Abstract
Chronic lymphocytic leukemia (CLL) is associated with a variety of immunologic disturbances. Hypogammaglobulinemia and autoimmune phenomena are both often present in this disease. In contrast, humoral or cellular antitumor responses are rarely observed. It has been previously shown that antigens detected in patients with malignant diseases can provide information regarding intracellular molecules engaged in the transformation process and can identify tumor antigens that may be useful for development of immunotherapeutic strategies. Serologic identification by recombinant expression cloning (SEREX) has been demonstrated to be a useful method to detect tumor and tumor-associated antigens in a variety of malignancies. Although this approach is complicated in CLL, we used a modified SEREX approach and identified 14 antigens (KW-1 to KW-14) using this methodology. Several clones showed a restricted expression pattern in normal tissues. Moreover, distinctive expression of splice variants and aberrant gene expression in malignant tissue were detected. In this study, 6 antigens were detected exclusively in patients with CLL. Eight antigens were detected also in lymphoma patients. Healthy donors showed antibody responses against only 3 of the identified antigens. T cells with specific cytotoxicity against peptides derived from the 2 antigens tested could be generated from healthy donors. These findings demonstrate that humoral and cellular immune responses against CLL-associated antigens can be detected. Ongoing experiments investigate their potential for the development of immunotherapeutic strategies.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in Western countries. It is increasing in frequency and is being increasingly diagnosed in younger patients. CLL remains incurable using currently available therapies, and younger patients almost invariably die of their disease.1,2 There is therefore a need for better understanding of the disease pathogenesis as well as a need for novel treatment strategies. CLL is in many regards an ideal candidate to study novel immunotherapeutic approaches. The disease has a long natural course, which would allow development of immune responses against the tumor cells, and most patients with CLL are advanced in age and therefore not suitable for aggressive chemotherapeutic regimens. Recent advances in the use of donor lymphocytes following allogeneic stem cell transplantation and the use of nonmyeloablative conditioning regimens have illustrated the existence of a graft-versus-leukemia effect in this disease, demonstrating that the disease can be a target of an ongoing T-cell–mediated immune response.3 However, there are several hurdles to develop immunotherapy in CLL. Patients with CLL often suffer from a variety of immune dysregulations and immunosuppression that can be worsened by conventional therapy with purine analogs and alkylating agents. Although autologous T-cell responses can be generated against the patient's own CLL cells using immunoglobulin (Ig)–derived peptides as target antigens,4,5 Ig-derived vaccine approaches have had limited clinical application. In addition, autologous cytotoxic immune responses are weak and only generated in a minority of patients. Thus, new tumor antigens are required that might represent potential targets for an effective antitumor immune response in CLL and that might also give a broader basis for polyvalent immunization strategies to prevent the escape of antigen loss variants.6
Ideal tumor antigen candidates should be selectively expressed in tumor tissue and detectable in most patients. However, most tumor antigens do not have these ideal characteristics and might be limited in their use by specific but low expression in tumor cells, by low antigenicity, or by broader expression in healthy tissue. Cancer-testis antigens such as NY-ESO-1 are expressed only in tumor cells and testis, making them among the more ideal candidates for immunotherapeutic approaches.7 However, clinical studies targeting these antigens are limited to date.8 Most antigens have broader expression in healthy tissue. The risk of using these antigens as targets of an immune response is induction of autoimmune phenomena. Although this risk must be taken very seriously, several animal studies have shown that there might be a difference in the susceptibility of normal and tumor tissue to the effector arms of the immune response.9-11 Antibody therapy with Her2/neu, which has a broader expression profile, has been demonstrated to increase remissions in patients with breast cancer, whereas no major autoimmune phenomena have been observed so far.12 13
Many tumor antigens have been identified either by cellular and/or humoral responses. SEREX is an approach to detect novel tumor antigens serologically.14 This method is based on the detection of antigens within recombinantly expressed phage libraries by autologous antibodies. It has been used successfully to identify new tumor antigens in several malignancies.7 15-25 There are several obstacles to the use of SEREX in CLL. Many patients with CLL suffer from hypogammaglobulinemia and other immune defects, aggravating the detection of serologic immune responses against potential tumor antigens in these patients. Patients with CLL often have autoantibodies against normal cellular proteins, so that antigens detected by SEREX might represent normal cellular components and not be suitable for developing immunotherapeutic strategies in this disease. In addition, CLL cells express immunoglobulin (Ig). Thus, false-positive results will arise by binding the secondary antibody to Ig expressed by the transfected Escherichia coli. However, we were largely able to overcome these obstacles. Fourteen antigens were identified following recognition by autologous and allogeneic sera of CLL patients. Many of the antigens described here are expressed in a variety of normal tissues. However, some show enhanced expression in hematopoietic cells, tumor samples, and testis. All antigens have been previously described as genes; however, in 6 of them protein function has not investigated experimentally. Of note, in several cases novel splice variants have been detected. Seven of the antigens are novel, whereas 7 have been previously described, 5 of them using SEREX in other malignancies. Six antigens have been exclusively recognized by patients with CLL, whereas 8 have been identified also by patients with other lymphoma. Sera from healthy donors were reactive in some cases against only 3 antigens. This study therefore identifies tumor antigen candidates in CLL that might be useful for the development of immunotherapeutic strategies in this disease.
Patients, materials, and methods
Patient samples
Cell and serum samples were collected with the informed consent of patients and healthy donors and after receiving institutional review board approval from Dana-Farber Cancer Institute and Brigham and Women's Hospital. All patients had diagnosis of CLL confirmed by morphology and flow cytometric analysis. CLL cells and sera were simultaneously collected by leukapheresis performed on 5 different CLL patients who had not been previously treated. These 5 patients were selected as representative of the CLL patient population. Three patients had early-stage disease, and 2 patients had more advanced diseases (Table 1). Purified CLL cells (> 95%) were freshly used to create cDNA libraries, and excess cells were cryopreserved. Additional serum samples were obtained during routine diagnostic procedures from 25 patients with CLL and 5 patients with other lymphomas (2 follicular lymphomas, 1 mantle cell lymphoma, 1 diffuse large cell lymphoma, and 1 Hodgkin lymphoma). Normal sera were collected from 14 healthy donors. All serum samples were processed in exactly the same way. Tumor cells and patient sera were isolated by Ficoll-Hypaque (Amersham, Uppsala, Sweden) density-gradient centrifugation immediately after collection. Sera were stored at −80°C until use.
Clinical characteristics of patients included for generation of CLL-cell cDNA libraries
| Patient . | Age, y . | Sex . | Stage . | Time from diagnosis to collection, y . | Prior treatment . | White blood count, × 109/L . | Autoimmune phenomena . | Isolated antigens . |
|---|---|---|---|---|---|---|---|---|
| 1 | 61 | M | Rai I/BinetA | 5 | None | 48 | None | 3 |
| 2 | 45 | M | Rai II/BinetA | 1 | None | 72 | None | 6 |
| 3 | 50 | M | Rai II/BinetA | 3 | None | 28 | None | 4 |
| 4 | 46 | F | Rai II/BinetB | 3 | None | 54 | None | 1 |
| 5 | 50 | M | Rai IV/BinetC | 8 | None | 466 | AIHA, ITP | 1 |
| Patient . | Age, y . | Sex . | Stage . | Time from diagnosis to collection, y . | Prior treatment . | White blood count, × 109/L . | Autoimmune phenomena . | Isolated antigens . |
|---|---|---|---|---|---|---|---|---|
| 1 | 61 | M | Rai I/BinetA | 5 | None | 48 | None | 3 |
| 2 | 45 | M | Rai II/BinetA | 1 | None | 72 | None | 6 |
| 3 | 50 | M | Rai II/BinetA | 3 | None | 28 | None | 4 |
| 4 | 46 | F | Rai II/BinetB | 3 | None | 54 | None | 1 |
| 5 | 50 | M | Rai IV/BinetC | 8 | None | 466 | AIHA, ITP | 1 |
Purified CLL cells were used for generation of 5 different cDNA expression libraries. Patient characteristics, clinical data, and number of isolated clones are shown. One antigen, KW-5, was selected independently from 2 different libraries.
AIHA indicates autoimmune hemolytic anemia; ITP, idiopathic thrombocytopenic purpura.
RNA extraction and polyA-RNA enrichment
Total RNA was extracted from tumor cells using standard methodology according to the manufacturer's recommendation (Trizol reagent, Life Technologies, Rockville, MD). PolyA-RNA was enriched from total RNA using the Messagemaker reagent assembly kit (Life Technologies). Quality of total RNA and polyA-RNA was assessed using formaldehyde gels.
Construction of cDNA expression libraries
The cDNA expression libraries were constructed from 5 μg polyA-RNA. First-strand cDNA synthesis was performed using an oligo(dT) primer that contained an internal XhoI restriction site. The cDNA was ligated to EcoRI adaptors and digested withXhoI before being size selected, cloned directionally into ZAP expression vector (Stratagene, La Jolla, CA), packaged into phage particles, and transfected into XL1-blue E coli. Libraries containing between 6.2 × 106 and 12.8 × 107 recombinants were amplified and used for immunoscreening.
Immunoscreening of the cDNA libraries
The cDNA libraries were screened with autologous and allogeneic sera in a modification of that previously described.24Serum was diluted 1:10, preabsorbed with transfected E colilysate, and further diluted 1:20 (final serum dilution 1:200). E coli transfected with recombinant λZAPII phages were plated on agar plates. After emergence of visible plaques, nitrocellulose filters, soaked in isopropyl β-d-thiogalactosidase (IPTG) for protein induction, were placed on plates and cultured for 3 hours. After colony lifting, filters were washed in TBST (50 mM Tris [tris(hydroxymethyl)aminomethane], 138 mM NaCl, 2.7 mM KCl, 0.05% wt/vol Tween 20, pH 8.0) and blocked overnight with 1% wt/vol nonfat dry milk in triethanolamine-buffered saline (TBS). The next day the filters were washed and then incubated with alkaline phosphatase–conjugated goat antihuman Fc secondary antibodies (Jackson ImmunoResearch Labs, West Grove, PA), and the reactive phage plaques were visualized by incubating with 5-bromo-4-chloro-3-indolyl-phosphate and nitroblue tetrazolium (Promega, Madison, WI). False-positive clones were not further processed. After prescreening, nitrocellulose membranes were washed again and then incubated with the preabsorbed sera overnight at 4°C. Serum antibodies binding to recombinant proteins expressed in lytic plaques were detected by repeated incubation with the alkaline phosphatase–conjugated goat anti–human IgG antibodies (Jackson ImmunoResearch Labs) and visualization with 5-bromo-4-chloro-3-indilyl phosphate and nitroblue tetrazolium (Promega).
Sequence analysis of the reactive clones
The clones reactive with the sera but not by incubation with secondary antibody alone were subcloned for isolation, purified, and the phagemid cDNA insert excised from the λZAP vector with an ExAssist helper phage system (Stratagene, La Jolla, CA). Plasmid DNA was prepared using the Quiaprep Spin Miniprep kit (Qiagen, Hilden, Germany). Clones were sequenced by the molecular biology core facility at the Dana Farber Cancer Institute using an ABI Prism 373 DNA sequencer (Applied Biosystems, Foster City, CA). Sequences were further analyzed for homology with known genes and proteins using public databases.
RACE-PCR for complete cloning of KW-13
To complete the 5′-sequence of clone KW-13, the RACE-PCR (rapid amplification of cDNA ends–polymerase chain reaction) approach was performed using the SMART RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to the manufacturer's recommendations. The antisense primer sequence was CCGTAGCACC TGCGTGGACT CCTGAACTCG C.
RT-PCR
To evaluate the mRNA expression pattern of the cloned cDNA in normal and malignant tissues, exon-overlapping oligonucleotide primers for PCR were designed to amplify cDNA segments. All primers were commercially synthesized (Life Technologies; Invitrogen, Carlsbad, CA). Reverse transcription (RT)–PCR was performed using 30 amplification cycles in a thermal cycler (Perkin-Elmer, Shelton, CT) at primer-specific annealing temperatures. PCR products were analyzed on ethidium bromide agarose gels, and cDNA panels of healthy and malignant tissues were purchased from Clontech.
Northern blot analysis
Northern blot analysis was performed using standard techniques. Briefly, 10 μg total RNA per lane was dissolved in loading buffer containing formamide and formaldehyde, heated at 65°C, separated on a 1% agarose gel with 16% formaldehyde, and transferred to a nitrocellulose membrane. Subsequent hybridization to the32P-labeled probe (NEN, Boston, MA) and washing were performed. RNA integrity of the membranes was determined using a probe for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript, and mRNA blots from healthy and malignant tissue were purchased from Clontech.
Phage plaque assay
To investigate the antigenicity of detected clones in CLL patients or other malignancies compared with healthy donors, a phage plaque assay was performed. Phages from positive clones were mixed with nonreactive phages of the cDNA library as internal negative controls and used to transfect XL1-blue E coli. IgG antibodies in 1:200 diluted E coli absorbed sera from patients with CLL, other lymphoma, and healthy donors were detected with the immunoscreening assay as described above.
Statistics
The number of detected clones in different patients was correlated with their clinical data (therapy, presence of hypogammaglobulinemia, disease stage, presence of autoimmune phenomena, CD38 expression on CLL cells, and Ig mutational status) using the Fisher exact test. In addition, detection of clones by CLL patients compared with healthy donors was analyzed by the Fisher exact test.
Peptide prediction analysis and bioinformatics
In the case of KW-2 and KW-13, clone-specific sequences were reviewed for peptides 9 and 10 amino acids in length that could potentially bind to major histocompatibility complex (MHC) class I molecules. Two independent computer prediction analyses were used (http://bimas.dcrt.nih.gov/molbio/hla_bind andhttp://syfpeithi.bmi-heidelberg.com) to examine the binding of native peptides to HLA-A*0201 molecules. Analysis was performed for HLA-A*0201 because this MHC class I allele is expressed in approximately 50% of our patients.
Peptide synthesis
Peptides deduced from the amino acid sequence of KW-2 and KW-13 were generated by fluorenylmethoxycarbonyl (Fmoc) synthesis (Sigma-Genosys Biotechnologies, The Woodlands, TX). Purity was determined by reverse phase high-performance liquid chromatography (HPLC) and verified by mass spectral analysis. Peptides were dissolved in PBS at a concentration of 2 mg/mL and stored at −70°C until use.
Screening for peptide-specific HLA-restricted cytotoxic T cells
For screening peptide-specific cytotoxic T-lymphocyte (CTL) responses, we used a previously established system.4,26 Briefly, purified HLA-A2 plus CD8 plus T cells of healthy donors were primarily stimulated with irradiated monocyte-derived peptide-pulsed dendritic cells and restimulated weekly with irradiated, peptide-pulsed CD40-activated B cells.27The cytotoxic activity of CTL lines was assessed after 4 to 5 stimulations using peptide-pulsed CD40-activated B cells, control-peptide pulsed CD40-activated B cells, and K562 cells in chromium (51Cr)–release assays. The concentration of peptide used for pulsing of the CD-40–activated B cells resulted in maximal killing of the targets. Specific lysis is presented as the percentage of specific 51Cr release, calculated from the following formula: (E − S/T − S) × 100, where E is experimental51Cr release, S is the spontaneous 51Cr release, and T is the total 51Cr release by 2% Triton X-100.
Results
Isolation of immunoreactive clones from CLL cell libraries by SEREX
We generated cDNA libraries from CLL cells isolated from 5 patients with different stages of CLL (Table 1). Using the sera from patients with CLL, other lymphomas, and healthy donors, we identified 24 independent clones representing 14 different genes (KW-1 to KW-14) after screening of more than 30 × 106 clones using the SEREX methodology (Table 2). All clones have high homologies to previously described genes (Table3).28-39 However, in 6 of them, the protein function has not been investigated experimentally.
Identification of 14 CLL-associated antigens by SEREX
| Clones . | Accession no. . | No. of selected clones . | Size, bp . |
|---|---|---|---|
| KW-1 | AF432208/AF432209 | 2 | 665/1342 |
| KW-2 | AF432215 | 1 | 3440 |
| KW-3 | AF432216 | 1 | 2008 |
| KW-4 | AF432217 | 2 | 3240 |
| KW-5 | AF432218 | 3 | 2376 |
| KW-6 | AF432219 | 1 | 2800 |
| KW-7 | AF432220 | 2 | 3076 |
| KW-8 | AF432221 | 1 | 1280 |
| KW-9 | AF432222 | 1 | 2100 |
| KW-10 | AF432210 | 4 | 2952 |
| KW-11 | AF432211 | 3 | 1380 |
| KW-12 | AF432212 | 1 | 579 |
| KW-13 | AF432213 | 1 | 3435 |
| KW-14 | AF432214 | 1 | 2593 |
| Clones . | Accession no. . | No. of selected clones . | Size, bp . |
|---|---|---|---|
| KW-1 | AF432208/AF432209 | 2 | 665/1342 |
| KW-2 | AF432215 | 1 | 3440 |
| KW-3 | AF432216 | 1 | 2008 |
| KW-4 | AF432217 | 2 | 3240 |
| KW-5 | AF432218 | 3 | 2376 |
| KW-6 | AF432219 | 1 | 2800 |
| KW-7 | AF432220 | 2 | 3076 |
| KW-8 | AF432221 | 1 | 1280 |
| KW-9 | AF432222 | 1 | 2100 |
| KW-10 | AF432210 | 4 | 2952 |
| KW-11 | AF432211 | 3 | 1380 |
| KW-12 | AF432212 | 1 | 579 |
| KW-13 | AF432213 | 1 | 3435 |
| KW-14 | AF432214 | 1 | 2593 |
CLL-associated antigens have high homologies to previously described genes
| Clones . | Homology to known genes . | Accession no. . | Chromosomal localization . | Family motifs/domains . | (Suggested) function . |
|---|---|---|---|---|---|
| KW-1 | KIAA1641 | XM047794 | 2q11.2 | bZIP, DUF 164 | (DNA binding/transcription regulation) |
| KW-2 | PIPMT | AY028423 | 8q12.1 | Met-10-like protein, putative RNA-methylase | Methylase |
| KW-3 | FosB | NM006732 | 19q13.2 | bZIP | DNA binding/transcription regulation |
| KW-4 | ZNF268 | AF317549 | 12q24.34 | Zinc finger | (DNA binding/transcription regulation) |
| KW-5 | SEBD4 | X75314 | 20q13.2 | RNA recognition motif | (RNA binding) |
| KW-6 | Ikaros | NM006060 | 7p11.2 | Zinc finger | DNA binding/transcription regulation |
| KW-7 | p75/LDEGF | NM021144 | 9p22.3 | AT-hook/PWWP | DNA binding/transcription regulation/growth factor |
| KW-8 | CHIP | NM005861 | 16p13.3 | Tetratricopeptide repeat region | Proteosomal induction |
| KW-9 | PYGB | NM002862 | 20p11.21 | Brain-type glycogen phosphorylase | Carbohydrate phosphorylase |
| KW-10 | ZNF148 | NM021964 | 3q21.2 | Zinc finger | DNA binding/transcription regulation |
| KW-11 | KIAA0336 | XM017779 | 2q12.3 | Translin, GBP, bZIP, lipoprotein, GRIP | (DNA binding/transcription regulation) |
| KW-12 | RPL11 | NM000975 | 1p36.11 | RPL-5 ribosomal L5-C terminus | Ribosomal protein |
| KW-13 | FMNL | XM038679 | 17q21.31 | FH1, FH2, FH3 | (Formin-like protein) |
| KW-14 | HGRG8 | NM016258 | 1p35.3 | No known family motifs present | (Chaperone) |
| Clones . | Homology to known genes . | Accession no. . | Chromosomal localization . | Family motifs/domains . | (Suggested) function . |
|---|---|---|---|---|---|
| KW-1 | KIAA1641 | XM047794 | 2q11.2 | bZIP, DUF 164 | (DNA binding/transcription regulation) |
| KW-2 | PIPMT | AY028423 | 8q12.1 | Met-10-like protein, putative RNA-methylase | Methylase |
| KW-3 | FosB | NM006732 | 19q13.2 | bZIP | DNA binding/transcription regulation |
| KW-4 | ZNF268 | AF317549 | 12q24.34 | Zinc finger | (DNA binding/transcription regulation) |
| KW-5 | SEBD4 | X75314 | 20q13.2 | RNA recognition motif | (RNA binding) |
| KW-6 | Ikaros | NM006060 | 7p11.2 | Zinc finger | DNA binding/transcription regulation |
| KW-7 | p75/LDEGF | NM021144 | 9p22.3 | AT-hook/PWWP | DNA binding/transcription regulation/growth factor |
| KW-8 | CHIP | NM005861 | 16p13.3 | Tetratricopeptide repeat region | Proteosomal induction |
| KW-9 | PYGB | NM002862 | 20p11.21 | Brain-type glycogen phosphorylase | Carbohydrate phosphorylase |
| KW-10 | ZNF148 | NM021964 | 3q21.2 | Zinc finger | DNA binding/transcription regulation |
| KW-11 | KIAA0336 | XM017779 | 2q12.3 | Translin, GBP, bZIP, lipoprotein, GRIP | (DNA binding/transcription regulation) |
| KW-12 | RPL11 | NM000975 | 1p36.11 | RPL-5 ribosomal L5-C terminus | Ribosomal protein |
| KW-13 | FMNL | XM038679 | 17q21.31 | FH1, FH2, FH3 | (Formin-like protein) |
| KW-14 | HGRG8 | NM016258 | 1p35.3 | No known family motifs present | (Chaperone) |
Detection of genetic diversity by novel open reading frames, splice variants, and possible mutations in detected clones
Sequence analysis of the detected clones KW-1, KW-4, KW-6, KW-11, KW-13, and KW-14 identified novel splice variants that have not previously been described (Table4).
Identification of novel splice variants in CLL-associated antigens
| Clone . | Gene . | Suggested splice variant-specific protein alteration . |
|---|---|---|
| KW-1 | KIAA1641 | Deletion of an uncharacterized protein motif (DUF164) |
| KW-4 | ZNF286 | Exclusion of the transcription-repressing KRAB domain |
| KW-6 | Ikaros | Loss of 10 aa at the activation site of Ikaros |
| KW-11 | KIAA0336 | Additional exon codes for a GTP binding site |
| KW-13 | FMNL | Full-length cloning of the gene shows 3 formin motifs (FH1, FH2, FH3) as well as 2 splice variants varying in 30 bp at the 5′ end |
| KW-14 | HGRG8 | Additional exon with unknown protein function |
| Clone . | Gene . | Suggested splice variant-specific protein alteration . |
|---|---|---|
| KW-1 | KIAA1641 | Deletion of an uncharacterized protein motif (DUF164) |
| KW-4 | ZNF286 | Exclusion of the transcription-repressing KRAB domain |
| KW-6 | Ikaros | Loss of 10 aa at the activation site of Ikaros |
| KW-11 | KIAA0336 | Additional exon codes for a GTP binding site |
| KW-13 | FMNL | Full-length cloning of the gene shows 3 formin motifs (FH1, FH2, FH3) as well as 2 splice variants varying in 30 bp at the 5′ end |
| KW-14 | HGRG8 | Additional exon with unknown protein function |
Detection of novel splice variants by sequencing positive clones detected by SEREX; in silico analysis of the protein sequence suggests alterations of the protein function.
Of note, most of these splice variants suggest alterations of protein function. KW-4 was found by sequencing of 2 independent clones. These clones represent a Kruppel-like zinc finger protein.30 Splice variant 2 represents a previously undescribed splice variant missing the Kruppel-associated box (KRAB) domain. KW-6 represents the 3′ end of Ikaros mRNA within the open reading frame (ORF) containing part of exon 6 and the whole exon 7.40 The mRNA lacks 30 amino acids (aa) of the previously described Ikaros mRNAs, resulting in a loss of 9 aa at the end of exon 6 and the first aa of exon 7, where the activation site starts. KW-11 has been previously described as KIAA0336.61The protein function of the KIAA0336 gene is not known. The aa sequence codes for many protein family motifs including several bZIP domains, suggesting a function as a transcription factor. Our clone codes for a previously undescribed splice variant, including an additional exon extending the protein at the 5′ end. This splice variant introduces a motif homologous to the C-terminal domain of the guanylate-binding protein GBP141 and gives additional homology to Translin.42 KW-13 has 99% homology to the previously described human Formin-like (FMNL) mRNA or C17ORF1, containing an FH2 domain.43 However, this mRNA contains only 1789 bp. Our clone contains 3435 bp with an open reading frame of 2976 bp. The 5′ end has been determined by RACE-PCR and shows an ORF of 3177 bp with an amino acid sequence of 1059 aa (Figure1). This protein has 85.4% homology to the mouse FRL (formin-related gene in leukocytes).38 In consensus with the mouse protein it shows an FH1 and FH3 domain in addition to the FH2 domain, suggesting similar functional properties as the mouse homolog.
Full-length amino acid sequence of the gene corresponding to KW-13.
Amino acid sequence of the gene corresponding to KW-13 with homology to the murine FRL is shown. The FH3-homolog region is double underlined, the FH1-homolog region is dot-dot-dash underlined, and the FH2-homolog region is single underlined. Peptides inducing peptide-specific cytotoxic immune responses are in squares. The methionine showing the starting sequence of the previously described human FMNL is underlined in bold.
Full-length amino acid sequence of the gene corresponding to KW-13.
Amino acid sequence of the gene corresponding to KW-13 with homology to the murine FRL is shown. The FH3-homolog region is double underlined, the FH1-homolog region is dot-dot-dash underlined, and the FH2-homolog region is single underlined. Peptides inducing peptide-specific cytotoxic immune responses are in squares. The methionine showing the starting sequence of the previously described human FMNL is underlined in bold.
Expression of identified antigens in healthy and malignant tissue
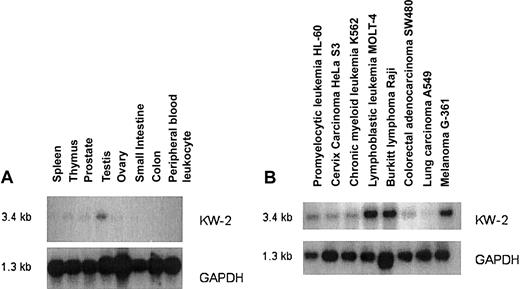
Expression of detected antigens in healthy and malignant tissue was investigated using RT-PCR. Exon-overlapping primer pairs were constructed to avoid amplification of genomic DNA. By RT-PCR, several genes demonstrated a wide tissue expression. However, 6 antigens (KW-2, KW-3, KW-4, KW-6, KW-7, KW-13) had restricted tissue expression and could not be demonstrated in several different healthy tissues by RT-PCR. For quantitative expression we performed Northern blot analysis. KW-2 has a cancer-testis–like expression profile. It has low expression in most tissues with the exception of testis. It is highly expressed in lymphoblastic leukemia MOLT-4, Raji cells, and the melanoma cell line G361 (Figure 2). We observed low levels of expression in 3 of 6 CLL samples examined but did not detect any expression in normal tonsilar B cells (data not shown).
Northern blot analysis of gene expression for the antigen KW-2 and GAPDH.
Normal tissue (A) and tumor tissue (B).
Northern blot analysis of gene expression for the antigen KW-2 and GAPDH.
Normal tissue (A) and tumor tissue (B).
The splice variant of KW-4, including the KRAB domain (splice variant number 1), is expressed in most normal tissues, whereas splice variant number 2, which is missing the KRAB domain, is not expressed in most normal tissue but is expressed in all CLL cases tested by RT-PCR (Figure 3). It also shows expression in healthy peripheral blood mononuclear cells by RT-PCR (data not shown).
Tissue expression of different splice variants of KW-4 by RT-PCR.
RT-PCR with splice variant–specific primers for KW-4: several normal tissues as well as 8 different CLL samples are shown.
Tissue expression of different splice variants of KW-4 by RT-PCR.
RT-PCR with splice variant–specific primers for KW-4: several normal tissues as well as 8 different CLL samples are shown.
Northern blot experiments show expression of the larger protein in normal tissue. However, the smaller protein missing the KRAB domain is highly expressed in several tumor cell lines (Figure4).
Northern blot analysis of gene expression for the antigen KW-4 and GAPDH.
Normal tissue (A) and tumor tissue (B).
Northern blot analysis of gene expression for the antigen KW-4 and GAPDH.
Normal tissue (A) and tumor tissue (B).
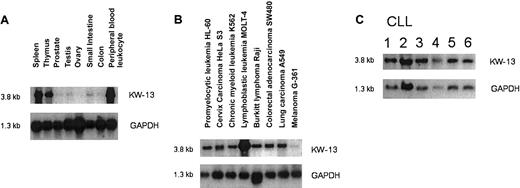
KW-13 is highly expressed in peripheral leukocytes, spleen, and thymus. In contrast to the mouse homolog, which has a much more widespread expression, it has only very low expression in normal tissue and is not expressed in colon. However, it shows high expression in several tumor cell lines, including colon carcinoma, and has high expression in CLL (Figure 5).
Northern blot analysis of gene expression for the antigen KW-13 and GAPDH.
Normal tissue (A), tumor tissue (B), and CLL cells (C).
Northern blot analysis of gene expression for the antigen KW-13 and GAPDH.
Normal tissue (A), tumor tissue (B), and CLL cells (C).
Prevalence of antibodies against antigens identified by sera from CLL patients, patients with other lymphomas, and healthy donors
To examine whether the identified antigens were recognized exclusively by autologous sera or also by allogeneic sera from CLL patients, other lymphoma patients, or from that of healthy donors, a phage plaque assay was performed as described above. Diluted, preabsorbed sera from patients with CLL, other lymphoma, and healthy donors were examined (Table 5).
Allogeneic serum responses against CLL-associated antigens
| Serum . | Clone . | CLL . | Lymphoma . | Healthy . | Detection as antigen in other studies . |
|---|---|---|---|---|---|
| Only CLL | KW-1 | 2 of 12 | 0 of 5 | 0 of 12 | |
| KW-3 | 2 of 16 | 0 of 5 | 0 of 12 | ||
| KW-4 | 3 of 12 | 0 of 5 | 0 of 12 | ||
| KW-2 | 3 of 12 | 0 of 5 | 0 of 12 | AF286340 | |
| KW-8 | 4 of 23 | n.d. | 0 of 12 | 16 | |
| KW-12 | 2 of 16 | 0 of 5 | 0 of 12 | 21 | |
| CLL + lymphoma | KW-6 | 2 of 12 | 1 of 5 | 0 of 12 | |
| KW-10 | 2 of 24 | 2 of 5 | 0 of 12 | ||
| KW-7 | 3 of 23 | 1 of 5 | 0 of 12 | 44 | |
| KW-11 | 4 of 23 | 3 of 5 | 0 of 12 | 17.25 | |
| KW-14 | 3 of 16 | 1 of 5 | 0 of 12 | 17 | |
| CLL + lymphoma + healthy donors | KW-5 | 20 of 30 | 2 of 5 | 2 of 12 | |
| KW-9 | 14 of 15 | 5 of 5 | 10 of 12 | ||
| KW-13 | 7 of 16 | 2 of 5 | 3 of 12 |
| Serum . | Clone . | CLL . | Lymphoma . | Healthy . | Detection as antigen in other studies . |
|---|---|---|---|---|---|
| Only CLL | KW-1 | 2 of 12 | 0 of 5 | 0 of 12 | |
| KW-3 | 2 of 16 | 0 of 5 | 0 of 12 | ||
| KW-4 | 3 of 12 | 0 of 5 | 0 of 12 | ||
| KW-2 | 3 of 12 | 0 of 5 | 0 of 12 | AF286340 | |
| KW-8 | 4 of 23 | n.d. | 0 of 12 | 16 | |
| KW-12 | 2 of 16 | 0 of 5 | 0 of 12 | 21 | |
| CLL + lymphoma | KW-6 | 2 of 12 | 1 of 5 | 0 of 12 | |
| KW-10 | 2 of 24 | 2 of 5 | 0 of 12 | ||
| KW-7 | 3 of 23 | 1 of 5 | 0 of 12 | 44 | |
| KW-11 | 4 of 23 | 3 of 5 | 0 of 12 | 17.25 | |
| KW-14 | 3 of 16 | 1 of 5 | 0 of 12 | 17 | |
| CLL + lymphoma + healthy donors | KW-5 | 20 of 30 | 2 of 5 | 2 of 12 | |
| KW-9 | 14 of 15 | 5 of 5 | 10 of 12 | ||
| KW-13 | 7 of 16 | 2 of 5 | 3 of 12 |
Phage plaque assay using multiple sera was performed. Screening results of multiple sera from patients with CLL, other lymphoma, and healthy donors for presence of antibodies against identified clones are shown. Previous detection of CLL-associated antigens in other studies is indicated.
In our experiments, 6 antigens have been detected only by CLL patients, whereas 8 of the 14 antigens were detected also in other lymphoma patients and 3 antigens were detected by sera from a subset of healthy donors. Three of the 6 antigens identified only in CLL patients in this study (KW-2, KW-8, KW-12) as well as 3 of the other antigens (KW-7, KW-11, KW-14) have been previously identified.16,17,21,25,44 62 However, KW-1, KW-3, and KW-4 are novel antigens, so far detected only in serum from patients with CLL. KW5 and KW13 were detected in a larger number of CLL patients. Both have also been recognized in some healthy donors. However, KW-5 is significantly more likely recognized by patients with CLL than in healthy donors (P = .01). It shows enhanced expression in most CLL cells compared with normal tonsilar B cells. However, it is also expressed in most normal tissues at least at low levels (data not shown). Clinical data of the majority of patients whose sera were used for the phage plaque assay are shown in Table6. Although patients with hypogammaglobulinemia and previous therapy seemed to have fewer humoral responses against identified antigens, this did not achieve statistical significance (hypogammaglobulinemia P = .08; previous therapy P = .1).
Clinical data of patients with CLL detecting 0 to 7 clones
| Patient sera . | Detected clones . | Rai stage . | Hypogamma-globulinemia . | Autoimmune phenomena . | Prior therapy . | CD38, % expression on CLL cells . | IgH mutation . | Cytogenetics . |
|---|---|---|---|---|---|---|---|---|
| 1 | 0 | I | Low | No | No | 2 | Yes | 13q del |
| 2 | 0 | II | Low | No | Yes | n.d. | Yes | Normal |
| 3 | 0 | II | Low | Yes | Yes | 85 | Yes | Normal |
| 4 | 1 | I | Normal | No | No | 17 | Yes | 13q del |
| 5 | 1 | IV | Normal | No | Yes | 83 | No | Normal |
| 6 | 1 | I | Low | No | No | 12 | Yes | Normal |
| 7 | 1 | II | Low | Yes | No | 15 | No | +12 |
| 8 | 1 | I | Low | No | No | 4 | Yes | Normal |
| 9 | 1 | II | Normal | Yes | Yes | 39 | Yes | 11q del |
| 10 | 1 | II | Low | Yes | Yes | 14 | No | Normal |
| 11 | 2 | II | Normal | Yes | Yes | 79 | Yes | Normal |
| 12 | 2 | IV | Low | No | Yes | 86 | Yes | Normal |
| 13 | 2 | IV | Normal | No | No | n.d. | Yes | n.d. |
| 14 | 3 | I | Normal | No | No | 10 | Yes | Normal |
| 15 | 3 | I | Normal | No | No | 0 | No | 13q del |
| 16 | 4 | II | Normal | No | No | 16 | Yes | +12, +18 |
| 17 | 7 | II | Normal | No | No | 7 | Yes | 13q del |
| Patient sera . | Detected clones . | Rai stage . | Hypogamma-globulinemia . | Autoimmune phenomena . | Prior therapy . | CD38, % expression on CLL cells . | IgH mutation . | Cytogenetics . |
|---|---|---|---|---|---|---|---|---|
| 1 | 0 | I | Low | No | No | 2 | Yes | 13q del |
| 2 | 0 | II | Low | No | Yes | n.d. | Yes | Normal |
| 3 | 0 | II | Low | Yes | Yes | 85 | Yes | Normal |
| 4 | 1 | I | Normal | No | No | 17 | Yes | 13q del |
| 5 | 1 | IV | Normal | No | Yes | 83 | No | Normal |
| 6 | 1 | I | Low | No | No | 12 | Yes | Normal |
| 7 | 1 | II | Low | Yes | No | 15 | No | +12 |
| 8 | 1 | I | Low | No | No | 4 | Yes | Normal |
| 9 | 1 | II | Normal | Yes | Yes | 39 | Yes | 11q del |
| 10 | 1 | II | Low | Yes | Yes | 14 | No | Normal |
| 11 | 2 | II | Normal | Yes | Yes | 79 | Yes | Normal |
| 12 | 2 | IV | Low | No | Yes | 86 | Yes | Normal |
| 13 | 2 | IV | Normal | No | No | n.d. | Yes | n.d. |
| 14 | 3 | I | Normal | No | No | 10 | Yes | Normal |
| 15 | 3 | I | Normal | No | No | 0 | No | 13q del |
| 16 | 4 | II | Normal | No | No | 16 | Yes | +12, +18 |
| 17 | 7 | II | Normal | No | No | 7 | Yes | 13q del |
Stage, serum Ig, autoimmune phenomena, therapy prior to serum harvest, CD38 expression level on CLL cells, presence of somatic hypermutation in the Ig rearrangement, and cytogenetic abnormalities are shown.
n.d. indicates not done.
Detection of cellular responses against identified antigens
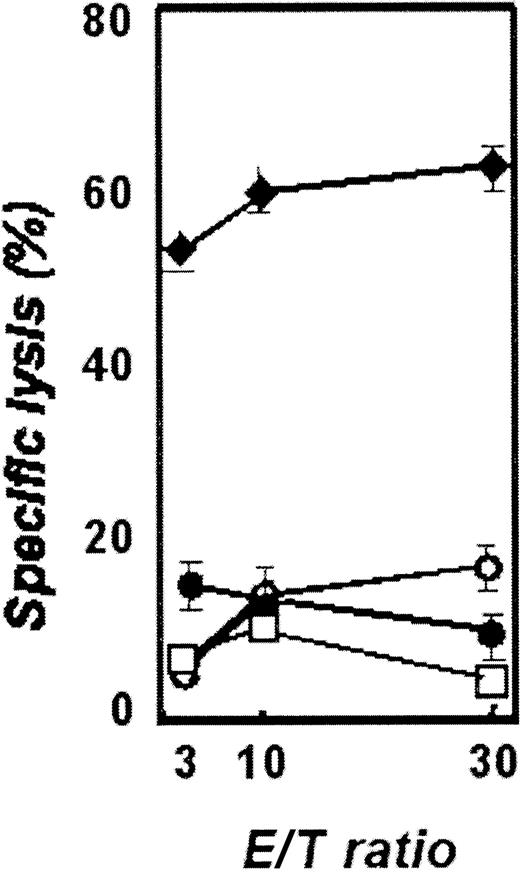
The clones KW-2, KW-4, KW-5, and KW-13 appeared most interesting for investigation as potential targets for immunotherapeutic approaches either by their expression profile or by their antigenic potential. So far we identified peptides from the proteins corresponding to KW-2 and KW-13 that could potentially bind to HLA-A2+ donors. Twelve nonamer peptides with varying affinity to HLA-A2 molecules derived from clones KW-2 and KW-13 were synthesized and used for stimulation of peripheral T cells of HLA-A2+ healthy donors. Three peptides, KW-2/2: ALARNNAEV, KW-13/2: TLLHYLVKV (Figure 6), and KW-13/5: AAPPPPPPL, were repeatedly able to induce cytotoxic T-cell responses against peptide-pulsed autologous CD40-activated B cells. In contrast, CD40-activated B cells pulsed with an irrelevant peptide or unpulsed were not lysed. However, in keeping with our previous data that CLL cells are poor targets of CTLs generated against class I peptides,5 no peptide-specific CTL lines could be generated that recognized and lysed HLA-A2+ CLL cells. However, these data demonstrate that these clones do represent antigens for both T-cell and B-cell pathways.
Cytoxicity of autologous T cells of healthy donors after stimulation with antigen-presenting cells pulsed with KW-13–derived peptide 2.
Targets are CD40-activated B cells pulsed with peptide 2 of KW-13 (diamonds), pulsed with an unrelated peptide (closed circles), unpulsed CD40-activated B cells (open circles), and unstimulated CLL cells (squares).
Cytoxicity of autologous T cells of healthy donors after stimulation with antigen-presenting cells pulsed with KW-13–derived peptide 2.
Targets are CD40-activated B cells pulsed with peptide 2 of KW-13 (diamonds), pulsed with an unrelated peptide (closed circles), unpulsed CD40-activated B cells (open circles), and unstimulated CLL cells (squares).
Discussion
CLL is in several regards a tempting but difficult target for the development of immunotherapeutic strategies. The fundamental question arises if cancer-specific tumor antigens exist in CLL and if they can be recognized by the autologous host. Following CD40 activation, CLL cells can be recognized by the autologous immune system, making it likely that tumor-associated antigens are present. Although cytotoxic T-cell responses can be generated against idiotypic peptides,4,5 cytotoxic immune responses were weak. Additional tumor-associated antigens are likely needed to give a broader basis for polyvalent immunization strategies to prevent the escape of antigen loss variants.6
SEREX provides a powerful method to identify immunogenic tumor antigens in many malignancies.14 However, the application of SEREX has several disadvantages for its use in CLL. Hypogammaglobulinemia is common in CLL patients as well as T-cell defects, resulting in reduced T-cell help. This is also reflected by the reduced antibody response of CLL patients after standard vaccinations.45 However, we detected humoral immune responses against 14 antigens expressed in cDNA libraries from 5 CLL primary tumor samples after screening of more than 30 million clones. The relatively low number of positive clones detected in this study after extensive screening compared with studies in other malignancies likely reflects the impaired immunologic responsiveness in CLL. Four CLL patients showed immune responses against 3 or more clones, suggesting that these patients had stronger immune recognition. These patients were untreated, had no hypogammaglobulinemia, early-stage disease, and low CD38 expression on CLL cells. Although the data are certainly preliminary, it is of interest to speculate that an ongoing immune response might even contribute to the low stage of these patients. In contrast, patients recognizing 2 or fewer antigens were mostly either treated with immunosuppressive agents or had hypogammaglobulinemia and higher percentage of CD38 expression on CLL cells.
The majority of antigens were recognized only in the minority of CLL patients. Molecular heterogeneity of this disease might contribute to this fact. However, in most SEREX studies selected antigens are detected only in the minority of tested patients. In a survey by Stockert et al,46 NY-ESO-1 was recognized in 12 of 127 melanoma patients and in 8 of 15 melanoma patients positive for NY-ESO-1. Antibodies against Her2/neu have been detected in 12 of 107 patients with early-stage breast cancer.47 MAGE-3 was detected only in 2 of 127 melanoma patients.46 However, clinical responses have been reported after immunizations with MAGE-3–derived HLA-A1–restricted peptides.48 Although experiences are limited, peptide vaccination has also shown promising results in a preliminary clinical trial using NY-ESO-1–related peptides.8 In this study, even 5 of the 7 vaccinated patients who were initially NY-ESO-1 antibody–negative developed stabilization or regression of individual metastases, and both humoral and cellular responses were observed.6
The antigens detected in this study might be tumor antigen candidates in CLL. Three antigens (KW-1, KW-3, KW-4) have been detected only in CLL patients so far. Examination of larger cohorts will be necessary to establish specificity of antibody responses against these antigens for this disease. The other antigens were also detected in sera from other lymphoma patients or patients with other malignancies according to other studies.16,17,21,25 44 These antigens might therefore be tumor associated rather than CLL associated. However, they might represent more universal targets for anticancer immunotherapeutic approaches.
Of particular interest, 4 antigens (KW-2, KW-4, KW-5, KW-13) have favorable properties in regard to antigenic potential and restricted tissue or overexpression in CLL. KW-2 has a cancer-testis–like expression profile and has been detected in 3 of 12 CLL patients. A splice variant of KW-4 has been identified that is selectively expressed in CLL compared with most normal tissues. KW-5 has a broader expression in healthy tissue. However, although it has been recognized in 2 healthy donors, it is significantly more recognized in the sera of CLL patients. The expression of KW-13 is restricted to the peripheral leukocytes, thymus, and spleen but is aberrantly expressed in several tumor cell lines and highly expressed in CLL. There are other antigens that are overexpressed in cancer cells but also expressed in normal tissue, including Her2/neu, α-fetoprotein, telomerase-catalytic subunit, G-250, Muc-1, carcinoembryogenic antigen, and p53.26,49-53 With regard to Her2/neu, no histologic evidence of lymphocyte infiltration or tissue destruction was seen in rats,54 and no autoimmune phenomena have been observed in humans after treatment with anti-Her2/neu antibody.13
We investigated 12 peptides for their potential to induce cytotoxic T-cell responses against 2 proteins (KW-2, KW-13) that might be suitable targets for immunotherapy. We were able to generate peptide-specific CTLs against these identified antigens. However, these CTLs were not able to lyse native CLL cells. There are many possible reasons for the lack of cytotoxicity. CLL cells might not present these peptides on their surface or are not able to provide sufficient costimulation. Further studies are under way to investigate this issue, including proteasome-mediated digestion analysis. However, conflicting data are available about a number of tumor antigen–derived peptides. One of the most antigenic peptides of HER-2/neu (p369-377) did not induce cytotoxic immune responses against native tumor cells in a study by Zaks and Rosenberg,55 whereas weak cytotoxic immune responses could be demonstrated by others.56 Using the 15 amino acids containing p369-384 as a putative helper peptide for vaccination, Knutson et al were able to show induction of cytotoxic peptide-specific T cells, which were able to lyse HLA-A2+tumor cells overexpressing HER2/neu protein.57SEREX-defined antigens may represent mostly T helper antigens. Nishikawa et al demonstrated that covaccination of mice with unmutated SEREX-defined antigens in combination with tumor-specific antigens mERK2 and HER2 induced specific cytotoxicity against HER2-neu+ tumors and inhibition of metastasis after late vaccination that could not be induced by vaccination with the tumor-specific antigens alone.58 These antigens might therefore provide help for tumor-specific immune responses. Humoral- or complement-mediated responses should not be underestimated in tumor eradication and protection. In transgenic mice a humoral response is essential for complete eradication of Her2/neu-expressing tumors.59 In addition, after idiotype-specific DNA vaccination in a murine B-cell lymphoma model, transfer of hyperimmune sera protected naive animals against tumor growth, whereas in vitro stimulation of immune splenocytes with tumor cells failed to induce idiotype-specific cytotoxicity.60Thus, although no suitable peptides have been identified so far to induce cytotoxic immune responses against native CLL cells, these antigens might provide antitumor help in this disease. Further studies are ongoing in vitro and in vivo to define the potential of the antigens presented in this study for immunotherapeutic strategies.
The authors are greatly thankful to G. J. Freeman and G. Dranoff for constructive discussion of the experimental work and manuscript as well as to Donna Neuberg and David Zahrieh for statistical analysis and Peter Varney for editorial assistance.
Prepublished online as Blood First Edition Paper, May 24, 2002; DOI 10.1182/blood-2002-02-0513.
Supported by grants from the Deutsche Krebshilfe and Dr Mildred Scheel Stiftung (A.M.K. and M.W.), a grant from the Berlex Oncology foundation (F.S.H.), grant UO1CA81534 from the National Cancer Institute (J.G.G.), and support from the Grayce B. Kerr Foundation.
A.M.K. and M.W. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John G. Gribben, Harvard Medical School, Adult Oncology, Dana Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: john_gribben@dfci.harvard.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal