protein 4.2 null patient.
The absence of CD47 from protein 4.2 null red cells suggests a specific association exists between the 2 proteins in the red cell. We can infer from this that CD47 has a functional role in the mature red cell and is not simply a “leftover” that participated in earlier events during erythroid maturation. The nature of this function in red cells has not been established but we can speculate on possible roles of CD47 based on its activity in other cells. In nonerythroid cells CD47 acts to initiate intracellular signaling pathways as a result of interactions with other cells and with matrix components in both integrin-dependent and integrin-independent fashions.18 The interaction with protein 4.2 gives CD47 the potential to signal between the extracellular environment and the red cell skeleton and thus modulate the mechanical and other properties of the red cell. One situation during which this system may operate is during the passage of red cells through the microcapillary system, where red cells need to be readily deformable to enter and travel through the microcapillaries. Interactions of red cell CD47 with the capillary endothelial cells or matrix, perhaps mediated by a member of the thrombospondin gene family, may act to signal the changes in the cytoskeleton required to adapt the mechanical properties of the red cell to this environment, and perhaps also regulate the other functional and transport properties of the cell.
We thank Prof David Anstee for monoclonal antibodies, Joyce Poole for Rh phenotyping, and Dr Kirstin Finning for advice with real-time PCR. We thank David Roper, Brian Green, Margaret Chetty, and Dr Barbara Wild for biochemical studies and mass spectrometry, Alexis Proust for help with DNA sequencing, and Dr Susan Kelly for referring the patient.
Prepublished online as Blood First Edition Paper, May 13, 2002; DOI 10.1182/blood-2002-03-0706.
Supported in part by grants from the Wellcome Trust, the Indo-French Center for the Promotion of Advanced Research (IFCPAR Project 1903.1, New Delhi, India), the Institut National de la Santé et de la Recherche Médicale (Unité 473), the Institut National de la Santé et de la Recherche Médicale jointly with the Association Française contre les Myopathies ('Réseaux de Recherche sur les Maladies Rares', Project 4MR09F), the Assistance Publique-Hôpitaux de Paris, the Faculté de Médecine Paris-Sud, the Swedish Medical Research Council (31X-14286, 06P-14098), the Swedish Society of Medicine, and The Sir Jules Thorn Trust.
Correspondence:Michael J. A. Tanner, Department of Biochemistry, School of Medical Sciences, University of Bristol, Bristol, BS8 1TD, United Kingdom; e-mail: m.tanner@bristol.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal