Abstract
Recent studies in humans have highlighted the importance of a distinct cellular entity, the plasmacytoid dendritic cell (PDC). To identify genes for which expression is restricted to human PDCs, a cDNA subtraction technique was applied using cDNA from activated monocyte-derived DCs (MDDCs) as competitor. In the 650 sequences analyzed, 25% were for B-cell transcripts. We also found lymphoid-related genes, immunoglobulinlike transcript 7 (ILT7), granzyme B (GrB), Spi-B, and the receptor tyrosine kinase Eph-B1. Granzyme B was up-regulated on activation, and protein was detected only in PDCs. Eph-B1 protein was expressed in the cytoplasm and the nuclei of PDCs and MDDCs, respectively. Interestingly, several novel molecules have been identified that were predicted to encode for a type 2 transmembrane protein (BRI3), a putative cytokine (C-15, a cysteine-rich–secreted protein), and a type 1 leucine-rich repeat protein (MAPA). The identification of genes expressed in PDCs provides new insights into their function and origin.
Introduction
Since their original identification, there has been increasing evidence that dendritic cells (DCs) represent a heterogeneous population of cells with distinct origins, stages of differentiation/maturation, and specific functions.1,2 In humans, a subset of immature CD11c− DCs with plasmacytoid morphology has been identified (plasmacytoid DCs [PDCs]) in the peripheral blood of adults and neonates,3,4tonsils,5 lymph nodes,6 and thymi.7 Unlike CD11c+ blood DCs, these cells are capable of producing high levels of type 1 interferon (IFN) in response to virus stimulation6,8 or to oligonucleotides containing particular CpG motifs.9,10 When compared with other DC subsets, PDCs are characterized by several unique features: cytokine requirements for their development, phenotype, response to pathogens correlating with a particular profile of pattern recognition and presentation receptors, cytokine production, migratory properties, and function.2,9,11 PDCs have a unique plasmacytoid morphology and distinct phenotypic features, such as expression of high levels of CD123/interleukin-3Rα (IL-3Rα), that correlate with their enhanced survival in response to IL-3.5 Of particular interest is the observation that PDCs can differentiate into mature DCs capable of priming naive CD4+ T cells toward either Th1 or Th2 responses, depending on the activation stimuli.6,12-14 IL-3– and CD40-activated PDCs induce a Th2 orientation (hence, their alternative name, pre-DC2/DC2), whereas virus-activated PDCs induce a Th1 profile. Functional PDCs (producing IFN-α) can be generated in vitro from CD34 progenitors isolated from fetal liver, bone marrow, or cord blood in the presence of Flt3 ligand for 2 to 3 weeks15 or from CD34+CD38− fetal liver progenitors cocultured for 1 week with a stromal murine cell line.16
To better characterize these cells, we performed a polymerase chain reaction (PCR)–based subtraction technique on cDNA from PDCs isolated from human tonsils (tester) and using cDNA from monocyte-derived DCs (MDDCs) as competitor. This approach is demanding because it requires high numbers of cells. MDDCs were chosen as competitor because they represent the prototype myeloid DCs, and they can be generated in high numbers. In addition, MDDCs have been shown to exert distinct effects compared with PDCs and thus were considered appropriate candidates for identifying genes differentially regulated in PDCs. Our aim was to identify transcripts preferentially overexpressed in PDCs, regardless of the stage of differentiation, compared with activated monocyte-derived DCs. One of the major drawbacks in the study of PDCs is the lack of markers specific for PDCs that are maintained and that remain specific for PDCs at a mature stage. For this purpose, we used a mixture of fresh, highly purified, fluorescence-activated cell sorter analysis (FACS)–sorted PDCs pooled from different donors and IL-3 + CD40L-activated PDCs as tester. In the current study we describe the cloning of Spi-B, ILT7, GrB, and Eph-B1 cDNA and the identification of novel transcripts such as BRI3, C-15, and monocyte and plasmacytoid activated molecule (MAPA). Comparison of the differential expression in MDDCs versus PDCs of those newly identified genes gives new insights into their origin and, interestingly, suggests uncovered functions of PDCs.
Materials and methods
Cell preparations
Discarded human surgical material (eg, tonsils and cord blood) was obtained anonymously according to the institutional regulations, in compliance with French law. Human blood was obtained anonymously from the Etablissement Français du Sang (Lyon, France) after the donor gave informed consent according to the Declaration of Helsinki specifically indicating the possible research use of the sample if it was not suitable for transfusion use. FACS-sorted PDCs (greater than 98% purity) were purified from human tonsils according to the method of Grouard et al.5 For reverse transcription-PCR (RT-PCR) analysis of activated PDCs, cells were cultured in the presence of IL-3 or of IL-3– and CD40L-transfected L cells (at a ratio of 5 PDCs to 1 L cell) for 16 or 48 hours. Activated PDCs used for the subtraction were harvested at different time points after culture in IL-3 + CD40L (24, 48, 72, and 96 hours). Freshly isolated tonsil PDCs did not express CD80, CD83, or CD86, whereas these molecules were up-regulated on IL-3 + CD40L activation. DCs were generated in vitro from monocytes (MDDCs) in the presence of granulocyte macrophage–colony-stimulating factor (GM-CSF) and IL-417 or from cord blood CD34 progenitors in the presence of GM-CSF and tumor necrosis factor (TNF).18Because of the quantities of material necessary for this study, we had to use pools of cells from different donors. Thus, for subtraction, a pool of tonsil PDCs from 12 donors and a pool of MDDCs derived from the monocytes of 10 blood donors were used. T cells, monocytes, and granulocytes were isolated from peripheral blood. B cells were isolated from tonsillar tissue. Purification of specific cell types was by negative selection with monoclonal antibody (mAb) and magnetic beads. Activation of granulocytes was by treatment with phorbol 12-myristate 13-acetate and ionomycin for T cells by anti-CD3 (5 μg/mL coated on plastic) (Becton Dickinson, Mountain View, CA) and anti-CD28 (1 μg/mL) (Becton Dickinson) for 24 hours and by coculture in the presence of CD40L-transfected L cells for 24 hours for B cells and 16 or 48 hours for DCs, as stated. We used Jurkat,19Silvanus,20 U937,21 and cancer cell lines.
RNA preparation
Cells were lysed, and total RNA was made as described.22 The integrity of the RNA was confirmed by denaturing agarose gel electrophoresis. PolyA+ messenger RNA was selected by oligo dT-coupled magnetic beads (Dynabeads Oligo [dT]25; Dynal ASA, Oslo, Norway).
Construction of a subtracted PDC cDNA library
To identify genes specifically expressed in PDCs, a subtracted hybridization technique was applied, as previously described,23 to 5 × 106 PDCs (tester) (composed of equal proportions freshly isolated and activated with IL-3 + CD40L) and to 108 CD40-activated MDDCs as competitor (driver). PDC mRNA (80 ng) was used as tester, and 1.5 μg MDDC mRNA was used as driver. Complementary DNA synthesis and subtraction were performed according to the PCR-select kit (Clontech, Palo Alto, CA) using Advantage KlenTaq polymerase (Clontech). The primary PCR reaction was for 28 cycles, and the second (nested) PCR was for 15 cycles on a thermal cycler 480 (Perkin-Elmer, Norwalk, CT). To clone subtracted PDC cDNA, 10 nested reactions were pooled and resolved on a 2% low-melting agarose. Aiming for individual bands, 12 gel slices in the 0.3- to 1.2-kb range were excised, DNA eluted, and cloned into a T/A-vector (pCRII; Invitrogen, San Diego, CA).
DNA sequencing and analysis
Inserts were sequenced in both strands by automatic sequencing (373 automated DNA sequencer; Applied Biosystems, Foster City, CA). Comparisons against GenBank and dbEST databases and protein homology prediction were obtained by BLAST.24Sequences were analyzed using Sequencher (Genecodes, CA) and Lasergene (DNASTAR, London, United Kingdom). Prediction of signal sequences was made using pSORTII.25
Analysis of gene expression by RT-PCR analysis
First-strand cDNA was prepared using the Superscript kit (Pharmacia, Orsay, France). PCR reaction was subjected to 35 cycles of denaturing (30 seconds, 94°C), annealing (30 seconds), and extension (2 minutes, 72°C). Quantification of cDNA was performed by comparison with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH)–PCR products, as follows—GAPDH: sense, ACCACAGTCCATGCCATCAC; antisense, TCCACCACCCTGTTGCTGTA; CD62L: sense, GGACCTGAGACCCTTGTGCT; antisense, CCCATAGTACCCCACATCAC; CD123/IL-3Rα: sense, ATGCCGACTATTCTATGCCG; antisense, TGTCTCTGACCTGTTCTGTG; CXCR4: sense, TGCTGACTATTCCCGACTTCATCT; antisense, GAATGTCCACCTCGCTTTCCTTTG; λ light chain: sense, CTCTGCAGTGAACATTCTGCAGGGGCCACCGT; antisense, GGGAATTCATGAGGCCAGGGACAGGCCAG; germline κ light chain: sense, CAGCTGACCCAGGACTCTGT; antisense, CTAACACTCTCCCCTGTTGAAGCTCTTTGTGACGGG; J chain: sense, GGGAGTCCTGGCGGTTTTTA; antisense, AGGCATCTGGGGTTAAGGCT; Spi-B: sense, ACCATGCTCGCCCTGGA; antisense, GGCTAGCGAAGTTCTCC; ILT7: sense, GCTACGGCTATGAAAACAACACCCC; antisense, CAGTTGGGGAAGTGTCTGCTGAGAA; Eph-B1: sense, TTTAGGGGAAGGCTGATGAA; antisense, GGGTGCCACTCCAGAATGAT; Granzyme B: sense, ACCTCTCCCAGTGTAAATCT; antisense, GCGGTGGCTTCCTGATACAA; CD19: sense, CGAGTTCTATGAGAACGACTC; antisense, ACTGGAAGTGTCACTGGCATG; and VH-Cμ: sense, GACACGGCCGTGTATTACTG; antisense, ATCCGCGGCCGCGGAATTCTCACAGGAGACGA.
Northern blot analysis
Hybridization of Northern blots (human immune system MTN blot 7768-1 and cancer cell line tissue blot 7757-1; Clontech) was with a 413-bp (C-15), a 515-bp (MAPA), and a 185-bp (BRI 3) DNA fragment corresponding to the original clone labeled with [α-32P]dCTP, using ready-to-go beads (Amersham Pharmacia Biotech, Orsay, France).
Chromosomal localization
Chromosomal localization of MAPA was performed with the Standford G3 RH medium resolution panel (Research Genetics, Huntsville, AL). PCR was as described above using oligonucleotides flanking a known intron (sense, CGAGAGCTTCCAGTGACCTTCT; antisense, CATGTTCTCATAGTCGGGAGTG). Results were scored manually, and analysis was performed with the Rhmapper program (http://shgc-www.stanford.edu).
FACS analysis and immunostaining
HSL2, HSL11, and HSL96 were kindly provided by Dr Karasuyama.26 HSL11 mAb and HSL96 mAb recognize λ-like and V–pre-B proteins, respectively, whereas HSL2 mAb does not bind to either component of the pre-BCR but binds only the completely assembled pre-BCR complex. Anti-granzyme B–phycoerythrin (PE)–labeled antibody (clone CLB-GB11) was from Research Diagnostics (INC, Flanders, NJ). Eph-B1 goat polyclonal antibody generated against an intracytoplasmic peptide of Eph-B1 (M19:sc-9319) and its blocking peptide (sc-9319P) were from Santa Cruz Biotechnology (San Diego, CA). Cytospin slides were first stained with anti–Eph-B1, washed, incubated with secondary biotinylated antigoat antibody (Santa Cruz Biotechnology), and stained by Vectastain ABC reagent (Vectastain ABC peroxidase kit; Vector Laboratories, Burlingame, CA). Control slides were made with unrelated antibody. Blocking peptide was added at the same time as antibody. For intracytoplasmic stainings, cells were permeabilized with saponin (0.1%) and paraformaldehyde (4%).
Results and discussion
Genes identified by subtracted hybridization
To identify genes specifically expressed in PDCs, a subtracted hybridization technique was applied because it combines normalization and subtraction in a single procedure.23,27 Thus, the resultant PCR products should represent restriction fragments of cDNA from PDCs absent, or at least rare, in MDDC used as competitor. PDC cDNA fragments were cloned (n = 736 clones), and clones with more than 300 bp cDNA insert were sequenced (n = 650), representing 80 different genes. Among these, 30% (n = 24 genes) were novel sequences. Three of these—C-15, MAPA, and BRI3—were retained for further analysis. Most of the known genes were not previously associated with PDCs. Surprisingly, approximately 25% of the sequences were for B-cell transcripts, such as immunoglobulin κ and λ (2 of 12 clones rearranged) light chains, the surrogate κ light chain,28 and the pentamer immunoglobulin M (IgM)–joining component J chain (Table1). They also included genes encoding for cell surface molecules (CD62L, CD123/IL-3Rα, immunoglobulinlike transcript ILT7, and receptor tyrosine kinase Eph-B1) and protein kinases involved in signal transduction (such as Tyk2), MyD88 involved in the IL-1 receptor and TLR transduction pathways, enzymes (napsin, cathepsin B, scramblase), transcription factors (Spi-B, EIF3), and genes involved in apoptosis (DR6, granzyme B) and migration (CXCR4) (Table 1). The subtracted hybridization technique was not completely efficient because some genes highly coexpressed in PDCs and MDDCs were not totally subtracted (eg, major histocompatibility complex class 2). In addition, 259 clones for which we were unable to identify an open reading frame (ORF) were excluded from this analysis (Table 1, miscellaneous).
Summary of the genes expressed in PDC after subtraction against MD-DC
| Family of genes . | Name . | Clone . |
|---|---|---|
| Surface molecules | CD62L | 14 |
| CD123/IL-3Rα | 7 | |
| ILT7 | 29 | |
| Eph-B1 | 4 | |
| HLA-DR | 2 | |
| Signaling molecules | Tyk 2 | 3 |
| MyD88 | 2 | |
| Transcription factors | EIF3 | 2 |
| Spi-B | 8 | |
| Apoptosis | DR6 (TNF receptor family) | 2 |
| Enzymes | Granzyme B | 64 |
| Cathepsin B | 2 | |
| Napsin | 40 | |
| Scramblase | 2 | |
| S-adenosyl hydrolase | 6 | |
| Chemokine receptor | CXCR4 | 11 |
| Immunoglobulin-related genes | Immunoglobulin κ chain | 18 |
| Immunoglobulin λ chain | 12 | |
| J chain | 6 | |
| Surrogate κ light chain | 62 | |
| Miscellaneous | Carnitine | 1 |
| Cytochrome c | 2 | |
| Novel | C15 | 84 |
| MAPA | 4 | |
| BRI3 | 4 |
| Family of genes . | Name . | Clone . |
|---|---|---|
| Surface molecules | CD62L | 14 |
| CD123/IL-3Rα | 7 | |
| ILT7 | 29 | |
| Eph-B1 | 4 | |
| HLA-DR | 2 | |
| Signaling molecules | Tyk 2 | 3 |
| MyD88 | 2 | |
| Transcription factors | EIF3 | 2 |
| Spi-B | 8 | |
| Apoptosis | DR6 (TNF receptor family) | 2 |
| Enzymes | Granzyme B | 64 |
| Cathepsin B | 2 | |
| Napsin | 40 | |
| Scramblase | 2 | |
| S-adenosyl hydrolase | 6 | |
| Chemokine receptor | CXCR4 | 11 |
| Immunoglobulin-related genes | Immunoglobulin κ chain | 18 |
| Immunoglobulin λ chain | 12 | |
| J chain | 6 | |
| Surrogate κ light chain | 62 | |
| Miscellaneous | Carnitine | 1 |
| Cytochrome c | 2 | |
| Novel | C15 | 84 |
| MAPA | 4 | |
| BRI3 | 4 |
Lymphoid-related gene expression in PDCs
In a first set of experiments, we selected a panel of genes of immunological interest to verify by RT-PCR whether their expression pattern was restricted to PDCs. The same samples of cDNA were used for PDCs and MDDCs as those used as the starting material before subtraction. We confirmed that subtraction allowed us to amplify transcripts exclusively expressed in PDCs rather than in MDDCs such as CD62L, λ light chain, Spi-B, ILT7, and EphB1 and transcripts preferentially expressed in PDCs such as surrogate κ light chain, J chain, and granzyme B. However, genes such as CD123/IL-3Rα and CXCR4 were not eliminated during subtraction, even though they were apparently equally expressed in the tester and the driver (Figure1). CD62L is expressed at the protein level on blood PDCs, whereas this protein is cleaved on PDC entry through high endothelial venules in secondary lymphoid organs.6 Thus, tonsil PDCs do not express CD62L at the protein level, whereas our data indicated that CD62L mRNA is maintained in PDCs and not expressed in MDDCs. Unlike the surface protein expressed on PDCs but not on MDDCs, CD123/IL-3Rα mRNA was also detected in MDDCs (Figure 1). Of note, the expression of CXCR4 in PDCs is in agreement with their potent ability to respond to its ligand, SDF-1.29 We also confirmed that PDCs expressed strong levels of immunoglobulin transcripts, such as the λ light chain, the J chain, and the surrogate κ light chain previously identified in normal pre-B cells30 (Table 1, Figure 1). That this expression was not caused by the presence of contaminating B cells during preparation was assessed by the absence of transcripts for CD19 and VH-Cμ (Figure 1).
Differential mRNA expression of selected genes between PDCs and MDDCs.
CD62L, CD123/IL-3Rα, CXCR4, λ light chain, surrogate κ light chain, J chain, Spi-B, ILT7, Eph-B1, and granzyme B expression patterns were compared with reverse-transcribed RNA from PDC (a mix of 50% freshly isolated PDCs and 50% of IL-3/CD40L-activated PDCs) and MDDCs (CD40-activated MDDCs) that was kept from the original material used for the subtraction. RT-PCR consisted of 35 cycles of denaturing (30 seconds, 94°C), annealing (30 seconds), and extension (2 minutes, 72°C). Lack of B-cell contamination in PDCs and MDDCs was verified using CD19 and VHconsensus-Cμ primers previously validated on cDNA from tonsil B cells. GAPDH-specific oligonucleotides were used on the same populations.
Differential mRNA expression of selected genes between PDCs and MDDCs.
CD62L, CD123/IL-3Rα, CXCR4, λ light chain, surrogate κ light chain, J chain, Spi-B, ILT7, Eph-B1, and granzyme B expression patterns were compared with reverse-transcribed RNA from PDC (a mix of 50% freshly isolated PDCs and 50% of IL-3/CD40L-activated PDCs) and MDDCs (CD40-activated MDDCs) that was kept from the original material used for the subtraction. RT-PCR consisted of 35 cycles of denaturing (30 seconds, 94°C), annealing (30 seconds), and extension (2 minutes, 72°C). Lack of B-cell contamination in PDCs and MDDCs was verified using CD19 and VHconsensus-Cμ primers previously validated on cDNA from tonsil B cells. GAPDH-specific oligonucleotides were used on the same populations.
Even though we detected the expression of J chain and surrogate κ light chain mRNA in myeloid DCs, the levels were much lower than those observed in PDCs. PDCs were reported to express other lymphoid-related transcripts such as pre-Tα,31 λ-like,7and V–pre-B (N.B.-V., unpublished data, April 1999). Thus, we tested whether PDCs could express the pre-BCR complex at their surfaces. HSL11 mAb and HSL96 mAb recognize λ-like and V–pre-B proteins, respectively, whereas HSL2 mAb does not bind to either component of the pre-BCR but binds only the assembled pre-BCR complex.26 None of these antibodies stained the PDC cell surface (not shown), suggesting that the presence of those lymphoid transcripts may represent residual nontranslated products of their development from a common lymphoid progenitor.
Among the known genes, we focused our attention on Spi-B, ILT7, Eph-B1, and granzyme B, for which we found 8, 29, 4, and 64 clones, respectively (Table 1). Spi-B, previously identified as a lymphoid-restricted transcription factor,32 was uniquely expressed in PDCs (Figure 1). Moreover, unlike pre-Tα, another lymphoid-related transcript that was found to be highly expressed by PDCs,31 Spi-B was not down-regulated during PDC differentiation (not shown). Based on these observations, we analyzed the expression of Spi-B in different human thymic DC populations and proposed that, within the thymus, PDCs may differentiate into mature/interdigitating DCs that retain Spi-B expression but that thymic myeloid DCs do not express Spi-B.7 Overall, the high numbers of immunoglobulin-related genes found clearly indicate a link between human PDCs and the lymphoid lineage. We are investigating the role of Spi-B in the development of PDCs versus that of T, B, and NK cells.
Expression of ILT7, Eph-B1, and granzyme B
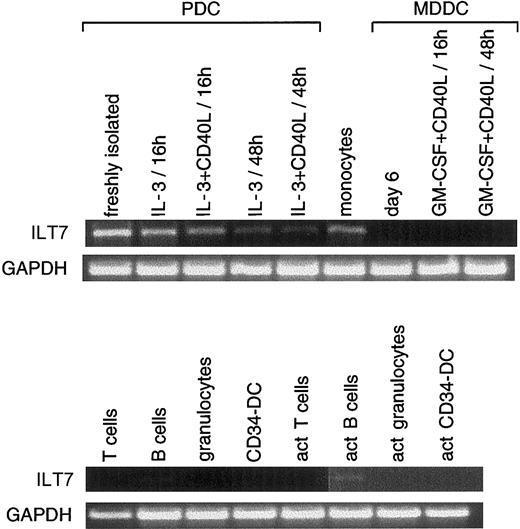
We then tested ILT7, Eph-B1, and granzyme B expression on different cell types and on PDCs and MDDCs at different stages of activation. For RT-PCR analysis presented in Figures2-7, 8 and 10 different donors were used for tonsil PDCs and for MDDCs, respectively. These donors were not the same as those used for subtraction. ILT7 mRNA was highly expressed in PDCs but was rapidly down-regulated by IL-3 with or without CD40 activation (Figure 2). Except for PDCs, monocytes, and activated B lymphocytes, ILT7 mRNA was absent from the other cell types tested. Weak expression was observed on cDNA from thymus (not shown). In our hands, MDDCs (activated or not) did not express detectable levels of ILT7 mRNA in contrast to previous reports.33ILT7 belongs to a family of ILT molecules that are highly homologous and functionally related to a group of NK cell receptors for HLA class 1 molecules. Because no antibodies specific for ILT7 were available, we could not ascertain whether PDC expressed ILT7 at their surfaces. ILT7 has a charged arginine amino acid in the transmembrane domain and a short cytoplasmic domain that is postulated to associate with the FcεRγ chain to transduce stimulatory signals.33 It will thus be important to determine whether ILT7 can transduce an activating stimulus in PDCs.
Restricted expression profile of ILT7.
RT-PCR (35 cycles) was performed using ILT7-specific oligonucleotides (amplifying a product of 973 bp) on first-strand cDNA from freshly isolated PDCs or activated (act) at different time points (from donors different from the one used for the subtraction; see “Materials and methods”); monocytes; MDDCs nonactivated (day 6) or activated (in the presence of CD40L-transfected cell line and GM-CSF) at different time points; on resting or activated T cells, B cells, and granulocytes; and on CD34-derived DCs. ILT7 was expressed mainly in PDCs, monocytes, and activated B cells. Results are representative of 1 of 2 experiments.
Restricted expression profile of ILT7.
RT-PCR (35 cycles) was performed using ILT7-specific oligonucleotides (amplifying a product of 973 bp) on first-strand cDNA from freshly isolated PDCs or activated (act) at different time points (from donors different from the one used for the subtraction; see “Materials and methods”); monocytes; MDDCs nonactivated (day 6) or activated (in the presence of CD40L-transfected cell line and GM-CSF) at different time points; on resting or activated T cells, B cells, and granulocytes; and on CD34-derived DCs. ILT7 was expressed mainly in PDCs, monocytes, and activated B cells. Results are representative of 1 of 2 experiments.
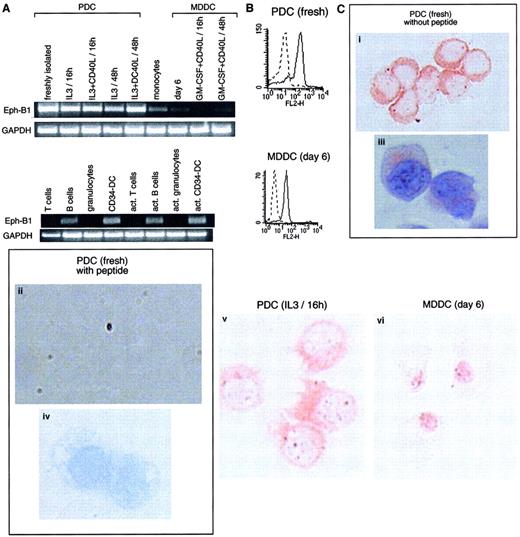
Cells of the immune system express Eph-B1.
(A) RT-PCR (35 cycles) was performed using Eph-B1–specific oligonucleotides (amplifying a product of 580 bp) on the same populations as those detailed in Figure 2. Eph-B1 is highly expressed on PDCs and present in B cells and CD34-derived DCs. Results are representative of 1 of 4 experiments. (B) FACS analysis expression of intracytoplasmic Eph-B1 protein on freshly isolated PDCs and MDDCs. (C) Eph-B1 stainings in PDCs (freshly isolated and cultured overnight in IL-3) and MDDCs on cytospin with (iii,iv) or without (i,ii,v,vi) hematoxylin counterstaining. Specific inhibition in the presence of Eph-B1 peptide (ii,iv) of the labeling using a rabbit polyclonal antibody generated against an intracytoplasmic peptide of Eph-B1 (i,iii). Original magnifications: (i,ii,v,vi) × 400; (iii,iv) × 1000.
Cells of the immune system express Eph-B1.
(A) RT-PCR (35 cycles) was performed using Eph-B1–specific oligonucleotides (amplifying a product of 580 bp) on the same populations as those detailed in Figure 2. Eph-B1 is highly expressed on PDCs and present in B cells and CD34-derived DCs. Results are representative of 1 of 4 experiments. (B) FACS analysis expression of intracytoplasmic Eph-B1 protein on freshly isolated PDCs and MDDCs. (C) Eph-B1 stainings in PDCs (freshly isolated and cultured overnight in IL-3) and MDDCs on cytospin with (iii,iv) or without (i,ii,v,vi) hematoxylin counterstaining. Specific inhibition in the presence of Eph-B1 peptide (ii,iv) of the labeling using a rabbit polyclonal antibody generated against an intracytoplasmic peptide of Eph-B1 (i,iii). Original magnifications: (i,ii,v,vi) × 400; (iii,iv) × 1000.
High levels of granzyme B mRNA in PDC and activated T cells.
(A) RT-PCR (35 cycles) was performed using granzyme B–specific oligonucleotides (amplifying a product of 676 bp). Granzyme B is weakly expressed on monocytes, activated MDDCs, B cells, and granulocytes. Results are representative of 1 of 4 experiments. (B) FACS analysis expression of intracytoplasmic granzyme B protein (solid lines) versus isotype-matched control (dashed line). Mean fluorescence intensity of granzyme B staining minus that obtained with the isotype matched control is indicated. Granzyme B protein was clearly detected on PDCs (activated or not) and was weakly expressed in blood monocytes and MDDCs.
High levels of granzyme B mRNA in PDC and activated T cells.
(A) RT-PCR (35 cycles) was performed using granzyme B–specific oligonucleotides (amplifying a product of 676 bp). Granzyme B is weakly expressed on monocytes, activated MDDCs, B cells, and granulocytes. Results are representative of 1 of 4 experiments. (B) FACS analysis expression of intracytoplasmic granzyme B protein (solid lines) versus isotype-matched control (dashed line). Mean fluorescence intensity of granzyme B staining minus that obtained with the isotype matched control is indicated. Granzyme B protein was clearly detected on PDCs (activated or not) and was weakly expressed in blood monocytes and MDDCs.
Distribution of BRI3 by RT-PCR analysis.
(A) RT-PCR (35 cycles) was performed using BRI3-specific oligonucleotides (amplifying a product of 185 bp). BRI3 was essentially expressed in different types of DCs and granulocytes. Results are representative of 1 of 4 experiments. (B) Northern blots of human tissues and cancer cell lines were analyzed with a probe corresponding to the BRI3 PCR product labeled with [α-32P]dCTP.
Distribution of BRI3 by RT-PCR analysis.
(A) RT-PCR (35 cycles) was performed using BRI3-specific oligonucleotides (amplifying a product of 185 bp). BRI3 was essentially expressed in different types of DCs and granulocytes. Results are representative of 1 of 4 experiments. (B) Northern blots of human tissues and cancer cell lines were analyzed with a probe corresponding to the BRI3 PCR product labeled with [α-32P]dCTP.
Hematopoietic cells express C-15.
(A) Predicted amino acid sequences of human and murine C-15. The predicted signal peptide is underlined and conserved cysteines are boxed. Conserved aa are highlighted in yellow. (B) RT-PCR (35 cycles) was performed using C-15–specific oligonucleotides (amplifying a product of 413 bp). Results are representative of 1 of 4 experiments. (C) Northern blots of human tissues were analyzed with a probe corresponding to the C-15 PCR product labeled with [α-32P]dCTP.
Hematopoietic cells express C-15.
(A) Predicted amino acid sequences of human and murine C-15. The predicted signal peptide is underlined and conserved cysteines are boxed. Conserved aa are highlighted in yellow. (B) RT-PCR (35 cycles) was performed using C-15–specific oligonucleotides (amplifying a product of 413 bp). Results are representative of 1 of 4 experiments. (C) Northern blots of human tissues were analyzed with a probe corresponding to the C-15 PCR product labeled with [α-32P]dCTP.
Hematopoietic cells expressed MAPA.
(A) Predicted amino acid sequence of MAPA. The predicted signal peptide and the transmembrane domain are boxed in dotted lines. The putative PI3 kinase interaction motif (solid black outline) and the ITAM motif (YXXI/L (7/8) YXXI/L) (dashed underlining) are boxed. Conserved cysteines are circled in red. Potential N-linked glycosylation sites (N) are boxed. Conserved aa are highlighted in yellow. (B) Genomic structure of MAPA gene. Schematic diagram illustrating the relative positions of introns and exons. (C) RT-PCR (35 cycles) was performed using MAPA-specific oligonucleotides (amplifying a product of 515 bp). Results are representative of 1 of 4 experiments. (D) Northern blots of human tissues were analyzed with a probe corresponding to the MAPA PCR product labeled with [32P]dCTP.
Hematopoietic cells expressed MAPA.
(A) Predicted amino acid sequence of MAPA. The predicted signal peptide and the transmembrane domain are boxed in dotted lines. The putative PI3 kinase interaction motif (solid black outline) and the ITAM motif (YXXI/L (7/8) YXXI/L) (dashed underlining) are boxed. Conserved cysteines are circled in red. Potential N-linked glycosylation sites (N) are boxed. Conserved aa are highlighted in yellow. (B) Genomic structure of MAPA gene. Schematic diagram illustrating the relative positions of introns and exons. (C) RT-PCR (35 cycles) was performed using MAPA-specific oligonucleotides (amplifying a product of 515 bp). Results are representative of 1 of 4 experiments. (D) Northern blots of human tissues were analyzed with a probe corresponding to the MAPA PCR product labeled with [32P]dCTP.
Receptor tyrosine kinase Eph-B1 mRNA was abundantly detected in PDCs whatever their stage of activation (Figure3A). Eph receptors and their membrane-associated ligands (ephrins) mediate developmental vascular assembly and direct axonal guidance during central nervous system development.34-36 Weak mRNA expression was also observed in monocytes and at much lower intensity in MDDCs. B cells and CD34-derived DCs (activated or not) also expressed Eph-B1 mRNA. In addition, using a polyclonal antibody generated against an intracytoplasmic peptide of Eph-B1, we observed an expression on both permeabilized freshly isolated PDCs and nonactivated MDDCs by FACS analysis (Figure 3B). Eph-B1 protein labeling was further confirmed by immunohistochemistry on cytospins of PDCs and MDDCs and was specifically inhibited in the presence of Eph-B1 peptide (Figure 3C). Eph-B1 expression in PDCs was clearly seen in the cytoplasm of PDCs and in the dendritic pseudopods of activated PDCs. Granular staining within the nucleus was revealed in MDDCs (Figure 3C), but no staining was obtained using unrelated goat IgG (not shown). E-cadherin has been shown to be necessary for surface expression of certain EphR, whereas EphR remained perinuclear in cells that did not express E-cadherin.37 Thus, the differential staining for Eph-B1 in PDCs versus MDDCs might be related to the fact that PDCs, but not MDDCs, express E-cadherin.6 Activation of Eph receptors has been recently reported to regulate integrin-mediated cell adhesion,38 39 and it remains to be determined whether those receptors could be involved in the migratory process of PDCs.
Regarding granzyme B mRNA, comparable high levels of mRNA were expressed in resting and activated PDCs, as observed in the positive control using activated T cells (Figure4A). A faint band was observed in monocytes and activated MDDCs; however, expression was barely detectable in nonactivated MDDCs. In vitro CD34-derived DCs do not express granzyme B mRNA. Weak expression could be seen in resting T cells, B cells, and activated granulocytes (Figure 4A). Furthermore, we showed that freshly isolated PDCs expressed intracytoplasmic granzyme B protein by FACS analysis. Higher expression was seen after overnight activation in IL-3 and CD40 or at a greater level with IL-3 alone that reached the intensity observed on NK cells (not shown). Conversely, only weak intensity of granzyme B staining was always detected in monocytes or in nonactivated or CD40-activated MDDCs (Figure 4B). Granzyme B is usually codeposited with perforin in cytoplasmic granules contained in immune cellular effectors. The granule secretory pathway is one of the mechanisms by which NK cells or cytotoxic T lymphocytes exert their cellular toxicity. We have been unable to show the expression of perforin in PDCs; thus, the physiological role of granzyme B in PDCs remains to be established.
Identification of a putative type 2 cell surface receptor, BRI3, expressed in PDCs
Four sequences were found to be identical to BRI3, a recently identified receptor, formerly known as E25A orIMT2A, that encodes for a protein that shares homology with the mouse E25B and the chicken E3-16 proteins.40The nucleotide sequence of this transcript is 1256 bp long and contains an ORF of 900 nt. pSORT predicted a type 2 transmembrane protein, C-terminal extracellular and N-terminal cytosolic, with an uncleaved signal anchor sequence (from approximately nt 57-74). A mouse homolog of BRI3 expressed in brain was identified in GenBank (accession number AB030199) encoding for a putative 312 aa. Protein alignments indicated that the human and mouse sequences shared approximately 91% homology at the amino acid level (not shown). Levels of expression by RT-PCR of human BRI3 were higher in PDCs than in MDDCs and monocytes, and a faint band could be observed in CD34-derived DCs (Figure 5A). In the different leukocyte populations, essentially only granulocytes expressed this mRNA. Northern blot analysis showed one band at approximately 2.1 kb that was strongly expressed in appendix, peripheral blood leukocytes, bone marrow, fetal liver, and, to a lesser extent, in spleen, lymph nodes, and thymus. In cancer cell lines, weak expression was found in melanoma (G361) and chronic leukemia (MOLT4) cell lines (Figure 5B). The BRI3 gene belongs to a family of type 2 transmembrane proteins containing at least 3 members (BRI1, BRI2, BRI3).40They represent conserved proteins for which no known motifs can be detected but that are rich in tyrosine in the extracellular domain. Mutations in BRI2 have been associated with dementia illnesses similar to Alzheimer disease,41 whereas BRI1 appears to be developmentally regulated in chondrogenesis, osteogenesis, and T-cell development.40,42The role of such genes, and eventually that of BRI3, might be to regulate the expression of molecules that could contribute to the interactions between PDCs and their stromal partners during PDC development.16
Identification of C-15, a putative novel-secreted molecule
C-15 was represented by 84 of 650 sequenced clones. Contiguous sequences were identified in GenBank dbEST by BLAST, leading to a putative complete nucleotide sequence of 728 bp that contained an ORF of 348 nt. pSORT predicted a secreted protein without a transmembrane domain but with a cleavable signal peptide. The deduced polypeptide of human C-15 was composed of 112 aa, including a 23-aa signal peptide, and it contained 16 cysteines, 15 of which were conserved in 115 aa of the predicted mouse homolog. This sequence was identified based on the alignment of more than 80 ESTs from GenBank, corresponding to the mouse ortholog of C-15. Alignments indicated that the mouse protein has 83% homology to the human protein at the amino acid level (Figure6A). Human C-15 was expressed in PDCs and in monocytes but was weakly expressed in MDDCs. Expression of C-15 seemed to be down-regulated on activation, particularly in DCs generated in vitro from CD34 progenitors. The expression pattern of this gene was not restricted to DCs because all leukocyte populations tested expressed C-15 mRNA (Figure 6B). Northern blot analysis showed one major band at 0.7 kb, detected at extremely high levels, and 2 bands with higher molecular weights at approximately 4.4 and 7 kb in different organs of the immune system (spleen, lymph nodes, peripheral blood leukocytes, bone marrow, and, to a lesser extent, in thymus, appendix, and fetal liver) (Figure 6C). The 0.7-kb band corresponds to the predicted cDNA sequence, whereas the 2 other bands could represent nonspliced nuclear pre-mRNA. By homology to the human BAC cloneAC073840, we could localize the C-15 gene to chromosome 4q13-4q21. The gene contains 5 exons, all of which obey the GT-AG U2-type splice rules43 (data not shown). This novel gene encodes a putative cytokine because it can be secreted and is expressed in different hematopoietic cells. It could also be a member of the defensinlike molecules given that defensins are a family of small peptides with 3 or 4 intramolecular cysteine disulfide bonds.44
Identification of a novel member of the leucine-rich repeat family, MAPA
MAPA was represented by 4 sequences. An additional group of 42 sequences was identified within the GenBank database, which allowed an extension of 823 bp 5′ and 117 bp 3′, leading to a nucleotide sequence that contained an ORF of 918 bp. The deduced polypeptide of human MAPA was composed of 305 aa. A mouse homolog was predicted from the ESTsW85307, BG276490, and AI430301 and was completed by analysis of the mouse genomic clone AC073795. Protein alignments indicated that the mouse and human sequences share 41% identity. We were also able to identify 7 ESTs (accession numbers: AW477641, AW668983, AW427493,AW632293, BE752242, BF600409, and BI540463) from Bos taurusthat appear to encode an ortholog of MAPA (Figure7A). The genomic structure of human MAPA could be predicted from the human BAC clone AC008397. The gene has 3 exons, 2 of which are coding exons (Figure 7B). Interestingly, like other members of the LRR family, the extracellular domain and the transmembrane domain are coded by a single exon. The mouse gene has a similar organization. The human gene spans 6464 bp because of the insertion of Alu-type repeats in intron 2, in contrast to the mouse gene, which is 3895 bp in length. By RT-PCR, human MAPA was expressed in PDCs and in MDDCs but down-regulated in CD40-activated MDDCs. Expression of MAPA also seemed to be down-regulated during the activation of DCs generated in vitro from CD34 progenitors. The expression pattern of this gene was not restricted to DCs because granulocytes, monocytes, and B lymphocytes expressed MAPA mRNA (Figure7C). Northern blot analysis showed 2 bands of 2.2 and 2.6 kb in the different organs of the immune system (peripheral blood leukocytes, spleen, bone marrow, and, to a lesser extent, lymph nodes, fetal liver, and appendix but not in thymus) (Figure 7D). In human ESTs that corresponded to MAPA, most were not spliced for intron 1. This difference of 480 bp probably accounts for the 2 observed mRNAs. None of the cancer cell lines tested expressed human MAPA (data not shown). Localization of the human MAPA gene to chromosome 19p13.2 to 19p12, in a region known as the leukocyte-receptor cluster, was identified by homology to human BAC clone AC008397 and was confirmed by radiation hybrid mapping. The predicted protein has 4 leucine-rich repeats often associated with protein–protein interactions, and 2 tyrosine-based motifs, one for interaction with phosphatidylinositol-3 (PI3) kinase (YENM) and an immunoreceptor tyrosine-based activation motif (ITAM; YXXI/L (7/8) YXXI/L).33 45 To our knowledge, this is the first example of a leucine-rich repeat protein with an ITAM motif. Therefore, MAPA may represent a novel type of pattern-recognition receptor involved in the activation of cells of innate and acquired immunity.
In conclusion, using a subtracted hybridization technique, we have identified (1) genes previously characterized but never described as expressed in PDCs (eg, lymphoid-related transcripts, Spi-B, Eph-B1, granzyme B), (2) novel genes that encode for putative proteins for which the functions on PDCs remain to be established (eg, BRI3), and (3) novel sequences for which exact functions are not yet established (eg, C-15, MAPA). This is the first report on a genomic PDC approach. In spite of the technical difficulties of working with human PDCs, we have been able to characterize genes whose expression pattern was restricted mainly to PDCs. We also found genes overexpressed in PDCs (activated or not) and absent in MDDCs (eg, Spi-B). These may provide further understanding of the biological function of PDCs in the host defense system and their origin.
Nucleotide sequences data have been submitted to the EMBL Nucleotide Sequence Database, and accession numbers have been assigned:Homo sapiens mRNA for C15 protein (AJ422147), Mus musculus mRNA for C15 protein (AJ422151), H sapiensmRNA for MAPA protein (AJ422148), M musculus mRNA for MAPA protein (AJ422149), and Bos taurus mRNA for MAPA protein (AJ422150).
We thank the doctors and colleagues from hospitals and clinics in Lyon who provided us with tonsils and umbilical cord blood samples. We also thank Isabelle Durand for FACS sorting and Olivier Clear and Bernadette Michat for maintenance support. We thank Dr Pierre Garrone and Pr Giorgio Trinchieri for their comments on the manuscript and Dr Karasuyama for his generous gift of anti–pre-BCR antibodies.
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-02-0638.
Supported by a grant from Fondation Marcel Mérieux, Lyon, France (N.B.-V.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Francine Brière, Laboratory for Immunological Research, Schering-Plough, 27 Chemin des Peupliers, BP11, 69571 Dardilly, France; e-mail: francine.briere@spcorp.com.



![Fig. 5. Distribution of BRI3 by RT-PCR analysis. / (A) RT-PCR (35 cycles) was performed using BRI3-specific oligonucleotides (amplifying a product of 185 bp). BRI3 was essentially expressed in different types of DCs and granulocytes. Results are representative of 1 of 4 experiments. (B) Northern blots of human tissues and cancer cell lines were analyzed with a probe corresponding to the BRI3 PCR product labeled with [α-32P]dCTP.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/9/10.1182_blood-2002-02-0638/4/m_h82123346005.jpeg?Expires=1763471421&Signature=QeqeK7NSBovl8SbX25zOHWya~uVjvICZlVm24c8UCfnJ2ZYo8uJep8IszBk-sSPlL7SJ1-TXlYHkQVfUWgh9HKWgfyjLf6wr1hYAwaY1M9VkoF923PTWaSEaiE1fnF4FsPsVam~WKK45AHHtkOBanNCncO1PQnAlMqpPB5Ms6atGrIbxT78vKIQkCVMFKJdKy1EcvzzKEolHk4f8dU5ux58EhxEr37kWWOJbeTfmxxChicg8OTGdkHYJTWyx2Un5tgxA~nUwywpHfy8u5zBzcj67lVwlnY6GgNdyKY2F4ZqrDFVv9qdPzeGqSeR7~2geeJloxjn1KblzTCUxfucgXg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Hematopoietic cells express C-15. / (A) Predicted amino acid sequences of human and murine C-15. The predicted signal peptide is underlined and conserved cysteines are boxed. Conserved aa are highlighted in yellow. (B) RT-PCR (35 cycles) was performed using C-15–specific oligonucleotides (amplifying a product of 413 bp). Results are representative of 1 of 4 experiments. (C) Northern blots of human tissues were analyzed with a probe corresponding to the C-15 PCR product labeled with [α-32P]dCTP.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/9/10.1182_blood-2002-02-0638/4/m_h82123346006.jpeg?Expires=1763471421&Signature=bX22t2H8rKani48o4qH8gPNg4LF6rx2aj6WcYSTwZmLK2A0ewLB-7qqOsdRkpKUc7pjd7IWckh-u5IDjo-g~OZAjYXzgSuyUArbwStR7wMD0M~hO8~yKABJA~U9HE1mTHdHikceDiaRG9IVu2H0VDYoD2A7Pz3EgC6hmCXhyvrbuQT6hWL80cmEGahPJ-ynS2mIWP05QBGa8xHmqznoAmpG0wkuLWgnnrr74FwTJmaGYIj1JOKOkgl1s9saEt2PrSDQjYC4DeDIeF8XORKbWVabyL1ONFIZcns~GayRRKVcAbP3VaIRWdqDpHhPOPsGI3H9YBZCMDYPWxfPOp60G3A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Hematopoietic cells expressed MAPA. / (A) Predicted amino acid sequence of MAPA. The predicted signal peptide and the transmembrane domain are boxed in dotted lines. The putative PI3 kinase interaction motif (solid black outline) and the ITAM motif (YXXI/L (7/8) YXXI/L) (dashed underlining) are boxed. Conserved cysteines are circled in red. Potential N-linked glycosylation sites (N) are boxed. Conserved aa are highlighted in yellow. (B) Genomic structure of MAPA gene. Schematic diagram illustrating the relative positions of introns and exons. (C) RT-PCR (35 cycles) was performed using MAPA-specific oligonucleotides (amplifying a product of 515 bp). Results are representative of 1 of 4 experiments. (D) Northern blots of human tissues were analyzed with a probe corresponding to the MAPA PCR product labeled with [32P]dCTP.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/9/10.1182_blood-2002-02-0638/4/m_h82123346007.jpeg?Expires=1763471421&Signature=ufV3NO7K1LvhSX1d1S6KJwK5Pie-q51N3KNAjxtG0BGkdfD~x8Q8XmTXhxfj57SKTQw7lgcSYFPqOppMtgeLyjqmyhlEGen3KfSEQPBp1S5TOO3yuNKoiyTidX2n0EncGgnB~URBukQ~20RwlIU3WQlMSjM5LbSOOBeO8A2itrkYSusHqDfvzCHZblo3SjzV6A9sTak67vnCagZd8VRsLUtHBIpqMIjwcd2tEbCX9ZPSum6Y0WDZjTrdMMAbNQT~is5xjWDAYUMPf5jEQpa02QwEQy~JOk3caPynLVhjZ5aFB6aaaKBZHeqCFsXO9f-bTfdxnPEQrpmlHl~vER7vJQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal