Recently, Roman-Gomez et al1 reported an incidence of p21CIP1 methylation of 41% in 124 patients with acute lymphocytic leukemia (ALL). Most importantly, they observed that p21CIP1 methylation was an independent predictor of poor prognosis both in adults and children with this disease.
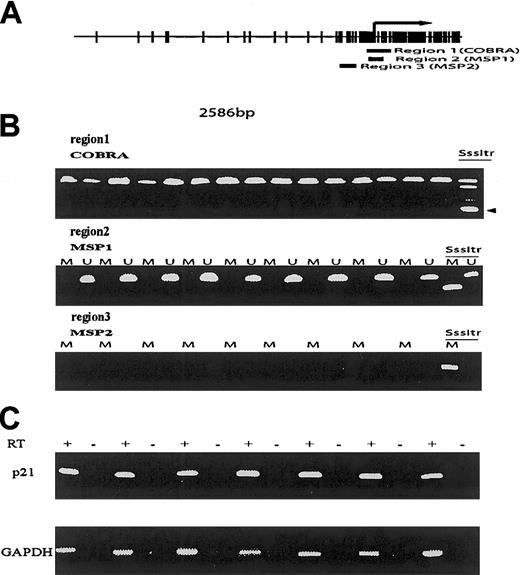
To follow these observations, we have analyzed the methylation status of p21CIP1 in a cohort of patients with ALL who were previously studied for methylation of multiple genes.2,3 We studied a total of 31 patients (19 male; median age, 39 years [range, 7-77 years]; 6 Philadelphia (Ph) chromosome positive). For methylation analysis, we used widely accepted methods based on bisulfite modification of DNA because these assays are sensitive and have a low rate of false positivity. We used 2 different bisulfite polymerase chain reaction (PCR) methods to assess methylation of 3 different regions in or in close proximity to the area studied by Roman-Gomez et al. Primer location is shown in Figure 1. The methylation status of region 1 was analyzed using the combined bisulfite restriction analysis assay (COBRA).4 Regions 2 and 3 were analyzed using the more sensitive methylation-specific PCR assay (MSP).5 A positive control consisting of genomic DNA methylated in vitro by SssI methylase was used in all these assays to verify the validity of the procedures. Using these 2 techniques, DNA methylation was not observed in any of the patients studied in any of the 3 regions analyzed (Figure 1). In contrast, methylation of many other genes, including ER,MDR1, THBS1, THBS2, p15,p73, Myf3, c-abl, andCD10, was observed previously in some of these cases.2 To analyze p21CIP1expression status, we have performed reverse transcriptase (RT)–PCR analysis in 8 cases. In contrast to the results reported by Roman-Gomez et al, all the cases studied had evidence ofp21CIP1 RNA expression (Figure 1).
Methylation and expression analysis ofp21CIP1 in acute lymphocytic leukemia.
(A) Map of the p21CIP1 CpG island. Each vertical mark indicates a CpG pair. The arrow indicates the initiation of the transcription start site. Region 1 indicates de location of COBRA primers. Region 2 of MSP1 primers, and region 3 of the MSP 2 primers. Note that region 3 corresponds to the area studied by Roman-Gomez et al. (B) Examples of COBRA (region 1), MSP1 (region 2), and MSP2 (region 3) assays. SssItr indicates the methylated positive control. The arrow indicates the restricted methylated band. M indicates methylation-specific MSP reactions, U, unmethylated reactions. (C) RT-PCR reactions. + indicates reactions with reverse transcriptase; –, without.
Methylation and expression analysis ofp21CIP1 in acute lymphocytic leukemia.
(A) Map of the p21CIP1 CpG island. Each vertical mark indicates a CpG pair. The arrow indicates the initiation of the transcription start site. Region 1 indicates de location of COBRA primers. Region 2 of MSP1 primers, and region 3 of the MSP 2 primers. Note that region 3 corresponds to the area studied by Roman-Gomez et al. (B) Examples of COBRA (region 1), MSP1 (region 2), and MSP2 (region 3) assays. SssItr indicates the methylated positive control. The arrow indicates the restricted methylated band. M indicates methylation-specific MSP reactions, U, unmethylated reactions. (C) RT-PCR reactions. + indicates reactions with reverse transcriptase; –, without.
Our results are in contrast with those of Roman-Gomez et al. No other investigator has reported evidence ofp21CIP1 methylation in ALL or other leukemias. Indeed, Kikuchi et al6 did not find evidence ofp21CIP1 methylation in 19 neoplastic cell lines, including 6 of hematopoietic origin. It is possible that the patients studied by Roman-Gomez et al are markedly different from patients in the United States, as geographic variation in methylation patterns has recently been reported.7 However, we suggest that the differences observed reside in the technique used by Roman-Gomez et al to assess methylation. In their experiments, they digested DNA with a methylation-sensitive restriction enzyme followed by PCR amplification. Unrestricted PCR-amplified fragments thus represented methylated alleles. Because all cases had evidence of an amplified (methylated) band, samples were considered methylated based on a normalized mean ratio of p21CIP1 to β-actin PCR amplification. This method is prone to false-positive results because unmethylated but incompletely digested DNA may amplify and give a positive reading. The authors did not show controls, nor did they confirm the methylation status of p21CIP1by Southern blot analysis or bisulfite-based methods.8These 2 last methods are considered more reliable.
In summary, we have found no evidence of p21CIP1methylation in our cohort of patients. Other investigators have reported similar findings in an extensive study of neoplastic cell lines.6 We believe that Roman-Gomez et al should reanalyze their samples using either Southern blot or bisulfite-based methods to confirm their important observations.
Hypermethylation of the p21 gene in acute lymphoblastic leukemia
Shen et al analyzed the methylation status of the p21gene in a cohort of ALL patients using methods based on bisulfite modification of DNA. They found lack of p21 methylation in their patients. This result is in disagreement with our recent report.1-1 However, the study by Shen et al contains a number of weak points and several problems.
First, their study included 31 acute lymphocytic leukemia (ALL) patients (probably selected by the availability of preserved DNA or other unknown reasons), which is a very small number of patients when the goal is to reach a definitive conclusion regarding the frequency of this molecular event. By comparison, our study included 124 consecutive ALL patients. It has been suggested that there are important geographic variations in the methylation patterns of several malignancies. For example, in our series of 150 consecutive Spanish ALL patients (manuscript in preparation), we have observed methylation of the E-cadherin and p73 genes in 19% and 18% of patients respectively, whereas E-cadherinmethylation has been observed in 53% of patients in the United States and in 76% of patients in Australia.1,2 By the same token, p73 methylation has been detected in 32% of patients in the United States.1-4-1-5 However, we think that these differences do not depend only on geographic variations but also on the premature release in high-impact journals of a plethora of descriptive methylation studies based on small and very selected groups of patients. These preliminary reports have reached conclusions that have been accepted by the scientific community as absolute truths when they are probably very far from the real incidence of this molecular finding.
Second, Southern blot methylation analysis using a U64 probe (encompassing p21 promoter from nucleotide [nt] – 571 to + 518) revealed the same results obtained after DNA digestion with HpaII followed by polymerase chain reaction (PCR). Moreover, normal bone marrow and peripheral blood showed incomplete methylation patterns, with bands corresponding to both methylated and unmethylated states. Similar results have been obtained analyzing peripheral lymphocytes by Chen et al.1-6Therefore, it is surprising that Shen et al did not find p21methylation when even healthy individuals show a partially methylated status. The p21 gene has a complex promoter and while some of its CpG sites are methylated, other HpaII sites that span the CpG-rich region of this gene (ie, −1004, −62, +15, +93, +288, and +553) are unmethylated in controls and ALL patients. Therefore, a careful choice of the primers for the methylation-specific PCR (MSP) method is necessary in order to obtain reliable results. A bad choice will lead to underestimation of the methylation pattern of the p21 gene.
Third, Shen et al suggest that our method is prone to false-positive results because unmethylated but incompletely digested DNA may amplify and give a false-positive reading. To avoid this problem, we had already examined 30 healthy individuals (a similar number of controls to the patients reported by Shen et al) and did not find hypermethylation in any of them. Moreover, we have extended ourp21 methylation analysis to other hematologic malignancies (manuscript submitted) and no p21 hypermethylation was observed among 179 chronic-phase chronic acute myeloid leukemia (CML) patients, 55 acute stage CML patients, or 72 acute myelogeneous leukemia (AML) patients. Therefore, it is improbable that false-positive results should appear only in ALL patients and not in more than 300 patients affected by other types of leukemias. It is more likely that this result seems to indicate some kind of technical problem in the method used by Shen et al. In fact, it is well recognized that study of the methylation status of theCDKI genes belonging to the KIP family is sometimes difficult and needs very sensitive technology. For example, it is necessary to perform the amplification with enzymes that permit raising the annealing temperature up from 68°C to 70°C with an extension temperature more than 74°C. Thus, special DNA polymerases must be used (ie, Pfu DNA polymerase) instead of conventionalTaq DNA polymerase. Another sensitive method employed for amplification of KIP family is nested PCR (ie,p57KIP2).
Finally, Shen et al studied p21 expression in only 8 patients using reverse transcriptase (RT)–PCR. They found evidence of p21 mRNA in all the cases. This result is not surprising because, in any case, most of ALL patients show p21expression regardless of the methylation status of the gene. However, hypermethylated patients disclosed decreased levels ofp21 transcripts as assessed by a semiquantitative RT-PCR method. Therefore, a quantitative method is mandatory in these circumstances. The qualitative RT-PCR performed by Shen et al is unsatisfactory for this purpose. Moreover, in addition to ALL, decreased levels of p21 have been recently reported in other hematologic tumors.1-7-1-8
For all of these reasons, the title “Lack ofp21CIP1 DNA methylation in acute lymphoblastic leukemia” is both adventurous and premature. We recommend that Shen et al revise some aspects of their technology and patient selection.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal