Lymphoplasmacytic lymphoma (LPL) is characterized by t(9;14)(p13;q32) in 50% of patients who lack paraproteinemia. Waldenström macroglobulinemia (WM), which has an immunoglobulin M (IgM) paraproteinemia, is classified as an LPL. Rare reports have suggested that WM sometimes is associated with 14q23 translocations, deletions of 6q, and t(11;18)(q21;q21). We tested for these abnormalities in the clonal cells of WM patients. We selected patients with clinicopathologic diagnosis of WM (all had IgM levels greater than 1.5 g/dL). Southern blot assay was used to detect legitimate and illegitimate IgH switch rearrangements. In addition to conventional cytogenetic (CC) and multicolor metaphase fluorescence in situ hybridization (M-FISH) analyses, we used interphase FISH to screen for t(9;14)(p13;q32) and other IgH translocations, t(11;18)(q21;q21), and 6q21 deletions. Genomic stability was also assessed using chromosome enumeration probes for chromosomes 7, 9, 11, 12, 15, and 17 in 15 patients. There was no evidence of either legitimate or illegitimate IgH rearrangements by Southern blot assay (n = 12). CC (n = 37), M-FISH (n = 5), and interphase FISH (n = 42) failed to identify IgH or t(11;18) translocations. Although tumor cells from most patients were diploid for the chromosomes studied, deletions of 6q21 were observed in 42% of patients. In contrast to LPL tumors that are not associated with paraproteinemia and that have frequent t(9;14)(p13;q32) translocations, IgH translocations are not found in WM, a form of LPL tumor distinguished by IgM paraproteinemia. However, WM tumor cells, which appear to be diploid or near diploid, often have deletions of 6q21.
Introduction
Waldenström macroglobulinemia (WM) is a B-cell lymphoproliferative disorder characterized by immunoglobulin M (IgM) paraproteinemia and the accumulation of clonal lymphoplasmacytic cells in the bone marrow (BM).1,2 With advancing disease, patients may acquire organomegaly, anemia, and hyperviscosity. A serum monoclonal IgM level that exceeds a concentration of 1.5 g/dL is characteristic of WM and is useful in differentiating WM from other neoplasms with plasmacytoid differentiation.3 The diagnosis is based on the presence of the characteristic accompanying clinical features and the pathologic findings, most commonly from the bone marrow (Table 1). Because of its morphologic and immunophenotypic features, the pathologic designation for WM has been lymphoplasmacytic lymphoma (LPL), as proposed by the revised European-American lymphoma classification.3Because LPL is not always associated with paraproteinemia, it is possible that LPL without paraproteinemia and WM (LPL with IgM paraproteinemia) are distinct pathologic entities.
Clinical and laboratory features of patients
| Characteristic . | All patients . | |
|---|---|---|
| n = 74 . | % . | |
| Sex | ||
| Male | 46 | 63 |
| Female | 28 | 37 |
| Light-chain type | ||
| κ | 55 | 74 |
| λ | 18 | 24 |
| Biclonal | 1 | 2 |
| B2-microglobulin > 2.7 (mg/L) | 24 of 39 | 62 |
| Albumin < 3.5 (g/dL) | 23 of 46 | 50 |
| Labeling index 1* (%) | 1 | 2 |
| Hemoglobin < 11 | 47 of 73 | 64 |
| BM clonal involvement (%) | 50 (median) | 5-95 (range) |
| Characteristic . | All patients . | |
|---|---|---|
| n = 74 . | % . | |
| Sex | ||
| Male | 46 | 63 |
| Female | 28 | 37 |
| Light-chain type | ||
| κ | 55 | 74 |
| λ | 18 | 24 |
| Biclonal | 1 | 2 |
| B2-microglobulin > 2.7 (mg/L) | 24 of 39 | 62 |
| Albumin < 3.5 (g/dL) | 23 of 46 | 50 |
| Labeling index 1* (%) | 1 | 2 |
| Hemoglobin < 11 | 47 of 73 | 64 |
| BM clonal involvement (%) | 50 (median) | 5-95 (range) |
Includes plasma cells and lymphocyte clonal cells (defined by light-chain restriction).
It has been reported that the t(9;14)(p13;q32) translocation occurs in at least 50% of patients with LPL.4 The t(9;14)(p13;q32) results in the up-regulation of PAX-5,5,6implicating this gene as a putative oncogene in the pathogenesis of LPL.7-9 Other PAX genes are known to be involved in diverse human neoplasia.10PAX-5encodes for the transcription factor, B-cell–specific activation protein (BSAP), which is a 50-kDa protein critical for B-cell development. BSAP up-regulation appears to cause an increase in B-cell proliferation11 and is characteristically absent in normal and malignant plasma cells.12-15 BSAP/PAX-5 is known to down-regulate the Iε promoter and the IgH 3′α enhancer, with consequent down-regulation of IgH transcription.16-19In the original series of LPL tumors with the t(9;14)(p13;q32) translocation, none of the patients had paraproteinemia, a finding consistent with the possibility that the overexpression ofPAX-5 prevents the expression of high levels of immunoglobulin that result in paraproteinemia. If this hypothesis is correct, one would expect that the t(9;14)(p13;q32) translocation would not be present in WM, a form of LPL with IgM paraproteinemia.
The underlying genetic abnormalities responsible for WM have not been identified. From a limited number of conventional cytogenetic (CC) studies, no recurrent cytogenetic abnormalities have been detected in tumor cells from patients with WM. However, there are sporadic reports of WM tumors with IgH or t(11;18)(q21;q21) translocations or 6q abnormalities, each of which occurs in other kinds of B-cell tumors. Thus, we decided to test a cohort of patients with well-defined WM to address 3 issues. First, despite the expression of IgM in WM, does legitimate or illegitimate IgH switch recombination occur, and perhaps mediate IgH translocations, as previously shown in multiple myeloma and other B-cell tumors?20 Second, using a highly sensitive and specific interphase fluorescence in situ hybridization (FISH) assay, what is the incidence of the t(9;14)(p13;q32) or other IgH translocations in WM? Third, through a combination of CC, multicolor metaphase-FISH (M-FISH), and interphase FISH assays, what is the incidence of recurrent numeric or structural karyotype abnormalities—including the t(11;18)(q21;q21) translocation or abnormalities of 6q21—in WM?
Patients and methods
Patients and bone marrow samples
We identified patients who fulfilled the clinical and pathologic diagnosis of WM1 2 (Table 1). The study was conducted under institutional review board approval. Each patient was required to have an IgM paraproteinemia level of 1.5 g/dL or greater and clonal lymphoplasmacytic infiltration comprising at least 20% of the mononuclear cells in the bone marrow (BM), but BM involvement could be lower (10%-19%) if the patient's monoclonal protein level exceeded 3 g/dL. The cohort consisted of 74 patients who fulfilled these criteria (65% had prior chemotherapy exposure). Samples available for study included BM aspirate obtained from 33 patients at the time of routine clinical procurement (all studies with cIg-FISH), fixed cell pellets in Carnoy fixative and cultured for the purpose of clinical karyotype analysis from 17 patients, or both types of samples from 24 patients (cultured and fixed cell pellets and uncultured bone marrow aspirate).
In 19 patients, bone marrow research aspirates were further enriched by simultaneous CD138+ and CD19+ magnetic bead selection (Miltenyi Biotec, Auburn, CA). Owen et al21 22 have found that clonal cells in WM express CD19, CD138, or both. Because of the pleomorphic nature of the clonal process in WM, we thought that with the combination of CD138 and the B-cell marker CD19, we could enrich for the clonal cells of patients. When we only had stored BM samples previously cultured for metaphase analysis, it was not possible to perform magnetic bead selection or concurrent immunofluorescence staining of the cytoplasmic immunoglobulins for those patients.
This study was conducted under the approval of the Mayo Clinic institutional review board, and patients gave informed consent for the sample collection. The study was conducted in accordance with the Declaration of Helsinki for research with human subjects.
Southern blot analysis
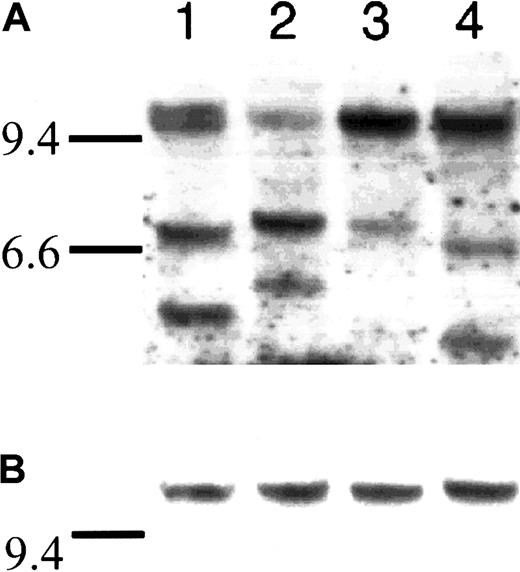
Southern blot analysis was performed according to our previously published technique.20 In brief, we used a JH probe to detect the germ line and clonal bands to ensure the presence of clonal DNA. We subsequently performed sequential hybridization with probes flanking the 5′ and the 3′ ends of the Sμ switch region. The presence of fully concordant 5′Sμ and 3′Sμ bands indicates the absence of IgH switch recombination. Alternatively, discordant 5′Sμ and 3′Sμ bands that do not hybridize with other IgH switch probes strongly suggest illegitimate IgH switch recombination caused by a chromosomal translocation.20 A total of 2.5 to 10 μg BM DNA (these samples were not magnetic microbead-enriched) was loaded into each well after complete digestion with restriction enzyme HindIII (Figure1)
Southern blot assay used to detect legitimate and illegitimate rearrangements at the IgH locus.
Southern blot of 4 WM samples that were fractionated on an agarose gel, blotted, and hybridized sequentially with a JH probe (A), and a 5′ Sμ probe (B). Horizontal bars at the left of each exposure correspond to λHindIII markers, with the 9.4-kb marker in each lane but the 6.6-kb marker included for panel A. Panel A shows the genomic DNA probes with a JH oligonucleotide probe and the corresponding clonal bands (variable size) and germ line fragments (approximately 10 kb). Panel B shows the constant size of the 5′ Sμ probe indicating lack of legitimate or illegitimate rearrangements. The total amount of genomic DNA loaded was 2.5 μg per lane.
Southern blot assay used to detect legitimate and illegitimate rearrangements at the IgH locus.
Southern blot of 4 WM samples that were fractionated on an agarose gel, blotted, and hybridized sequentially with a JH probe (A), and a 5′ Sμ probe (B). Horizontal bars at the left of each exposure correspond to λHindIII markers, with the 9.4-kb marker in each lane but the 6.6-kb marker included for panel A. Panel A shows the genomic DNA probes with a JH oligonucleotide probe and the corresponding clonal bands (variable size) and germ line fragments (approximately 10 kb). Panel B shows the constant size of the 5′ Sμ probe indicating lack of legitimate or illegitimate rearrangements. The total amount of genomic DNA loaded was 2.5 μg per lane.
Standard cytogenetic analysis
Conventional karyotypes performed for clinical evaluation were available for 37 patients. BM specimens were processed by direct and short-term culture techniques, as described previously.23At least 20 banded metaphases were analyzed for each patient, with representative karyotypes prepared from at least 2 metaphases from each clone. The karyotype was described according to the International System for Human Cytogenetics Nomenclature (ISCN, 1995).
Multicolor-FISH assay
M-FISH was performed as previously described.24 25Slides with informative karyotypes in 5 patients were processed using the Spectra Vysion (Vysis, Downer's Grove, IL) reagent. Probes for M-FISH were placed on the hybridization site, protected with a coverslip, sealed with rubber cement, and placed in the HYBrite (Vysis) for codenaturation. Slides were viewed with a Zeiss (Thornwood, NY) microscope powered by a 100-W mercury bulb. Filter sets for capturing M-FISH images and viewing metaphases were from Vysis.
Interphase FISH
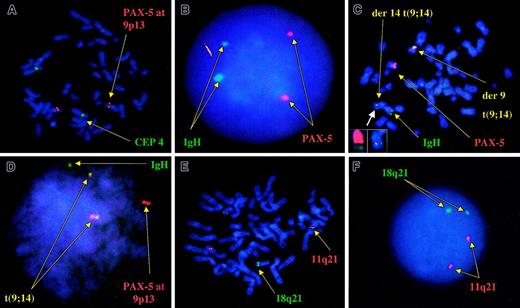
We used 2 different probe strategies to detect IgH rearrangements by interphase FISH. We first used a break-apart strategy (segregation), using a cosmid probe (VH) (labeled red) specific for the IgH variable region and a BAC clone (CH) specific for the IgH constant (labeled green).26 Under separate experiments and to detect the t(9;14)(p13;q32) translocation, we used a fusion strategy (colocalization), using a BAC clone (clone 112gO2; Incyte Genomics, Palo Alto, CA) specific for thePAX-5 gene at 9p13 (labeled red) (Figure2), together with the VH/CH probes (both labeled in green).26 To validate these probes, we tested them in the cell line KIS-1, which harbors t(9;14)(p13;q32) and was kindly provided by Dr Hitoshi Ohno (Kyoto University, Japan) and in a patient sample with t(9;14)(p13;q32) that was kindly provided by Dr Herve Avet-Loiseau (Nantes, France). In both cases our probes showed 2 fusion signals of the IgH probe and the PAX-5 BAC clone, on both derivative chromosomes in each case, indicating its usefulness for the detection of this translocation.
Correct localization of the probes used for the detection of t(9;14)(p13;q32) and t(11;18)(q21;q21).
(A) Localization of the PAX-5–specific probe to chromosome 9 at band p13 (red). The green signal is that of chromosome 4 centromeric probe. (B) Normal pattern of hybridization for the IgH/PAX-5 probes, consistent with 2 pairs of discrete green and red signals, respectively, without fusion signals. (C) Metaphase FISH assay in a patient with known t(9;14)(p13;q32) serving as the positive control. Metaphase shows 2 fusion signals indicating a balanced reciprocal translocation and the germ line configuration for the IgH probes (green) and the 9p13 PAX-5probe (red). Smaller boxes in the lower left corner depict an enlarged view of the der(14)t(9;14) chromosome, showing the fusion signal indicative of the translocation (right small box) and the same chromosome after stripping of the probes and hybridizing with whole chromosome paints (WCPs). Left small box: red indicates WCP 14, and green indicates WCP 9. (D) Interphase FISH results on the same patient, and 2 distinct fusion signals indicative of the balanced translocation. (E) Correct localization of the probes used for the detection of t(11;18)(q21;q21). (F) Normal interphase FISH pattern for these same 2 probes, again consistent with 2 pairs of discrete green and red signals, respectively, without resultant fusion signals.
Correct localization of the probes used for the detection of t(9;14)(p13;q32) and t(11;18)(q21;q21).
(A) Localization of the PAX-5–specific probe to chromosome 9 at band p13 (red). The green signal is that of chromosome 4 centromeric probe. (B) Normal pattern of hybridization for the IgH/PAX-5 probes, consistent with 2 pairs of discrete green and red signals, respectively, without fusion signals. (C) Metaphase FISH assay in a patient with known t(9;14)(p13;q32) serving as the positive control. Metaphase shows 2 fusion signals indicating a balanced reciprocal translocation and the germ line configuration for the IgH probes (green) and the 9p13 PAX-5probe (red). Smaller boxes in the lower left corner depict an enlarged view of the der(14)t(9;14) chromosome, showing the fusion signal indicative of the translocation (right small box) and the same chromosome after stripping of the probes and hybridizing with whole chromosome paints (WCPs). Left small box: red indicates WCP 14, and green indicates WCP 9. (D) Interphase FISH results on the same patient, and 2 distinct fusion signals indicative of the balanced translocation. (E) Correct localization of the probes used for the detection of t(11;18)(q21;q21). (F) Normal interphase FISH pattern for these same 2 probes, again consistent with 2 pairs of discrete green and red signals, respectively, without resultant fusion signals.
To detect t(11;18)(q21;q21) we used a set of probes, kindly provided by Dr David James (Mayo Clinic), that span both breakpoints in all reported t(11;18)(q21;q21) translocations and that result in a double fusion, as validated by Remstein et al27 (Figure 2). To screen for possible aneuploidy, we used commercially available centromere enumeration probes (CEPs) for chromosomes 7, 9, 11, 12, 15, and 17 (Vysis). To screen for 6q21 deletions, we used the clone RPCI 91C23 (obtained from Oakland Children's Hospital, Oakland, CA) with simultaneous hybridization for CEP 6. This same cohort of patients has also been studied for deletions at 13q14 and 17p13 by interphase FISH, and the results are compared with those obtained from this study.28
Noncommercial probes were directly labeled using standard nick translation with SpectrumRed or SpectrumGreen (Vysis). Slides and probes were co-denatured for 7 minutes at 80°C, placed in a humidified chamber, and allowed to hybridize in a 37°C oven overnight. Slides were then washed, an antifade mounting medium (Vectashield H-1000) was applied to each, and the slides were coverslipped. For slides not assessed for cytoplasmic immunoglobulin stain, DAPI (Vector Laboratories, Burlingame, CA) was added to the medium.
Cytoplasmic staining
In all patients (n = 33) for whom cytospin slides were available, we performed cytoplasmic staining of the IgM cytoplasmic protein using an AMCA (7-amino-4-methylcoumarin-3-acetic acid)–conjugated anti-IgM antibody (Vector Laboratories), in a variation of our previously published technique.29 If we could not use the cIg-FISH technique, or if the original cytospin slides were depleted, we used slides made from the cultured and fixed cells. Because of these limitations, these slides were scored on unselected cells without regard to cellular morphology.
Scoring statistics
Using a Leica epifluorescence microscope with fluorescein isothiocyanate, Texas red, and a DAPI filter, we attempted to score at least 100 clonal cells from each patient. In the cytospin slides, only cells identified by morphologic features or cytoplasmic anti-IgM staining were scored. Fusion of probe signals (colocalization) was defined as 2 probe signals making contact (Figure2). Break-apart of probe signals (segregation) was defined by a distance of more than 3 signal widths between 2 differently labeled probe signals. A sample was said to have an abnormal pattern if the percentage of cells exceeded the mean percentage background level plus 3 SD found for normal cells. The mean percentage background was determined in normal and abnormal samples, and at least 1000 cells were scored for each set of probes.
Results
Southern blot analysis
To detect IgH rearrangements we conducted a pilot study using Southern blot analysis, but we did not find evidence of legitimate or illegitimate IgH switch recombination fragments. In all cases the presence of clonal cell DNA was confirmed by the finding of clonal rearrangements by hybridization with a JH probe, but there was complete concordance of the 5′Sμ and the 3′Sμ bands in sequential hybridizations (n = 12) (Figure 1).
Conventional cytogenetics and M-FISH
We reviewed the karyotype results of 45 patients using conventional cytogenetic analysis performed for routine clinical purposes. Six patients were excluded because chromosomal analysis was performed at the time of diagnosis of secondary myelodysplasia or leukemia. Abnormal findings on BM examination and resultant abnormal karyotypes were consistent with the diagnoses. Of the remaining 39 patients, 35 had sufficient numbers of metaphases to be evaluable (Table 2). Of these 35 patients, 22 (63%) patients were found to have only normal metaphases, possibly originating from the normal myeloid elements in the BM, including 2 patients whose only abnormality was the loss of chromosome Y. Abnormal metaphases were obtained in 13 (37%) patients, 5 of whom were not previously treated. Structural and numerical chromosomal abnormalities were encountered in the abnormal karyotypes, but usually we observed single abnormalities in an otherwise normal karyotype (Table 2). For the 13 patients with abnormal karyotype, 4 (31%) had recurrent abnormalities of chromosome 13, with del(13)(q14) in 3 and monosomy 13 in one; 2 (15%) had del(6)(q13q21)(q13q25); 1 (8%) had an addition at chromosome 6q27 (add(6)(q27)); 2 (15%) had abnormalities of chromosome 17q25 and 1 (8%) had chromosome 17 monosomy; 2 had del(5q)(13q35) (one with and one without prior therapy); and 4 had diverse abnormalities in chromosome 8 (t(Y;8)(q11;p23), i8(q10), der(8;9)(q10;q10), and −8). As a follow-up to conventional cytogenetics in 5 of 13 patients with abnormal karyotype, M-FISH studies identified karyotypic abnormalities in 2 patients but provided no additional information in 3 patients (Table 2).
Karyotypic abnormalities in patients with informative karyotypes
| Patient . | Karyotype . | Additional information provided by M-FISH [no. metaphases analyzed] . |
|---|---|---|
| Patients not previously treated | ||
| 1 | ∼4n with fragmentation[5]/46,XX[20] | 48,XX,der(9),der(17)[8] |
| 2 | 47,X,t(Y;8)(q11.2;p23),del(5)(q13q31), del(6)(q13q21),der(19)t(19;?)(q13.1;?), +2mar[10]/46,XY [10]/10= | Not done |
| 3 | 46,XX,del(6)(q13q25)[1]/46,XX[29] | No further information [4] |
| 4 | 46,XY,del(13)(q14q22)[2]/46,XY[18] | 49X,−X+der(3),+4,der(9),+18,+19[2] |
| 5 | 47,X,−Y,add(3)(q21),add(4)(q21),add(6)(q27),add(7)(q11.2), i(8)(q10),+10,−13,−13,−15,add(17)(q25),+add(18)(q23),+19,−20,+3mar/46,XY[17] | No further information [4] |
| Patients previously treated | ||
| 6 | 47-48,XY,der(16)t(16;?)(q24;?),der(17)t(17;?)(q25;?),+1−2mar[5]/46,XY[14] | Not done |
| 7 | 45,X,−X,der(8;9)(q10;q10),+9,−17,−19,+2mar[1]/45,idem,t(7;18)(p15;q21)[1]/46,XX[28] | Not done |
| 8 | 45,XY,−7[7]/46,XY[23] | 45,XY,−7[3] |
| 9 | 46,XX,−12,+r[8]/46,XX[10] | Not done |
| 10 | 46,XX,del(13)(q14q22)[3]/46,XX[17] | Not done |
| 11 | 46,XX,del(13)(q12q14)[3]/46,XX[5] | Not done |
| 12 | 46,XY,+add(1)(p12),−15[1]/46,XY[29] | Not done |
| 13 | 47,XY,+8,?del(5)(q13q35),del(6)(p21.3?p23),tdic(13;?)[20] | Not done |
| Patient . | Karyotype . | Additional information provided by M-FISH [no. metaphases analyzed] . |
|---|---|---|
| Patients not previously treated | ||
| 1 | ∼4n with fragmentation[5]/46,XX[20] | 48,XX,der(9),der(17)[8] |
| 2 | 47,X,t(Y;8)(q11.2;p23),del(5)(q13q31), del(6)(q13q21),der(19)t(19;?)(q13.1;?), +2mar[10]/46,XY [10]/10= | Not done |
| 3 | 46,XX,del(6)(q13q25)[1]/46,XX[29] | No further information [4] |
| 4 | 46,XY,del(13)(q14q22)[2]/46,XY[18] | 49X,−X+der(3),+4,der(9),+18,+19[2] |
| 5 | 47,X,−Y,add(3)(q21),add(4)(q21),add(6)(q27),add(7)(q11.2), i(8)(q10),+10,−13,−13,−15,add(17)(q25),+add(18)(q23),+19,−20,+3mar/46,XY[17] | No further information [4] |
| Patients previously treated | ||
| 6 | 47-48,XY,der(16)t(16;?)(q24;?),der(17)t(17;?)(q25;?),+1−2mar[5]/46,XY[14] | Not done |
| 7 | 45,X,−X,der(8;9)(q10;q10),+9,−17,−19,+2mar[1]/45,idem,t(7;18)(p15;q21)[1]/46,XX[28] | Not done |
| 8 | 45,XY,−7[7]/46,XY[23] | 45,XY,−7[3] |
| 9 | 46,XX,−12,+r[8]/46,XX[10] | Not done |
| 10 | 46,XX,del(13)(q14q22)[3]/46,XX[17] | Not done |
| 11 | 46,XX,del(13)(q12q14)[3]/46,XX[5] | Not done |
| 12 | 46,XY,+add(1)(p12),−15[1]/46,XY[29] | Not done |
| 13 | 47,XY,+8,?del(5)(q13q35),del(6)(p21.3?p23),tdic(13;?)[20] | Not done |
Interphase FISH
Of the 31 patients studied, 30 had no evidence of IgH translocations by the VH/CH strategy, and in one patient 37% of cells harbored the abnormality. In addition, in none of these 31 patients could we find fusion signals indicative of t(9;14)(p13;q32) (Figure 2). When we extended our study to include 18 additional patients in whom we had CC cell suspension to study, we found that none of them had evidence of IgH translocations by the VH/CH break-apart strategy (n = 17) or the t(9;14)(p13;q32) translocation by the fusion strategy (n = 18). One patient had 3 or more signals from the PAX-5 BAC clone in 69% of the cells but no fusions (17% with 3 signals, and 52% with 4 signals). This same patient had evidence of trisomy for chromosome 9 in the metaphase analysis. None of the 24 patients studied had evidence of t(11;18)(q21;q21) as detected by FISH.
To screen for possible aneuploidy, tumor cells from 15 patients were studied by interphase FISH using CEP probes for chromosomes 7, 9, 11, 12, 15, and 17. With the exception of one patient, all chromosomes were normal; one patient had only one signal for chromosome 9, indicative of monosomy in 26% of the clonal cells. Although some patients showed abnormalities in the karyotype consistent with aneuploidy (Table 2), interphase FISH showed that this is the exception, and patients with karyotype abnormalities likely have aggressive variants of the disease. There is probably an inherent bias in the patient population that is subjected to scrutiny of karyotype analysis (ie, worsening disease). In addition, it is highly likely, as in multiple myeloma, that the ability to obtain informative metaphases is closely related to tumor burden and proliferative activity.30
Deletions of 6q
We also tested 24 patients (in whom we had metaphase culture slides but could not perform cIg-FISH assay) for evidence of 6q21 deletions and found that 10 (42%) had abnormalities in more than 25% of the cells. In addition 5 (21%) more patients had between 10% and 25% abnormal cells. Because in many of these latter patients the percentage of clonal involvement of the bone marrow was between 10% and 20%, it is conceivable that they could also have had deletions at this site. Therefore, between 42% and 63% of patients had deletions of 6q21.
Correlation between interphase FISH and karyotype analysis
Three patients had chromosome abnormalities detected by metaphase analysis; we also performed specific interphase FISH. In one patient −13 was confirmed by FISH, and in another patient the deletion of 17p was confirmed by FISH. In yet another patient, del(6)(q13q25) was detected by our FISH probe.
Discussion
In this study we have found that clonal cells from patients with clinically defined WM do not have the t(9;14)(p13;q32) translocation, as previously found in 50% of patients with LPL but without paraproteinemia. Thus, WM, a type of LPL with IgM paraproteinemia, and LPL with no paraproteinemia differ not only in phenotype but also in the presence of the t(9;14)(p13;q32) translocation. This result is consistent with the hypothesis that the predicted biologic features of B-cell clones with PAX-5 up-regulation are incompatible with the phenotype of WM.17-19 Notably, BSAP is absent in plasma cells, and the overexpression of PAX-5/BSAP abrogates the production of the J peptide.13 This protein is an integral component of the IgM pentamers, which give rise to the hyperviscosity of patients with WM.31 In addition,PAX-5/BSAP negatively regulates the 3′α enhancer, with the likely consequent inhibition of transcription at the IgH locus,17-19,32,33 a result inconsistent with the IgM paraproteinemia observed in these patients. Finally, the inhibition of PAX-5 transcription with antisense oligonucleotide down-regulates immunoglobulin class switching,11 a result consistent with our failure to identify evidence for legitimate or illegitimate IgH isotype switch recombination in the clonal cells of IgM-producing WM.
Our finding that IgH translocations are not present in WM is consistent with the previously reported karyotypic abnormalities in these patients. In a series of 45 patients with WM reported by Louviaux et al,34 12 patients had abnormal metaphases, but no abnormalities of chromosome 14 were noted. In our review of the cytogenetic database at the Mayo Clinic, which we report here, we were unable to find any patient with 14q32 abnormalities. Unlike multiple myeloma20,35,36 and low-grade lymphomas,37,3814q32 translocations appear not to be initiation events for disease pathogenesis in WM. Scattered reports in the literature describe patients with WM and 14q32 translocations, including t(8;14)(q24;q32).39-42 Some of these reports are confounded either by the lack of consistent clinical features associated with a diagnosis of WM or by the samples originating from pleural effusions and, thus, of unknown relation to the original clone.39-42
There are rare reports of WM patients in whom tumor cells had a t(11;18)(q21;q21) translocation,43,44 an abnormality also seen in approximately 20% of extranodal marginal zone lymphomas.45 Because we have been unable to detect this translocation in the tumor cells of 24 WM patients, it must be, at best, rare.
Despite an apparent paucity of structural and numeric karyotypic abnormalities in tumor cells from our cohort of WM patients, we did identify a high prevalence of deletions of the long arm of chromosome 6. This is consistent with what has been previously reported in selected patients with WM46,47 and other B-cell neoplasias in which abnormalities in this region are common.48-50 This is also consistent with findings from our karyotype analysis that 3 patients had 6q deletions. Further work is under way to characterize the area of minimal deletion and the search for putative genes involved in disease pathogenesis. We suspect that the inactivation of a tumor suppressor gene at this locus, as is seen in other B-cell neoplasias, is likely to be of importance for clone immortalization.
Our data suggest less genomic instability in WM than in multiple myeloma, as determined by metaphase analyses for structural and numeric abnormalities and interphase FISH for numeric abnormalities. Moreover, we have recently reported that, unlike multiple myeloma and B-cell chronic lymphocytic leukemia (B-CLL), deletions of 13q14 are rarely observed at the time of diagnosis.28 As are deletions of 17p13.1 (see below), 13q14 deletions are seen mostly in advanced stages and in clonal cells in a small fraction (approximately 15%) of patients with WM at the time of diagnosis.28 Therefore, we can conclude that WM is clearly different from myeloma in that it lacks IgH translocations, has infrequent deletions of 13q14, and has limited numeric and structural karyotypic abnormalities. WM is also different from B-CLL in the lower incidence of 13q14 deletions (seen in approximately 50% of patients with B-CLL) and the low prevalence of trisomy 12 (seen in approximately 15% of patients with B-CLL).51 WM is also different from extranodal marginal zone lymphomas in that we could not detect any patient with t(11;18)(q21;q21). The lack of IgH translocations also differentiates WM from many kinds of B lymphomas (follicular, mantle cell, diffuse large cell).52 We thus conclude that the biologic nature of WM is different from that of multiple myeloma and most lymphomas, but it appears similar to postgerminal center B-CLL (Table3).
Genetic comparison between WM, myeloma, and B-CLL
| Feature . | WM . | Myeloma . | B-CLL . |
|---|---|---|---|
| Clinical course | Usually indolent | Aggressive | Usually indolent |
| Genomic stability (ploidy) | Diploid | Aneuploid | Diploid |
| Structural abnormalities (other than 6q) | Infrequent | Frequent, multiple | Infrequent |
| IgH translocations | Rare | 75% | Rare |
| Chromosome 13 abnormalities at diagnosis | Rare28 | ∼50% | ∼50% |
| Deletions of 17p13 | Rare at diagnosis28 | Rare (10%-15%) | Rare (5%) |
| Proliferation by the labeling index | Low/absent | Active | Not done |
| Somatic hypermutation | Yes | Yes | Variable |
| Feature . | WM . | Myeloma . | B-CLL . |
|---|---|---|---|
| Clinical course | Usually indolent | Aggressive | Usually indolent |
| Genomic stability (ploidy) | Diploid | Aneuploid | Diploid |
| Structural abnormalities (other than 6q) | Infrequent | Frequent, multiple | Infrequent |
| IgH translocations | Rare | 75% | Rare |
| Chromosome 13 abnormalities at diagnosis | Rare28 | ∼50% | ∼50% |
| Deletions of 17p13 | Rare at diagnosis28 | Rare (10%-15%) | Rare (5%) |
| Proliferation by the labeling index | Low/absent | Active | Not done |
| Somatic hypermutation | Yes | Yes | Variable |
The normal counterpart of the malignant cell in WM might be a postgerminal center IgM memory B cell that has undergone somatic hypermutation but has failed to undergo isotype class switching. Our Southern blot results, failing to identify legitimate or illegitimate IgH switch recombination rearrangements indicative of isotype class switching, and the presence of the IgM paraproteinemia are consistent with this. In addition, individual WM tumor cells show heterogeneous morphologic features consistent with variable differentiation from a B cell to a plasma cell but failure to fully differentiate into plasma cells. We speculate that the presumed genetic event(s) responsible for generating an immortalized clone of tumor cells in WM are directly or indirectly responsible for the failure of isotype class switching and the lack of differentiation to plasma cells. With the exception of frequent deletions of the long arm of chromosome 6, our knowledge of specific primary or secondary genetic events in WM remains elusive.
R.F. is supported by a research grant from the International Waldenström Macroglobulinemia Foundation and is a Leukemia and Lymphoma Society Translational Research Awardee. R.F. and P.R.G. are supported by the CI-5 Cancer Research Fund-Lilly Clinical Investigator Award of the Damon Runyon-Walter Winchell Foundation. Supported in part by Public Health Service grant R01 CA83724-01 from the National Cancer Institute (R.F.). P.R.G. and R.A.K. are supported in part by research grant P01 CA62242 from the National Cancer Institute. P.R.G. is supported by the ECOG grant CA21115-25C from the National Cancer Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Rafael Fonseca, Department of Hematology and Internal Medicine, Stabile 628, Mayo Clinic, Rochester, MN 55905; e-mail: fonseca.rafael@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal