FLT3 mutations, either internal tandem duplications (ITDs) or aspartate residue 835 (D835) point mutations, are present in approximately one third of patients with acute myeloid leukemia (AML) and have been associated with an increased relapse rate. We have studied FLT3 mutations in paired presentation and relapse samples to ascertain the biology of these mutations and to evaluate whether they can be used as markers of minimal residual disease. At diagnosis, 24 patients were wild-type FLT3, and 4 acquired a FLT3 mutation at relapse (2 D835+, 2 ITD+), with a further patient acquiring an ITD at second relapse. Of 20 patients positive at diagnosis (18 ITD+, 2 D835+), 5 who were all originally ITD+ had no detectable mutation at relapse, as determined by a sensitive radioactive polymerase chain reaction. One of these patients had acquired an N-Ras mutation not detectable at presentation. Furthermore, another patient had a completely different ITD at relapse, which could not be detected in the presentation sample. These results indicate that FLT3 mutations are secondary events in leukemogenesis, are unstable, and thus should be used cautiously for the detection of minimal residual disease.
Introduction
Approximately 80% of patients with acute myeloid leukemia (AML) younger than 55 years of age achieve complete remission (CR) following intensive induction therapy.1However, the actuarial survival at 5 years is only just over 40% because most patients relapse and die of their disease.1Considerable effort has therefore been directed at identifying molecular markers that can be used to detect residual disease or predict relapse at an earlier stage and lead to therapeutic intervention before overt hematological relapse occurs.
Ideally, a candidate marker for minimal residual disease (MRD) should always or frequently be present in a specific disease and it should be easy to assay and stable; that is, always present at relapse. In lymphoid malignancies, clonal rearrangements of either immunoglobulin or T-cell receptor genes are suitable for such analysis.2They occur in up to 90% of patients, can be detected using sensitive polymerase chain reaction (PCR) techniques, and reappear at relapse, although development of secondary rearrangements has been noted in some cases.3 Studies evaluating their usefulness in clinical practice as a predictor of relapse have shown that the presence and level of residual leukemia can correlate with the risk of early relapse in acute lymphoid leukemia.4 Similarly, the BCR/ABL fusion transcript is present in nearly all patients with chronic myeloid leukemia (CML), and a number of different approaches have been used to correlate this marker with disease status and outcome.5-8
In AML, the use of MRD has been limited by the lack of suitable specific molecular markers and the diversity of those that do exist. Although cytogenetic markers have proved to be significant prognostic indicators, fewer than 50% of patients have an identifiable cytogenetic abnormality.1 Some patients have partial or complete loss or gain of a chromosome, but such abnormalities can be identified only using relatively insensitive “whole cell” techniques, and even the more sensitive fluorescence in situ hybridization technique requires evaluation of thousands of cells.9 There are 3 molecular markers that have been used for MRD studies: PML/RARα fusion transcript arising from the t(15;17) translocation, AML1/ETO arising from t(8;21), and CBFB/MYH11 from inv(16).10-13 Quantitative assays are necessary, particularly because several groups have shown that patients who are positive for AML1/ETO using sensitive reverse transcription (RT)–PCR assays can remain in morphologic and clinical remission for many years.14,15 Nevertheless, reappearance of the marker, or persistence at a certain level after induction chemotherapy, does generally signify an increased risk of relapse.16 17
However, these 3 markers together account for only approximately one fourth of adult AML patients,1 and other markers are required to extend the potential application of MRD to more AML patients. Recent studies have demonstrated that mutations in theFLT3 gene occur in about one third of adult AML patients. In 22% to 27% of patients, there is an internal tandem duplication (ITD) in the juxtamembrane region between exons 14 and 1518,19(previously called exons 11 and 12).20 The extra sequence varies between 18 base pairs (bp) and 222 bp (as detected in one patient in the present study), but the resulting protein sequence remains in frame and is thought to lead to a constitutively activated tyrosine kinase receptor.21-23 Multivariate analysis has shown that presence of the mutation is the most significant factor predicting for relapse, and it can add additional prognostic information to a cytogenetic risk group.19 A further 7% of patients have a mutation of aspartate residue 835 (D835) in the activation loop of the second kinase domain, predominantly to tyrosine or valine, but occasionally to other amino acids.24 25This also leads to an activated receptor, although the clinical significance is unclear at present due to the small number of patients studied.
To gain further insight into the role of FLT3 mutations in AML and to determine whether they are suitable markers for MRD, we have therefore studied paired samples from patients at presentation and relapse for the presence of ITDs and D835 alterations.
Patients, materials, and methods
Patients
Paired peripheral blood or bone marrow samples at presentation and first relapse were available from 44 patients with de novo AML who were treated with protocols from the United Kingdom Medical Research council (MRC) AML 10 and 12 trials.1,19 A further sample at second relapse was obtained from one patient. Of these patients, 11 had AML French-American-British type M1; 7, M2; 5, M3; 13, M4; 7, M5; and 1 patient had RAEB-t. Median age at presentation was 42 years (range, 12-61 years), and median white cell count was 50.1 × 109/L (range, 0.8-541 × 109/L). DNA samples were used for analysis in 42 patients; in the remaining 2 patients, only RNA samples were available. Paired DNA samples from patients at presentation and first complete remission (CR) following induction chemotherapy were also studied in 13 AML patients who had a mutation in the FLT3 gene at diagnosis. CR was defined as a normocellular bone marrow containing fewer than 5% blast cells. Results of FLT3/ITD analysis at diagnosis have already been reported on some of the patients used in these studies.19
Detection of mutations in the FLT3 gene
ITDs
For FLT3/ITDs, genomic DNA was amplified as previously described to produce a fragment of 328 bp from wild-type (WT) alleles, which included exons 14 and 15 and the intervening intron.19 The same primers were used to analyze cDNA, but the fragment produced was 90 bps smaller. Semiquantitative PCR using a32P-labeled PCR primer was also carried out as previously described.19 To evaluate sensitivity of the technique, WT cells were mixed with cells from a patient carrying predominantly mutant alleles to create 9 different mixtures containing between 100% and 0.01% patient cells. DNA was extracted and assayed radioactively. Reproducibility was assessed by 5 separate analyses of 3 patient samples with high, intermediate, or low level of mutantFLT3.
D835
Mutations at residue D835 in exon 20 (previously exon 17) were detected as described by Yamamoto et al.24 Approximately 100 ng DNA was added to a reaction mix containing 1× buffer (16 mM (NH4)2SO4, 67 mM Tris HCl pH 8.8, 0.01% Tween 20), 1.0 mM MgCl2, 200 μM dNTPs, and 10 pmols each primer (Table 1) in a total volume of 19 μL. The mixture was heated to 95°C for 5 minutes and held at 85°C while 1 μL containing 0.5U BIOTAQ DNA polymerase (Bioline, London, United Kingdom) was added; then 35 cycles each of 95°C for 30 seconds, 63°C for 30 seconds, and 72°C for 30 seconds were performed, followed by 5 minutes at 72°C. Amplified products were digested with EcoRV then electrophoresed through 4% agarose gels and visualized under ultraviolet light with ethidium bromide staining. In alleles containing a D835 mutation, the 114-bp PCR fragment remained uncut, but in WT alleles it was digested to fragments of 68 and 46 bps.
Oligonucleotide primer used in PCR analysis
| Oligonucleotide primer . | Primer . | Sequence . |
|---|---|---|
| FLT3/ITD | F | 5′-GCAATTTAGGTATGAAAGCCAGC-3′ |
| FLT3/ITD | R | 5′-CTTTCAGCATTTTGACGGCAACC-3′ |
| FLT3/D835 | F | 5′-CCGCCAGGAACGTGCTTG-3′ |
| FLT3/D835 | R | 5′-GCAGCCTCACATTGCCCC-3′ |
| N-RAS exon 1 (DNA) | F | 5′-GACTGAGTACAAACTGGTGG-3′ |
| R | 5′-TGCATAACTGAATGTATACCC-3′ | |
| N-RAS exon 2 (DNA) | F | 5′-CAAGTGGTTATAGATGGTGAAACC-3′ |
| R | 5′-AAGATCATCCTTTCAGAGAAAATAAT-3′ | |
| N-RAS exons 1 + 2 (cDNA) | F | 5′-CTGTCCAAAGCAGAGGCAGTG-3′ |
| R | 5′-AGGCTTCCTCTGTGTATTTGCC-3′ | |
| KB9 | F | 5′-TGCAAAGGCTTGGAGGGCTGATG-3′ |
| R | 5′-ATCTCGGACAACAGCAGGCCTCG-3′ | |
| D21S270 | F | 5′-GAAATGTTTTAATAAATGGTGGTTA-3′ |
| R | 5′-ACAAAGTTATGGTCAAGGGG-3′ | |
| D21S65 | F | 5′-CCGAAAACTTACTGGAGAAC-3′ |
| R | 5′-GATCATCCAGGAATCACCAA-3′ |
| Oligonucleotide primer . | Primer . | Sequence . |
|---|---|---|
| FLT3/ITD | F | 5′-GCAATTTAGGTATGAAAGCCAGC-3′ |
| FLT3/ITD | R | 5′-CTTTCAGCATTTTGACGGCAACC-3′ |
| FLT3/D835 | F | 5′-CCGCCAGGAACGTGCTTG-3′ |
| FLT3/D835 | R | 5′-GCAGCCTCACATTGCCCC-3′ |
| N-RAS exon 1 (DNA) | F | 5′-GACTGAGTACAAACTGGTGG-3′ |
| R | 5′-TGCATAACTGAATGTATACCC-3′ | |
| N-RAS exon 2 (DNA) | F | 5′-CAAGTGGTTATAGATGGTGAAACC-3′ |
| R | 5′-AAGATCATCCTTTCAGAGAAAATAAT-3′ | |
| N-RAS exons 1 + 2 (cDNA) | F | 5′-CTGTCCAAAGCAGAGGCAGTG-3′ |
| R | 5′-AGGCTTCCTCTGTGTATTTGCC-3′ | |
| KB9 | F | 5′-TGCAAAGGCTTGGAGGGCTGATG-3′ |
| R | 5′-ATCTCGGACAACAGCAGGCCTCG-3′ | |
| D21S270 | F | 5′-GAAATGTTTTAATAAATGGTGGTTA-3′ |
| R | 5′-ACAAAGTTATGGTCAAGGGG-3′ | |
| D21S65 | F | 5′-CCGAAAACTTACTGGAGAAC-3′ |
| R | 5′-GATCATCCAGGAATCACCAA-3′ |
Polymorphic marker analysis
In cases where differences were observed between the results at presentation and relapse, 3 polymorphic markers were used to confirm that the DNA samples were from the same patient: KB9 on chromosome 19, and D21S270 and D21S65 on chromosome 21.26 27 One primer from each pair (Table 1) was 32P end-labeled, and PCR was performed as described above except that only 25 cycles of amplification were used and the annealing temperatures were 58°C for D21S270 and D21S65, and 65°C for KB9. Products were electrophoresed through denaturing polyacrylamide gels (7M urea, 6% polyacrylamide crosslinker ratio 37.5:1, 0.5× Tris-Borate-EDTA [ethylenediaminetetraacetic acid]), the gels were dried and exposed to Hyperfilm (Amersham, Little Chalfont, United Kingdom).
Cloning and sequencing of PCR products
PCR products from ITD+ patients either were sequenced directly or were cloned into the pGEM-T vector (Promega, Madison, WI) and selected clones sequenced using an Applied Biosystem 310 Analyzer with BigDye version 2 terminator chemistry (Applied Biosystems, Foster City, CA).
Detection of N-Ras gene mutations
Samples were screened for mutations in codons 12, 13, and 61 of the N-Ras gene using heteroduplex analysis (WAVE technology; Transgenomic, San Jose, CA). For DNA samples, a 241-bp fragment was amplified for exon 1 containing codons 12 and 13, and a 201-bp fragment for exon 2 containing codon 61. Thirty-five cycles of amplification were performed on approximately 100-ng genomic DNA using 0.625 units HotStarTaq (Qiagen, Crawley, West Sussex, United Kingdom) per 25 μL reaction, 1 × manufacturer's buffer, 200 μM dNTPs, and 12.5 pmol each primer (Table 1). After an initial denaturing cycle of 15 minutes at 95°C, each cycle was 30 seconds at 94°C, 60 seconds at 55.5°C, one minute at 72°C, followed by 10 minutes at 72°C. PCR products were denatured at 95°C for 5 minutes then cooled slowly to 25°C to allow heteroduplexes to form and be analyzed by denaturing high performance liquid chromatography (dHPLC). For cDNA a single fragment of 296 bp was amplified, which covered exons 1 and 2, and the annealing temperature was 64°C. The sensitivity of the dHPLC is such that at least 10% mutant DNA can be confidently detected in a sample (manuscript in preparation). Samples exhibiting an abnormal dHPLC profile were sequenced as above.
Results
Evaluation of semiquantitative PCR analysis
Sensitivity of the method was determined from analysis of mixtures of WT cells and cells from a patient with 94% FLT3/ITD alleles. The mutant could be detected when ≥ 0.5% of totalFLT3. Quantification of the different mixtures gave results that were in good agreement with the expected values: expected and observed values were 47% and 48%; 19% and 23%; 9.4% and 12.8%; 4.7% and 5%; 1% and 1.4%, respectively. The results were highly reproducible: 5 analyses of 3 samples with differentFLT3/ITD levels gave mean ± SD values of 94.4% ± 0.5% (range, 93%-95%), 53.5% ± 1.1% (52%-55%), and 12.8% ± 0.8% (12%-14%), respectively.
Paired presentation/remission samples
Remission samples from 13 patients who had a FLT3mutation at presentation were studied. All 13 patients had an ITD, median mutant level 44% of total FLT3 (range, 2%-90%), and 1 patient also had a D835 mutation (D835Y). In remission all patients lost their mutation(s), confirming that FLT3mutations are leukemia-specific.
Paired presentation/relapse samples
Patients without a FLT3 mutation at presentation.
Of the 24 patients studied who had only WT FLT3 alleles at presentation, 20 patients remained WT at first relapse and 4 patients had acquired a FLT3 mutation (Table2). Two patients acquired a D835 mutation, both G > T leading to substitution of tyrosine for aspartate, and 2 patients gained a FLT3/ITD. The D835 mutants accounted for approximately half of the total FLT3in the samples, and in the patients with ITDs, the mutant levels were 38% and 42%, respectively, suggesting that most cells at relapse were heterozygous for the mutant alleles. No ITD was detected in presentation samples from the latter 2 patients using the more sensitive radioactive PCR, and all 4 patients had high blast counts at diagnosis (patients 21-24, Table 2). These results suggested that aFLT3 mutation had not been missed at diagnosis and that its acquisition at relapse was evidence of clonal progression. In 3 of these 4 patients, sufficient DNA was available at both presentation and relapse to study the alleles at 3 loci known to be polymorphic for repeat sequences. In each case they confirmed that the 2 samples came from the same individual. The median time between remission and relapse was similar in the patients who remained WT and those who acquired anFLT3 mutation (367 days; range, 70-1779 days). One patient who had only WT FLT3 alleles at presentation and first relapse acquired an FLT3/ITD at second relapse with a mutant level of 45%. Polymorphic markers indicated that all 3 samples were from the same patient.
FLT3 status and relative percentage of mutant in PB or bone marrow from 44 AML patients at presentation and relapse
| No. . | Presentation . | Relapse . | ||||||
|---|---|---|---|---|---|---|---|---|
| FLT3 . | % Mutant . | Sample . | % Blasts . | FLT3 . | % Mutant . | Sample . | % Blasts . | |
| 1 | WT | — | PB MNC | 90 | WT | — | NA | 76 |
| 2 | WT | — | BM | 97 | WT | — | PB MNC | NA |
| 3 | WT | — | PB MNC | 95 | WT | — | BM | NA |
| 4 | WT | — | PB MNC | 28 | WT | — | BM | 42 |
| 5 | WT | — | NA | 100 | WT | — | BM | 95 |
| 6 | WT | — | NA | 100 | WT | — | BM | 60 |
| 7 | WT | — | PB | NA | WT | — | BM | 85 |
| 8 | WT | — | BM | 98 | WT | — | PB | 52 |
| 9 | WT | — | PB | 95 | WT | — | PB | 80 |
| 10 | WT | — | BM | 89 | WT | — | BM | 96 |
| 11 | WT | — | PB | 99 | WT | — | PB | 99 |
| 12 | WT | — | PB | 80 | WT | — | BM | NA |
| 13 | WT | — | BM | 86 | WT | — | BM | 60 |
| 14 | WT | — | BM | 58 | WT | — | BM | 90 |
| 15 | WT | — | BM | 90 | WT | — | BM | 87 |
| 16* | WT | — | BM | 72 | WT (ITD+) | (45) | BM (PB) | 13 (87) |
| 17 | WT | — | BM | NA | WT | — | BM | 11 |
| 18 | WT | — | PB | 78 | WT | — | BM | 56 |
| 19 | WT | — | BM | 90 | WT | — | BM | 64 |
| 20 | WT | — | BM | 90 | WT | — | BM | 74 |
| 21 | WT | — | PB MNC | 90 | D835+ | ≈50 | PB MNC | NA |
| 22 | WT | — | BM | 87 | D835+ | ≈50 | BM | NA |
| 23 | WT | — | PB MNC | 90 | ITD+ | 42 | BM | 95 |
| 24 | WT | — | BM | 95 | ITD+ | 38 | BM | 95 |
| 25 | ITD+ | 20 | PB MNC | 53 | WT | — | BM | 55 |
| 26 | ITD+ | 6 | BM | 88 | WT | — | BM | 92 |
| 27 | ITD+ | 11 | PB | 80 | WT | — | BM | 82 |
| 28 | ITD+ | 28 | BM | 94 | WT | — | BM | 85 |
| 29 | ITD+ | 44 | BM | 95 | WT | — | BM | 44 |
| 30 | D835+ | ≈50 | BM | 95 | D835+ | ≈50 | BM | 96 |
| 31 | D835+ | ≈50 | BM | 60 | D835+ | ≈50 | BM | 60 |
| 32 | ITD+ | 13 | BM | 55 | ITD+ | 16 | BM | 96 |
| 33 | ITD+ | 39 | BM | 51 | ITD+ | 39 | BM | 60 |
| 34 | ITD+ | 44 | PB MNC | 77 | ITD+ | 45 | NA | NA |
| 35 | ITD+ | 82 | BM | 97 | ITD+ | 84 | BM | 92 |
| 36 | ITD+ | 86 | BM | 86 | ITD+ | 83 | PB | 93 |
| 37 | ITD+ | 91 | PB | 70 | ITD+ | 91 | BM | NA |
| 38 | ITD+ | 5 | BM | NA | ITD+ | 35 | PB | 97 |
| 39 | ITD+ | 23 | PB MNC | 86 | ITD+ | 88 | BM | 100 |
| 40 | ITD+ | 25 | PB | 79 | ITD+ | 50 | PB | 99 |
| 41 | ITD+ | 26 + 2 | BM | 90 | ITD+ | 56 + 14 | BM | 94 |
| 42 | ITD+ | 28 + 3 + 1 | BM | 80 | ITD+ | 41 | BM | 99 |
| 43 | ITD+ | 40 | PB MNC | 88 | ITD+ | 69 | PB MNC | NA |
| 44† | ITD+ | 44 | BM | 89 | ITD+ | 60 | PB | NA |
| No. . | Presentation . | Relapse . | ||||||
|---|---|---|---|---|---|---|---|---|
| FLT3 . | % Mutant . | Sample . | % Blasts . | FLT3 . | % Mutant . | Sample . | % Blasts . | |
| 1 | WT | — | PB MNC | 90 | WT | — | NA | 76 |
| 2 | WT | — | BM | 97 | WT | — | PB MNC | NA |
| 3 | WT | — | PB MNC | 95 | WT | — | BM | NA |
| 4 | WT | — | PB MNC | 28 | WT | — | BM | 42 |
| 5 | WT | — | NA | 100 | WT | — | BM | 95 |
| 6 | WT | — | NA | 100 | WT | — | BM | 60 |
| 7 | WT | — | PB | NA | WT | — | BM | 85 |
| 8 | WT | — | BM | 98 | WT | — | PB | 52 |
| 9 | WT | — | PB | 95 | WT | — | PB | 80 |
| 10 | WT | — | BM | 89 | WT | — | BM | 96 |
| 11 | WT | — | PB | 99 | WT | — | PB | 99 |
| 12 | WT | — | PB | 80 | WT | — | BM | NA |
| 13 | WT | — | BM | 86 | WT | — | BM | 60 |
| 14 | WT | — | BM | 58 | WT | — | BM | 90 |
| 15 | WT | — | BM | 90 | WT | — | BM | 87 |
| 16* | WT | — | BM | 72 | WT (ITD+) | (45) | BM (PB) | 13 (87) |
| 17 | WT | — | BM | NA | WT | — | BM | 11 |
| 18 | WT | — | PB | 78 | WT | — | BM | 56 |
| 19 | WT | — | BM | 90 | WT | — | BM | 64 |
| 20 | WT | — | BM | 90 | WT | — | BM | 74 |
| 21 | WT | — | PB MNC | 90 | D835+ | ≈50 | PB MNC | NA |
| 22 | WT | — | BM | 87 | D835+ | ≈50 | BM | NA |
| 23 | WT | — | PB MNC | 90 | ITD+ | 42 | BM | 95 |
| 24 | WT | — | BM | 95 | ITD+ | 38 | BM | 95 |
| 25 | ITD+ | 20 | PB MNC | 53 | WT | — | BM | 55 |
| 26 | ITD+ | 6 | BM | 88 | WT | — | BM | 92 |
| 27 | ITD+ | 11 | PB | 80 | WT | — | BM | 82 |
| 28 | ITD+ | 28 | BM | 94 | WT | — | BM | 85 |
| 29 | ITD+ | 44 | BM | 95 | WT | — | BM | 44 |
| 30 | D835+ | ≈50 | BM | 95 | D835+ | ≈50 | BM | 96 |
| 31 | D835+ | ≈50 | BM | 60 | D835+ | ≈50 | BM | 60 |
| 32 | ITD+ | 13 | BM | 55 | ITD+ | 16 | BM | 96 |
| 33 | ITD+ | 39 | BM | 51 | ITD+ | 39 | BM | 60 |
| 34 | ITD+ | 44 | PB MNC | 77 | ITD+ | 45 | NA | NA |
| 35 | ITD+ | 82 | BM | 97 | ITD+ | 84 | BM | 92 |
| 36 | ITD+ | 86 | BM | 86 | ITD+ | 83 | PB | 93 |
| 37 | ITD+ | 91 | PB | 70 | ITD+ | 91 | BM | NA |
| 38 | ITD+ | 5 | BM | NA | ITD+ | 35 | PB | 97 |
| 39 | ITD+ | 23 | PB MNC | 86 | ITD+ | 88 | BM | 100 |
| 40 | ITD+ | 25 | PB | 79 | ITD+ | 50 | PB | 99 |
| 41 | ITD+ | 26 + 2 | BM | 90 | ITD+ | 56 + 14 | BM | 94 |
| 42 | ITD+ | 28 + 3 + 1 | BM | 80 | ITD+ | 41 | BM | 99 |
| 43 | ITD+ | 40 | PB MNC | 88 | ITD+ | 69 | PB MNC | NA |
| 44† | ITD+ | 44 | BM | 89 | ITD+ | 60 | PB | NA |
MNC indicates mononuclear cells; BM, bone marrow; PB, peripheral blood; NA, not available.
Information in parentheses relates to second relapse.
Different mutations were detected at presentation and relapse.
Patients with an FLT3 mutation at presentation.
Twenty patients were studied who had an FLT3 mutation at presentation: 18 were ITD+ with median mutant level 28% (range, 5%-91%), and 2 had D835Y mutations at about the 50% mutant level (Table 2). The median time to relapse was 218 days (range, 38-716 days), which was less than in those patients who had not had anFLT3 mutation at presentation (P = .008, Student t test).
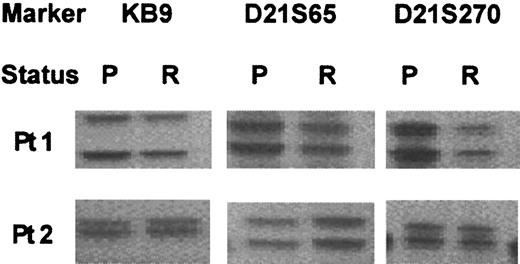
Five patients (25%) lost their FLT3 mutation at relapse. All had presented with an ITD, and the mutant levels were 6%, 11%, 20%, 28%, and 44%. Polymorphism analysis demonstrated that the paired samples were from the same individual in 4 cases from whom DNA samples were available (Figure 1); only RNA was available in the remaining patient. The loss was confirmed by radioactive PCR. In 4 of the 5 patients, the percentage of blast cells was similar in the presentation and relapse samples (nos. 25-28, Table2) and therefore it is unlikely that the mutation had been missed at relapse. In the remaining patient (no. 29), the blast cell count was lower at relapse (44%) than at presentation (95%). However, at diagnosis all cells appeared to be heterozygous for the mutation (mutant level, 44%), and therefore, even with the lower blast cell count, a heterozygous mutation would still have been well within the limits of detection of the technique.
Polymorphic marker analysis in 2 AML patients at presentation (P) and relapse (R).
Both patients presented with a FLT3/ITD, which was lost at relapse.
Polymorphic marker analysis in 2 AML patients at presentation (P) and relapse (R).
Both patients presented with a FLT3/ITD, which was lost at relapse.
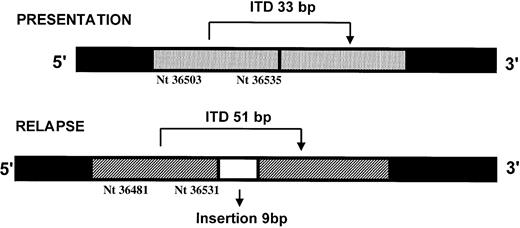
Fifteen patients had an FLT3 mutation at both presentation and relapse: 2 had D835Y mutations at both stages of their disease and 13 had ITDs. Sequencing and semiquantitative PCR showed that 6 patients relapsed with the same ITD at approximately the same level (nos. 32-37, Table 2). The median difference between presentation and relapse in the level of mutant was 0% (range, −3% to +3%). Of these patients, 3 had levels of mutant that were consistent with homozygosity or hemizygosity in most blast cells at both presentation and relapse (nos. 35-37). Six patients relapsed with the same ITD but at an increased level, where the median difference between presentation and relapse in the level of mutant was 29.5% (range, 13.5%-45%) (nos. 38-43, Table2). In 1 of these patients, 3 different ITDs were detected at diagnosis (mutant levels 28%, 3%, 1%, respectively), but only the predominant mutation was present at relapse (no. 42, Table 2). Another patient (no. 41, Table 2) had 2 different ITDs at diagnosis, mutant levels 26% and 2%, and both were increased at relapse, 56% and 14%, respectively. The remaining patient who was ITD+ at presentation, mutant level 44%, relapsed with a different ITD, mutant level 60% (#44, Table 2). The relapse mutation was not detectable in the presentation sample using radioactive PCR analysis. At presentation the ITD was 33 bp, nucleotides 36503-36535 from the DNA sequence (Genbank accession no. 13628652), whereas at relapse the ITD was 60 bp, 51 bp from nucleotides 36481-36531 plus an additional insertion of 9 bp (Figure2). Polymorphic markers confirmed that the samples were from the same individual.
The position of the FLT3/ITDs detected in one patient at presentation and relapse.
The position of the FLT3/ITDs detected in one patient at presentation and relapse.
Analysis of N-Ras mutations in selected patients
Recent studies indicated that the presence of both aFLT3 and an N-Ras mutation in the same individual is infrequent,18 suggesting that both mutations confer a proliferative or survival advantage through a common pathway. We therefore determined N-Ras mutational status at codons 12, 13, and 61 both at presentation and at relapse in those patients whoseFLT3 status had changed. Of the 4 patients who acquired aFLT3 mutation at first relapse, none had had anN-Ras mutation at presentation and one acquired anN-Ras codon 61 (position 2, A > G) mutation at relapse, coincident with the development of a D835Y mutation. Of the 5 patients who lost an ITD at relapse, only 1 had acquired anN-Ras mutation in codon 12 (position 2, G > A).
Discussion
In hematologic malignancies with disease-specific markers, the ability to test for MRD using sensitive molecular analyses is now a useful tool for predicting which patients are at risk for relapse. This information is also used in determining therapeutic strategies. For example, after allogeneic transplantation in patients with CML, intervention with donor lymphocyte infusions before the onset of hematologic relapse has been shown to be associated with an increased likelihood of antileukemic response and absence of bone marrow aplasia.28 Similarly, in acute promyelocytic leukemia, conversion to PCR positivity for PML/RARα in 2 successive bone marrow samples after consolidation chemotherapy is being used as a trigger to initiate salvage treatment.16 However, the clinical potential for such analysis in AML is restricted by the paucity of suitable markers. The recent identification of activating mutations in the FLT3 gene as the most common mutation in AML, occurring in up to one third of adult patients,18,19 24 suggests that the mutations may be a relevant marker for MRD.
To investigate this we first confirmed that the mutation is found only in leukemic cells. Analysis of a small group of 13 patients in morphologic remission demonstrated that neither the presentation ITDs nor D835 mutation could be detected in bone marrow or peripheral blood samples obtained after induction chemotherapy. Of the patients, 8 had had mutant levels of 40% to 46% of total FLT3, indicating that most cells carried the mutation (assuming heterozygosity), and 2 of the patients had had evidence for biallelic mutations or loss of one allele, with mutant levels of 74% and 90%, respectively.
For a leukemic marker to be clinically useful as an early predictor of relapse, it is important that the marker consistently reappears at relapse. However, 5 (25%) of the 20 patients who were positive for aFLT3 mutation at diagnosis lost their mutation at relapse. All had had ITDs, and the level of mutant suggests that in 3 of them at least half of the cells in the sample analyzed carried the mutation. This is consistent with a study by Nakano et al29 in which 1 of 6 ITD+ patients at presentation wereFLT3/WT at relapse, but differs from Schnittger et al,30 who reported that all 25 patients with a mutation at presentation relapsed with the same marker. The inability to detect a mutation at relapse in our study cannot be attributed to insensitivity of the technique used, because in all 5 cases the negative results from the cold PCR used bone marrow samples and were confirmed by the more sensitive radioactive PCR, which can detect ITDs at least at the 0.5% level. Furthermore, in 4 of the 5 patients the blast cell counts were similar to those at presentation. A number of other complexities arise that are relevant for MRD detection, particularly for ITDs. At diagnosis, 2 patients had evidence of more than 1 mutation, and in 1 of these patients only 1 of the 3 ITDs was detected at relapse. This indicated that the different mutants were present in separate subclones, only 1 of which survived or was selected after chemotherapy. In the other patient both mutants were detected at relapse at increased levels, although in different relative proportions. It is not possible to determine whether the mutants were in separate clones that both survived, or whether the minor mutant was acquired on the other allele in a cell that was already heterozygous for an ITD and neither clone was completely eliminated by the treatment. In addition, 1 patient with an ITD at diagnosis relapsed with a completely different ITD, which would present as a false-negative result if mutation sequence-specific primers were used to improve the sensitivity of the assays used. Analysis of FLT3 mutations as an early indicator of relapse should therefore be used with caution.
The results of this study also shed light on the role ofFLT3 mutations in the pathogenesis of AML. The findings show that these mutations may be present in only a minority of blast cells at presentation, and at relapse the ratio of mutant to WT allele frequently increases, as seen in 6 of 12 patients with the same ITD mutation at presentation and relapse (nos. 38-43, Table 2). Results in 3 of these patients were consistent with a greater proportion of ITD+ blast cells at relapse (nos. 39-41), and in 1 patient (no. 39) relapse appeared to be associated with the development of homozygosity or hemizygosity for the mutant in at least some of the cells. This indicates that the FLT3 alterations can be secondary mutations arising in an already malignant clone, with selection of the subclone containing the FLT3 mutation because of the growth or survival advantage it confers. Five (21%) of 24 patients who were WT at presentation acquired FLT3mutations for the first time in either first or second relapse, which is similar to the frequency found at presentation.18,19This is fully in accord with the fact that FLT3 mutations are secondary events. It also indicates that the FLT3 pathway is active in myeloid cells at the stage of differentiation equivalent to the clonogenic leukemic cell, as the presence of these random mutations occurring in an already transformed clone lead to its further selection. The observation that at relapse 1 of 20 patients had a different mutation and 5 had lost the FLT3 mutation is also compatible with this model. For this to occur, another subclone would need to develop from a leukemic cell that had been present at diagnosis but did not contain the FLT3 mutation. To outgrow the mutant FLT3+ cells, this subclone must have acquired alternative mutation(s) imparting a greater survival/growth advantage than that provided by the original FLT3 mutation. One possibility is a secondary mutation in N-Ras, as such mutations have been documented to arise in relapse when not present at diagnosis.29,31 This is a potential target as FLT3 activates N-Ras,32 and the rarity of FLT3mutations and N-Ras mutations in the same blast cells suggests that both mutations are predominantly using the same pathways.18 In the 5 patients we studied who lostFLT3 mutations at relapse, only one acquired anN-Ras mutation. Consequently, in the future it may be informative to screen for mutations in other candidate genes.
These results have implications for the therapeutic use of FLT3 kinase inhibitors in patients whose cells express FLT3mutations.33,34 35 As these mutations are secondary events, which is in marked contrast to the BCR/ABL translocation in CML, there will always be a high probability that leukemic subclones not containing the mutant will be present that will have a selection advantage in the presence of a FLT3 kinase inhibitor. Such inhibitors will therefore need to be used in combination with other agents.
The AML DNA and RNA bank is supported by the United Kingdom Medical Research Council and the Leukaemia Research Fund.
Prepublished online as Blood First Edition Paper, May 31, 2002; DOI 10.1182/blood-2002-02-0420.
P.D.K. and M.E.F. are supported by the Leukaemia Research Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Panagiotis D. Kottaridis, Department of Haematology, University College London, 98 Chenies Mews, London WC1E 6HX, United Kingdom; e-mail: p.kottaridis@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal