Abstract
The expression of cytotoxic T-lymphocyte antigen–4 (CTLA-4) molecule in human normal and neoplastic hematopoietic cells, both on the cell membrane and in the intracellular compartment, was evaluated. Flow cytometric analysis carried out with a panel of anti–CTLA-4 human single-chain fragment of variable domain (scFv) antibodies revealed that CTLA-4 was not expressed on the surface, whereas it was highly expressed within the cytoplasm, in freshly isolated peripheral blood mononuclear cells (PBMCs), T cells, B cells, CD34+ stem cells, and granulocytes. Various treatments with agents able to specifically activate each cell type induced CTLA-4 expression on the surface of these cells. Similarly, increased CTLA-4 expression was observed in different hematopoietic cell lines although they also expressed surface CTLA-4, at different degrees of intensity, before activation. Surprisingly, CTLA-4 RNA transcripts were detectable in such cell lines only after nested polymerase chain reaction (PCR) specific for CTLA-4 extracellular domain, suggesting a very fast CTLA-4 RNA processing accompanied by prolonged CTLA-4 protein accumulation. We further demonstrated surface expression of CTLA-4 in a variety of acute and chronic myeloid leukemias (AMLs and CMLs) and B- and T-lymphoid leukemias, either adult or pediatric. CTLA-4 was expressed in 25% to 85% of AMLs and CMLs depending on the leukemia subtype and the epitope analyzed, whereas in acute B- and T-leukemias CTLA-4 expression was mainly cytoplasmic. Chronic B leukemias appeared to express CTLA-4, both on the surface and in cytoplasm, whereas few cases tested of chronic T leukemias were negative. Two anti–CTLA-4 immunotoxins (scFvs-saporin) induced in vitro apoptosis of neoplastic cells from a representative AML, suggesting a novel immunotherapeutic approach to AML based on CTLA-4 targeting.
Introduction
Cytotoxic T-lymphocyte antigen–4 (CTLA-4) (CD152) is a T-cell costimulatory receptor that functions as a negative regulator of T-cell activation,1,2 in contrast to CD28, which delivers positive signals. The engagement by its ligands CD80 and CD86 expressed on antigen-presenting cells (APCs) inhibits the induction of interleukin 2 receptor (IL-2R) α-chain and CD69 expression, the production of IL-2, interferon-γ (IFN-γ), IL-4, and other cytokines.3 CTLA-4 is expressed mainly on activated CD4+ and CD8+ T cells where it is predominantly localized in intracellular compartments, while its membrane expression is maximal after 48- to 72-hour stimulation.2 Recently, it was shown that a subpopulation of peripheral human CD4+CD25+ T cells, with regulatory function, constitutively expresses intracellular CTLA-4, which can be induced on the surface upon activation.4 After T-cell receptor (TCR)–mediated stimulation, these regulatory T cells inhibit the activation of other CD4+ and CD8+ T cells by direct cell-cell contact.5 Relatively high levels of CTLA-4 have also been found expressed on activated B cells,6 including malignant B cells from non-Hodgkin lymphomas,7 placental fibroblasts,8 cultured muscle cells,9 and monocytes.10
Because of its dominant role in modulating T-cell activation, CTLA-4 has received considerable attention as a therapeutic agent in a variety of in vivo immune responses, including induction of transplantation tolerance.11 The chimeric CTLA-4–immunoglobulin (CTLA-4–Ig) protein,12 interferes with T-cell/APC interaction both in vitro and in vivo, preventing xenograft rejection13 and prolonging allograft survival.14 In contrast to strategies that interfere with the costimulatory CD28-CD80/CD86 pathway, the availability of reagents that completely abolish T-cell activation by targeting the CTLA-4 molecule might have important clinical implications. In this regard, we previously described the generation of human recombinant anti–CTLA-4 single-chain fragments of variable domain (scFvs)15 and their use as therapeutic reagents in transplantation, either as unconjugated antibodies16 or conjugated with saporin,17 a type-1 ribosome-inactivating protein (RIP).18 19
Since most of the available data on CTLA-4 expression are limited to human T cells, the goal of the present study was to analyze the distribution pattern of CTLA-4 in a panel of human normal and neoplastic hematopoietic cells (cell lines and leukemic myeloid and lymphoblastic cells). The analysis was performed by flow cytometry with the parallel use of 5 different human anti–CTLA-4 scFv antibodies to a commercial anti–CTLA-4 murine monoclonal antibody (mAb) and by reverse transcriptase–polymerase chain reaction (RT-PCR). In addition, we investigated the ability of 2 immunotoxins, consisting of anti–CTLA-4 scFv antibodies and saporin, to induce apoptosis of CTLA-4–expressing neoplastic cells from an acute myeloid leukemia.
Patients, materials, and methods
Monoclonal antibodies and immunotoxins
Recombinant anti–CTLA-4 scFv antibodies, namely scFvs 3, 40, and 83, were obtained by selecting the human Nissim scFv phage library, as described previously,15 whereas scFvs 55 and 67 were obtained by selecting the high-affinity human ETH-2 phage library20 with purified CTLA-4–Ig fusion protein. Specificity of scFvs was checked by enzyme-linked immunosorbent assay (ELISA), Western blot, and immunofluorescence. All the scFv antibodies showed different CDR3 regions upon DNA sequencing of the heavy chain variable (VH) regions. They were conjugated to fluorescein isothiocyanate (FITC) and used for direct immunofluorescence together with the commercially available FITC-conjugated anti–CTLA-4 BN13 mAb (Coulter Immunotech, Birmingham, United Kingdom) as positive control. The FITC-conjugated anti–bovine serum albumin (anti-BSA) scFv 26 (selected from the Nissim library, Medical Research Council Centre, Cambridge, United Kingdom) and a FITC-conjugated mouse IgG1 mAb (isotypic control; Coulter Immunotech) were used as negative controls.
Two scFvs, 67 and 83, were conjugated to the type-1 ribosome-inactivating protein (RIP) saporin-S6, and the resulting immunotoxins were tested for reactivity with activated T cells and for toxicity with hematopoietic precursors according to standard protocols.17
Cells and culture conditions
Peripheral blood mononuclear cells (PBMCs) were isolated from the buffy coats of 10 healthy donors by density gradient centrifugation over Ficoll/Biocoll (Biochrom, Berlin, Germany). Purification of T cells, B cells, stem cells, and monocytes was achieved from PBMCs by the use of magnetic cell sorting (MACS) CD3, CD19, CD34, and CD14 microbeads respectively, followed by magnetic separation columns (Miltenyi Biotec, Bergisch Gladbach, Germany). Purity was always greater than 95%, checked by direct immunofluorescence with FITC anti-CD3/CD2 mAbs (Coulter Immunotech) for T cells, FITC anti-CD19/CD20 mAbs (Coulter Immunotech) for B cells, FITC anti-CD34 HPCA-2 mAb (Becton Dickinson, San Jose, CA) for stem cells, and FITC anti-CD1a mAb (Pharmingen, San Diego, CA) for monocytes. Approval was obtained from the institutional review board of the Department of Internal Medicine, University of Genoa, and Department of Pediatrics, University of Padua, Italy for these studies. Informed consent was provided according to the Declaration of Helsinki.
PBMCs, T cells, B cells, and cell lines were activated by culturing them in RPMI 1640 (Biochrom) supplemented with 10% fetal calf serum (FCS) (Biochrom), antibiotics, and 2 mMl-glutamine in the presence of phorbol ester (PMA; Sigma, Milan, Italy) at 5 ng/mL and/or phytohemagglutinin (PHA; Life Technologies, Milan, Italy) at a final concentration of 2 μg/mL for 48 hours at 37°C. For B cells, other stimuli such as lipopolysaccharide (LPS, 1 μg/mL; Sigma) and anti-CD40 mAb (clone EA-5, 1 μg/mL; Biosource, Camarillo, CA) were used. Activation of stem cells derived from PBMCs of adult normal healthy donors, treated subcutaneously with 10 μg/kg/d glycosylated granulocyte colony-stimulating factor (G-CSF, Lenograstin; Rhone-Poulenc Rorer, Milan, Italy) for 5 to 6 days, was obtained by culturing CD34+ purified cells for 7 days in RPMI 1640 supplemented with 20% FCS and 50 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; Walter Occhiena, Turin, Italy). Activation of monocytes was performed by culturing 1 × 106 purified CD14+ cells for 5 to 7 days in 1 mL complete medium supplemented with 800 U/mL IL-4 (Schering Plough, Kenilworth, NJ) and 50 ng/mL GM-CSF as previously described.21
The following human transformed cell lines were used: CEM, Jurkat, and Molt-4, derived from T-lymphoblastic cells; Daudi and Raji, derived from a Burkitt lymphoma; HOM-2, an Epstein-Barr virus (EBV) B-lymphoblastoid cell line; HL60 and KG1a, 2 myeloid leukemia–derived cell lines; and K562, an erythroleukemia cell line. The murine cell line J558L is a myeloma transfected with human CTLA-4–Ig cDNA22 and was used as positive control.
Patients and isolation of leukemic cells
A total of 100 patients, either adult or pediatric, affected by different hematological malignancies have been studied, including 31 patients with acute myeloid leukemia (AML), 4 patients with chronic myeloid leukemia (CML), 53 patients with acute B-lymphocytic leukemia (B-ALL), 6 patients with acute T-lymphocytic leukemia (T-ALL), 4 patients with chronic B-lymphocytic leukemia (B-CLL) and 2 patients with chronic T-lymphocytic leukemia (T-CLL) large granular lymphocytic leukemia (LGL). The diagnosis was established according to the standard criteria of each leukemia23 at the Department of Internal Medicine, University of Genoa (Italy) for the adult leukemias and at the Department of Pediatrics, University of Padua Medical School (Italy) as far as the pediatric leukemias were concerned. None of the patients received therapy at the time of the study. Leukemic cells were obtained by density gradient centrifugation of peripheral blood or bone marrow aspirates and were freshly used for testing or stored in liquid nitrogen. In the latter case, cells were thawed and cultured at a final concentration of 5 × 105cells per milliliter in RPMI 1640 medium with l-glutamine and 20% FCS, overnight at 37°C, before testing. For kinetics studies of CTLA-4 and CD34 expression, AML and CML leukemic cells were cultured in the presence of GM-CSF for 7 days.
Immunofluorescence and flow cytometry
A direct immunofluorescence was performed for analyzing surface and cytoplasmic expression of CTLA-4 in normal and neoplastic cells. Briefly, a pellet of 4 × 105 cells, with or without fixation in 2% paraformaldehyde followed by permeabilization with 0.5% saponin, was incubated for 30 minutes at room temperature with different FITC anti–CTLA-4 scFvs (nos. 3, 40, 55, 67, 83), the FITC anti–CTLA-4 BN13 mAb, or the FITC-negative controls. CTLA-4 expression on CD34+ stem cells and on AML and CML leukemic cells was confirmed by double staining with FITC-83 scFv in combination with phycoerythrin (PE)–conjugated anti-CD33 mAb (Coulter Immunotech). The fluorescence intensity was measured by flow cytometry (EPICS Elite; Coulter Electronics, Hialeah, FL); at least 15 000 cells per sample were counted.
Analysis of CTLA-4 expression in granulocytes was performed with appropriate gating. Flow cytometry evaluation of CTLA-4 antibody reactivity with leukemic cells from pediatric leukemias was always carried out with the use of simultaneous 4-color staining for CTLA-4, CD7, CD19, and CD45 molecules; CTLA-4 expression was evaluated only in the leukemic cells identified by immunological gate (atypical CD45 expression and/or CD19). The intracytoplasmatic evaluation was performed by 4-color analysis (CD19, CD7, CD45 for surface and CTLA-4 for intracytoplasmatic expression) with the use of Fix and Perm permeabilization solution (Walter Occhiena; Caltag, Turin, Italy)
Apoptosis
In a series of 3 independent experiments, neoplastic cells derived from an adult AML patient were incubated with different concentrations of 67-saporin or 83-saporin immunotoxins, or with a mix of saporin and scFvs 67 or 83, in complete RPMI 1640 medium for 72 hours. Viable and dead cells were evaluated by double staining with FITC-annexin V and propidium iodide (kit from Bender Medsystem, Vienna, Austria), following the manufacturer's instructions. Analysis was performed by flow cytometry, with calculation of both the percentage of apoptotic (annexin V+/propidium iodide−) and necrotic (annexin V+/propidium iodide+) cells.
Cytospin and immunohistochemistry of AML cells
Cytospin preparations of CD34+ cells and neoplastic cells from an AML patient were air-dried overnight and fixed for 10 minutes in acetone at room temperature. The slides with AML cells were incubated with biotin-conjugated anti–CTLA-4 67 scFv for 30 minutes at room temperature and, after washing in phosphate-buffered saline (PBS), alkaline phosphatase (AP)–labeled streptavidin (Dako, Copenhagen, Denmark) was applied for 20 minutes. The slides with CD34+ cells were incubated with the BN13 mAb followed by incubation with alkaline phosphatase (AP)–labeled antimouse immunoglobulin antibody (Dako). Color development was detected with phosphatase substrate (Dako). Slides were counterstained with hematoxylin-eosin.
RNA preparation and cDNA synthesis
Total cellular RNA and cDNA were prepared as previously described.17
Polymerase chain reaction
PCRs were carried out in 50 μL volume, with 1/10 of the RT mixture (500 ng RNA). Specific amplifications of both CTLA-4 full-length and extracellular domain transcripts were performed on each cDNA sample with the use of the set of primers previously described.24 25 Both PCR reactions were run following the same cycle profile (denaturation at 94°C for 1 minute and elongation at 72°C for 1 minute for a total of 35 cycles), except for the annealing temperature (AT). Briefly, for CTLA-4 full-length amplification, the AT was at 60°C for 1 minute, while for CTLA-4 extracellular domain the AT was at 58°C for 1 minute. Both reactions were initially hot-started (94°C for 3 minutes) and were terminally extended at 72°C for 5 minutes. As internal control, β-actin gene amplification (β-actin forward primer, 5′-ATGGATGATGATATCGCCGCG-3′; β-actin reverse primer, 5′-CGGTTGGCCTTGGGGTTCAG-3′) was carried out for each cDNA sample with the use of 65°C of AT and the same reaction conditions as for CTLA-4 amplification. The obtained PCR products were analyzed by electrophoresis on a 2% agarose gel. The size and specificity of CTLA-4 PCR products were confirmed after direct sequencing analysis in both directions by means of an ABI-PRISM 377 Perkin-Elmer DNA Sequencer (Foster City, CA)
Nested PCR
A second round of PCR was performed amplifying 1 μL CTLA-4 full-length first PCR products with CTLA-4 extracellular domain primers as inner primers. Twenty-five more cycles were carried on at 58°C AT, as previously described. Negative as well as positive results were confirmed by repeating the assay with a second aliquot of each original total RNA sample. Reproducibility was almost 100% in negative cases and greater than 90% in positive cases. Adequate precautions to prevent cross-contamination and negative control reactions were performed routinely. The obtained nested-PCR products were analyzed and sequenced as described above.
Western blot analysis
Cells from different cell lines, either untreated or treated with PMA/PHA or PMA for 48 hours, were washed twice with PBS and then resuspended in lysis buffer (10 mM Tris-HCL [tris(hydroxymethyl)aminomethane], pH 7.5; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 0.1% sodium dodecyl sulfate [SDS], 1% Nonidet P-40) for 30 minutes on ice. Cell debris was removed by centrifugation, and equal amounts of cells (8 × 105) were loaded on a nonreducing 12% (wt/vol) SDS–polyacrylamide gel, electrophoresed at 100 V for 2 hours, and blotted onto nitrocellulose membrane by electrotransfer at 300 mA for 3.5 hours in 190 mM glycine and 25 mM Tris, pH 8.3. After blocking, the membrane was incubated overnight at 4°C with scFv 83 mixed to the mAb 9E10, which recognizes the C-terminal c-myc peptide tag linked to the scFv. After washing, horseradish peroxidase (HRP)–conjugated goat antimouse IgG was applied 1 hour at room temperature. After washing, the membrane was treated with electrochemiluminescence (ECL) reagents (Amersham Life Science, Buckinghamshire, United Kingdom) and exposed to Hyperfilm ECL (Amersham Life Science).
Results
CTLA-4 expression in human hematopoietic normal cells and cell lines
We have shown recently that CTLA-4 receptor is expressed on the surface of human cells of the lymphoid, myeloid, and erythroid lineages, on which it represents a target molecule suitable for apoptosis induction by immunotoxins.17 To further investigate the distribution pattern of surface and intracellular CTLA-4 molecule, we tested its expression levels in a variety of hematopoietic normal cells and cell lines by immunofluorescence using a panel of new and previously produced anti–CTLA-4 scFv antibodies. These scFvs, namely nos. 3, 40, 55, 67, and 83, were developed by selection of 2 different phage libraries and recognize different epitopes of CTLA-4 extracellular domain (data not shown). The commercially available anti–CTLA-4 BN13 mAb and the CTLA-4–transfected J558L murine myeloma were used as positive controls. Cells were analyzed by flow cytometry, with anti–CTLA-4 FITC-scFvs and FITC-BN13 mAb or anti-BSA FITC-scFv 26 and mouse IgG1 mAb as negative controls, either in resting conditions or after activation with different stimuli specific for each cell type. In particular, with the exception of monocytes, we detected a negative expression profile of surface CTLA-4 on resting cells, since the molecule was not constitutively expressed on freshly isolated PBMCs, T cells, B cells, granulocytes, or CD34+ stem cells, but it was induced upon 48-hour activation with PMA/PHA or PMA, GM-CSF (Table 1). In contrast, high CTLA-4 expression was detected in the cytoplasm of all these cells, except granulocytes, even in resting conditions. Weak CTLA-4 expression was observed on the cell surface of fresh monocytes, whereas the intracellular expression was higher and present in all the cells, differing from recent studies reporting CTLA-4 expression in only 20% of monocytes.10
Summary of CTLA-4 expression in human normal hematopoietic cells
| Hematopoietic normal cells . | Surface . | Cytoplasm . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| scFv no. . | mAbs . | scFv no. . | mAbs . | |||||||||||||
| 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | |
| PBMCs | − | − | − | − | − | − | − | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| PBMCs plus PMA/PHA | + | ++ | ++ | ++ | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| T cells | − | − | − | − | − | − | − | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| T cells plus PMA/PHA | + | + | ++ | ++ | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| B cells | − | − | − | − | − | − | − | − | + | ++ | + | ++ | ++ | − | ++ | − |
| B cells plus PMA | ± | + | + | + | + | − | + | − | + | ++ | ++ | ++ | ++ | − | ++ | − |
| Granulocytes | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Granulocytes plus G-CSF | + | + | + | + | + | − | + | − | + | + | + | + | + | − | + | − |
| Stem cells (CD34+) | − | − | − | − | − | − | − | − | + | ++ | ++ | + | + | − | ++ | − |
| Stem cells (CD34+) plus GM-CSF | + | + | + | + | ++ | − | ++ | − | + | ++ | ++ | ++ | ++ | − | ++ | − |
| Monocytes (CD14+) | − | − | ± | ± | ± | − | ± | − | + | + | + | + | + | − | + | − |
| Monocytes (CD14+) plus GM-CSF | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Hematopoietic normal cells . | Surface . | Cytoplasm . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| scFv no. . | mAbs . | scFv no. . | mAbs . | |||||||||||||
| 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | |
| PBMCs | − | − | − | − | − | − | − | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| PBMCs plus PMA/PHA | + | ++ | ++ | ++ | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| T cells | − | − | − | − | − | − | − | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| T cells plus PMA/PHA | + | + | ++ | ++ | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| B cells | − | − | − | − | − | − | − | − | + | ++ | + | ++ | ++ | − | ++ | − |
| B cells plus PMA | ± | + | + | + | + | − | + | − | + | ++ | ++ | ++ | ++ | − | ++ | − |
| Granulocytes | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Granulocytes plus G-CSF | + | + | + | + | + | − | + | − | + | + | + | + | + | − | + | − |
| Stem cells (CD34+) | − | − | − | − | − | − | − | − | + | ++ | ++ | + | + | − | ++ | − |
| Stem cells (CD34+) plus GM-CSF | + | + | + | + | ++ | − | ++ | − | + | ++ | ++ | ++ | ++ | − | ++ | − |
| Monocytes (CD14+) | − | − | ± | ± | ± | − | ± | − | + | + | + | + | + | − | + | − |
| Monocytes (CD14+) plus GM-CSF | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
Reactivity of different peripheral blood cells with a panel of FITC-conjugated anti-CTLA-4 scFvs and the commercial BN13 mAb by flow cytometry. Fluorescence intensity was scored as follows: −, negative; ±, weak; and + to ++, positive, with grading from 1 log to more than 1 log of histogram shift relative to the negative control. The FITC-conjugated anti-BSA scFv 26 and a FITC-conjugated mouse IgG1 mAb were used as negative controls for anti-CTLA-4 scFvs and BN13 mAb, respectively. All cell samples were tested either resting or in vitro activated, except granulocytes that were derived from in vivo G-CSF–treated donors, as described in “Patients, materials and methods.”
After B-cell activation, 52% of B cells were induced to express CTLA-4 on the surface by PMA, whereas other stimuli, such as anti-CD40 mAb and LPS, induced CTLA-4 expression in only 20% of B cells. In summary, the CTLA-4 expression pattern observed in fresh hematopoietic cells is consistent with the known T-cell surface and intracellular distribution of CTLA-4, which is primarly localized in vesicles of the Golgi compartment.26
CTLA-4 expression in leukemic cell lines appeared stronger on the surface of KG1a, Raji, and Jurkat cells, as detected by most of the scFvs and BN13 mAb, whereas it was weaker on CEM (except with scFv 83) and Daudi cells (Table 2). The expression was highlighted after stimulation with PMA/PHA or PMA. but in CEM (except with scFv 83) and Daudi cells, it remained at low levels. Strong cytoplasmic reactivity with both the anti–CTLA-4 scFvs and BIN13 mAb was observed in all the cell lines tested either resting or activated. Of all the scFv antibodies, 4 showed a similar reactivity pattern and 1, scFv 3, a consistently weaker or absent binding to surface, but not cytoplasmic, CTLA-4, suggesting that it might recognize an internal molecule epitope that requires permeabilization before being detected. The anti-BSA scFv 26 and the IgG1 isotype control mAb were consistently negative with either normal or neoplastic cells both at the surface and the cytoplasmic level.
Summary of CTLA-4 expression in human hematopoietic cell lines
| Hematopoietic normal cells . | Surface . | Cytoplasm . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| scFv no. . | mAbs . | scFv no. . | mAbs . | |||||||||||||
| 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | |
| Jurkat | − | + | ++ | + | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Jurkat plus PMA/PHA | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| CEM | − | ± | ± | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | + | − |
| CEM plus PMA/PHA | − | ± | ± | ± | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Molt-4 | − | ± | + | + | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Molt-4 plus PMA/PHA | − | + | ++ | ++ | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Raji | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Raji plus PMA | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Daudi | − | ± | ± | ± | ± | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Daudi plus PMA | − | ± | ± | ± | ± | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HOM-2 | − | + | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HOM-2 plus PMA | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HL-60 | − | ± | ++ | ± | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HL-60 plus PMA | + | + | ++ | + | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| KG1a | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| KG1 plus PMA | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| K562 | + | + | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| K562 plus PMA | + | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| J558L | ± | ± | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| J558L plus PMA | ± | ± | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Hematopoietic normal cells . | Surface . | Cytoplasm . | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| scFv no. . | mAbs . | scFv no. . | mAbs . | |||||||||||||
| 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | 3 . | 40 . | 55 . | 67 . | 83 . | 26 . | BN13 . | IgG1 . | |
| Jurkat | − | + | ++ | + | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Jurkat plus PMA/PHA | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| CEM | − | ± | ± | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | + | − |
| CEM plus PMA/PHA | − | ± | ± | ± | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Molt-4 | − | ± | + | + | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Molt-4 plus PMA/PHA | − | + | ++ | ++ | ++ | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Raji | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Raji plus PMA | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Daudi | − | ± | ± | ± | ± | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| Daudi plus PMA | − | ± | ± | ± | ± | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HOM-2 | − | + | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HOM-2 plus PMA | ± | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HL-60 | − | ± | ++ | ± | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| HL-60 plus PMA | + | + | ++ | + | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| KG1a | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| KG1 plus PMA | ++ | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| K562 | + | + | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| K562 plus PMA | + | ++ | ++ | ++ | ++ | − | ++ | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| J558L | ± | ± | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
| J558L plus PMA | ± | ± | + | + | + | − | + | − | ++ | ++ | ++ | ++ | ++ | − | ++ | − |
Reactivity of different cell lines with a panel of FITC-conjugated anti–CTLA-4 scFvs and the commercial BN13 mAb by flow cytometry. All cell lines were tested either resting or in vitro activated as described in “Patients, materials, and methods.” See Table 1 footnote for an explanation of fluoresence intensity scoring and a description of negative controls.
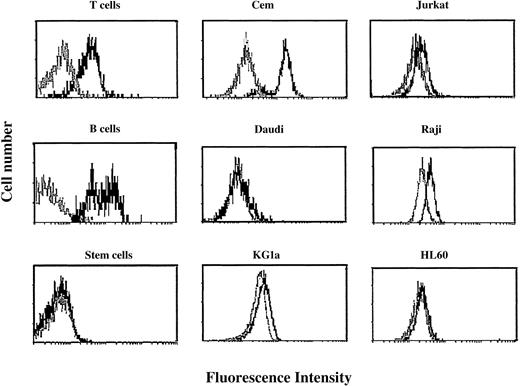
The flow cytometry profiles of surface CTLA-4 expression in 6 cell lines either before or after stimulation were compared with those of their normal counterparts (Figure1).
Flow cytometric profiles of surface CTLA-4 expression in peripheral blood T cells, B cells, stem cells, and cell lines.
Cells were tested before (gray histograms) and after (black histograms) activation for 48 hours as described in “Patients, materials, and methods.” Resting and activated cells were stained with FITC-conjugated anti–CTLA-4 scFv 83 and analyzed by flow cytometry. Results are expressed as fluorescence intensity.
Flow cytometric profiles of surface CTLA-4 expression in peripheral blood T cells, B cells, stem cells, and cell lines.
Cells were tested before (gray histograms) and after (black histograms) activation for 48 hours as described in “Patients, materials, and methods.” Resting and activated cells were stained with FITC-conjugated anti–CTLA-4 scFv 83 and analyzed by flow cytometry. Results are expressed as fluorescence intensity.
CTLA-4 expression in human acute and chronic leukemias
CTLA-4 expression was analyzed by flow cytometry with the use of the selected scFv antibodies, in a panel of 100 adult and pediatric hematologic malignancies, including AML, CML, B-ALL, T-ALL, B-CLL, and T-CLL (LGL), all containing leukemic cells at high frequency (greater than 75%) in peripheral blood or bone marrow aspirate samples. Few differences were observed in the surface reactivity of AML leukemias with the different antibodies, since the epitopes recognized by scFvs 67 and 83 (67- and 83-epitopes) were detected at high frequency (23 of 27 and 23 of 31, respectively) and intensity of staining; the same was true for the epitopes recognized by scFvs 40 and 55 (40- and 55-epitopes; 22 of 31 and 22 of 27, respectively), whereas the epitope recognized by scFv 3 (3-epitope) was detected at a lower frequency (16 of 31) (Table3). It is noteworthy that these AML samples belonged to different subtypes (M0-M5) according to the French-American-British (FAB) classification23 and were indifferently expressing the myeloid CD34 marker (data not shown). All the CTLA-4 epitopes consistently showed higher expression in the cytoplasm of the AML samples analyzed (Table 3).
Summary of CTLA-4 expression in human acute and chronic leukemias
| Leukemia . | ScFv no. (total no.) . | mAb BN13 . | ||||
|---|---|---|---|---|---|---|
| 3 . | 40 . | 55 . | 67 . | 83 . | ||
| AML | ||||||
| Surface | 16 (31) | 22 (31) | 22 (27) | 23 (27) | 23 (31) | 23 (27) |
| Cytoplasm | 30 (31) | 30 (31) | 25 (27) | 25 (27) | 31 (31) | 26 (27) |
| CML | ||||||
| Surface | 1 (4) | 3 (4) | 3 (4) | 4 (4) | 4 (4) | 4 (4) |
| Cytoplasm | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) |
| B-ALL | ||||||
| Surface | 2 (53) | 1 (53) | 1 (12) | 1 (15) | 8 (53) | 2 (10) |
| Cytoplasm | 24 (35) | 7 (35) | 8 (12) | 10 (15) | 34 (35) | 9 (10) |
| T-ALL | ||||||
| Surface | 1 (6) | 1 (6) | 1 (6) | 1 (6) | 1 (6) | 1 (2) |
| Cytoplasm | 2 (3) | 3 (3) | 2 (2) | 2 (2) | 3 (3) | 2 (2) |
| B-CLL | ||||||
| Surface | 3 (4) | 3 (4) | 3 (4) | 3 (4) | 3 (4) | 3 (4) |
| Cytoplasm | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) |
| T-CLL (LGL) | ||||||
| Surface | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) |
| Cytoplasm | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) |
| Leukemia . | ScFv no. (total no.) . | mAb BN13 . | ||||
|---|---|---|---|---|---|---|
| 3 . | 40 . | 55 . | 67 . | 83 . | ||
| AML | ||||||
| Surface | 16 (31) | 22 (31) | 22 (27) | 23 (27) | 23 (31) | 23 (27) |
| Cytoplasm | 30 (31) | 30 (31) | 25 (27) | 25 (27) | 31 (31) | 26 (27) |
| CML | ||||||
| Surface | 1 (4) | 3 (4) | 3 (4) | 4 (4) | 4 (4) | 4 (4) |
| Cytoplasm | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) |
| B-ALL | ||||||
| Surface | 2 (53) | 1 (53) | 1 (12) | 1 (15) | 8 (53) | 2 (10) |
| Cytoplasm | 24 (35) | 7 (35) | 8 (12) | 10 (15) | 34 (35) | 9 (10) |
| T-ALL | ||||||
| Surface | 1 (6) | 1 (6) | 1 (6) | 1 (6) | 1 (6) | 1 (2) |
| Cytoplasm | 2 (3) | 3 (3) | 2 (2) | 2 (2) | 3 (3) | 2 (2) |
| B-CLL | ||||||
| Surface | 3 (4) | 3 (4) | 3 (4) | 3 (4) | 3 (4) | 3 (4) |
| Cytoplasm | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) | 4 (4) |
| T-CLL (LGL) | ||||||
| Surface | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) |
| Cytoplasm | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) | 0 (2) |
Reactivity of leukemic cells with a panel of FITC-conjugated anti-CTLA-4 scFvs and the commercial BN13 mAb by flow cytometry. Positivity was considered when the histogram shift of the sample was greater than 1 log relative to the negative control. Data are presented as the number of samples that reacted positively.
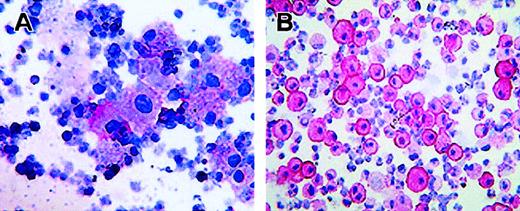
Moreover, alkaline-phosphatase staining with biotin-labeled scFv 67 performed in a cytospin preparation of AML neoplastic cells indicated a diffuse CTLA-4 cytoplasmic staining (Figure2A).
Intracellular localization of CTLA-4 in activated CD34+ stem cells and AML neoplastic cells.
(A) Immunostaining with the biotin-conjugated anti–CTLA-4 scFv67 and AP-labeled streptavidin of neoplastic cells from peripheral blood of an AML patient. (B) Immunostaining with the anti–CTLA-4 BN13 mAb and AP-labeled antimouse immunoglobulin antibody of CD34+ stem cells activated with GM-CSF for 7 days. Staining was performed on cytospins, visualized by the addition of phosphatase substrate, and counterstained by hematoxylin (original magnifications: panel A, × 70; panel B, × 110). Panel A and B stainings show cytoplasmic and membrane positivity for CTLA-4.
Intracellular localization of CTLA-4 in activated CD34+ stem cells and AML neoplastic cells.
(A) Immunostaining with the biotin-conjugated anti–CTLA-4 scFv67 and AP-labeled streptavidin of neoplastic cells from peripheral blood of an AML patient. (B) Immunostaining with the anti–CTLA-4 BN13 mAb and AP-labeled antimouse immunoglobulin antibody of CD34+ stem cells activated with GM-CSF for 7 days. Staining was performed on cytospins, visualized by the addition of phosphatase substrate, and counterstained by hematoxylin (original magnifications: panel A, × 70; panel B, × 110). Panel A and B stainings show cytoplasmic and membrane positivity for CTLA-4.
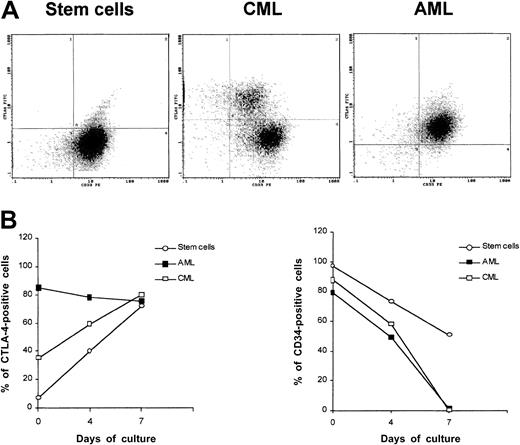
Surface expression of CTLA-4 was detected at various degrees in CML samples showing higher frequency of 67- and 83-epitopes (4 of 4) followed by 40- and 55-epitopes (3 of 4) and 3-epitope (1 of 4). All CML samples strongly expressed CTLA-4 epitopes in the cytoplasm (Table3). CML neoplastic cells were consistently found to express surface CTLA-4 epitopes at lower density compared with AML, as shown by double staining with FITC-scFv 83 and PE–anti-CD33 mAb (Figure3A). Positive staining with both antibodies also confirmed that CTLA-4–expressing cells from patients with AML and CML were likely to be leukemic cells.
CTLA-4 expression in CD34+ stem cells and AML and CML neoplastic cells.
(A) Double staining of peripheral blood normal CD34+ stem cells and neoplastic cells from AML and CML patients with FITC-conjugated anti–CTLA-4 scFv 83 and PE-conjugated anti-CD33 mAb by flow cytometry. (B) Kinetics of CTLA-4 surface expression after treatment of normal CD34+ stem cells and neoplastic cells from AML and CML patients resting or stimulated with GM-CSF. Cells were analyzed by flow cytometry to determine the percentage of positive cells. These results are representative of 3 independent experiments.
CTLA-4 expression in CD34+ stem cells and AML and CML neoplastic cells.
(A) Double staining of peripheral blood normal CD34+ stem cells and neoplastic cells from AML and CML patients with FITC-conjugated anti–CTLA-4 scFv 83 and PE-conjugated anti-CD33 mAb by flow cytometry. (B) Kinetics of CTLA-4 surface expression after treatment of normal CD34+ stem cells and neoplastic cells from AML and CML patients resting or stimulated with GM-CSF. Cells were analyzed by flow cytometry to determine the percentage of positive cells. These results are representative of 3 independent experiments.
CTLA-4 epitopes were present on either the surface or the cytoplasm of B-CLL (3 of 4 and 4 of 4, respectively), whereas they were lacking on T-CLL LGL (0 of 2).
Flow cytometry analysis of the same leukemia samples was also carried out with the control BN13 mAb, resulting in overlapping flow cytometric profiles, and with scFv and isotypic control mAbs, consistently resulting in negative staining (data not shown).
As far as B-ALL is concerned, 3- and 83-epitopes were expressed on the surface (2 of 53 and 8 of 53, respectively) and more frequently in the cytoplasm (24 of 35 and 34 of 35, respectively), whereas the other epitopes were expressed mainly in the cytoplasm. These findings suggest that CTLA-4 expression might be differently regulated in classical B-origin ALL independently of the CD34 expression and the stage of differentiation (data not shown). In only 1 of 6 cases of T-ALL studied was CTLA-4 expressed on the surface, although it was expressed more frequently in the cytoplasm. All the CTLA-4+ T-ALLs were adult T-ALLs, whereas all the pediatric leukemias never demonstrated CTLA-4 expression. This could fit with a different degree of differentiation in the adult and the pediatric cases.
CTLA-4 expression in human CD34+ normal and neoplastic blood cells
We determined whether, either at the time of collection or following in vitro stimulation with GM-CSF for 7 days, CTLA-4 was differentially expressed in CD33+/CD34+ stem cells and neoplastic cells from AML and CML leukemias by flow cytometric analysis (Figure 3A). Results confirmed that CTLA-4 was not expressed by freshly isolated stem cells, whereas it was highly expressed by AML and, to a lesser extent, by CML cells (Figure 3A). During culture stimulation, the frequency of CTLA-4–expressing cells was increased up to 56% for CML cells and to 91% for stem cells at day 7, whereas it remained quite similar for AML cells. Alkaline-phosphatase staining of CD34+-activated cells carried out with the BN13 mAb showed cytoplasmic and, to a major extent, membrane CTLA-4 localization (Figure 2B).
To determine whether this expression correlated with the expression of the CD34 marker, we simultaneously analyzed the kinetics of CD34 expression in the 3 cell populations. The frequency of CD34-expressing cells followed a time course opposite to the one observed for CTLA-4, resulting in a 47% and 98% decrease, respectively, for stem cells and AML/CML cells (Figure 3B). These findings suggest that up-regulation of CTLA-4 expression might be inversely associated with cell differentiation.
Effect of activation on CTLA-4 RNA expression
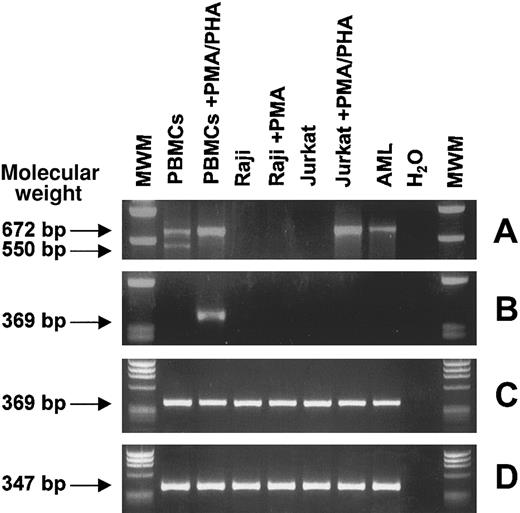
Expression of CTLA-4–specific transcripts was investigated by RT-PCR with the use of 2 sets of primers specific for CTLA-4 full-length coding sequence24 and CTLA-4 extracellular domain.25 The first primer set detected 2 main RNA transcript variants of 672 and 550 bp in resting PBMCs, corresponding to the membrane CTLA-4 transcript and to the spliced CTLA-4del transfer membrane (TM) transcript respectively, as previously described.24 Stimulation of PBMCs with PMA/PHA resulted in the disappearance of the CTLA-4delTM band, whereas the membrane CTLA-4 band increased in intensity (Figure 4A). The same RT-PCR analysis was performed on 2 representative hematopoietic resting and activated cell lines (Raji and Jurkat) and 1 AML leukemia sample, highly expressing CTLA-4 by flow cytometry.
RT-PCR analysis of CTLA-4 transcripts from normal and neoplastic, resting and activated, hematopoietic cells.
Total RNA from the indicated cells was reverse transcribed and PCR amplified with primers specific for CTLA-4 full-length coding sequence (A) and for CTLA-4 extracellular domain (B). Nested PCR was performed on the first-round CTLA-4 full-length PCR product as template with CTLA-4 extracellular domain inner primers (C). As internal control, β-actin gene amplification was carried out (D). PBMCs are freshly isolated peripheral blood mononuclear cells; Raji and Jurkat are B- and T-lymphoblastic cell lines, respectively; and AML is an adult acute myeloid leukemia sample. Molecular weights are expressed as base pairs (bp).
RT-PCR analysis of CTLA-4 transcripts from normal and neoplastic, resting and activated, hematopoietic cells.
Total RNA from the indicated cells was reverse transcribed and PCR amplified with primers specific for CTLA-4 full-length coding sequence (A) and for CTLA-4 extracellular domain (B). Nested PCR was performed on the first-round CTLA-4 full-length PCR product as template with CTLA-4 extracellular domain inner primers (C). As internal control, β-actin gene amplification was carried out (D). PBMCs are freshly isolated peripheral blood mononuclear cells; Raji and Jurkat are B- and T-lymphoblastic cell lines, respectively; and AML is an adult acute myeloid leukemia sample. Molecular weights are expressed as base pairs (bp).
A clear 672-bp band was detected only for PMA-stimulated Jurkat cells, whereas an identical but faint band was detected for the AML sample. The second primer set detected a 369-bp band corresponding to the CTLA-4 extracellular domain only in PMA/PHA–stimulated PBMCs (Figure4B). To explain the discrepancy between CTLA-4 expression detected at protein level and that detected at RNA level, a nested PCR assay was developed, in which the first-round CTLA-4 full-length PCR product was amplified with CTLA-4 extracellular domain inner primers. Surprisingly, all the samples tested showed a sharp 369-bp band (Figure4C). These results demonstrate that the failure in detecting CTLA-4 transcripts by conventional PCR was probably due to the low amount of specific CTLA-4 RNA molecules present in the cell lines, thus explaining the low sensitivity of a single-round PCR.
Effect of activation on CTLA-4 protein expression
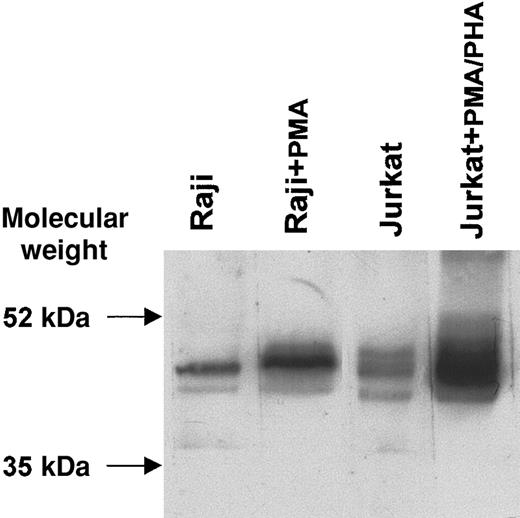
CTLA-4–specific bands of expected molecular weight (46 to 50 kDa)12 were observed by Western blot under nonreducing conditions in the lanes containing 2 representative cell lines (Jurkat and Raji) in either resting or activated conditions. A highly significant increase in CTLA-4 protein levels was observed after activation with PMA/PHA or PMA, by immunoblotting with the scFv 83 mixed to the 9E10 mAb recognizing the peptide tag linked to the scFv fragment (Figure 5). These results confirm the increased CTLA-4 expression observed by flow cytometry after activation, although this was undetectable at transcription level, thus supporting the concept of CTLA-4 protein intracellular accumulation.
Western blot analysis of CTLA-4 protein from resting and activated B-lymphoblastic (Raji) and T-lymphoblastic (Jurkat) cell lines.
Proteins from cell lysates were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) on a nonreducing 12% (wt/vol) polyacrylamide gel, transferred to nitrocellulose membrane, and probed with scFv 83 mixed to the anti–c-myc tag 9E10 mAb. The reaction was visualized by ECL.
Western blot analysis of CTLA-4 protein from resting and activated B-lymphoblastic (Raji) and T-lymphoblastic (Jurkat) cell lines.
Proteins from cell lysates were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) on a nonreducing 12% (wt/vol) polyacrylamide gel, transferred to nitrocellulose membrane, and probed with scFv 83 mixed to the anti–c-myc tag 9E10 mAb. The reaction was visualized by ECL.
Effect of anti–CTLA-4 immunotoxins on the induction of apoptosis in AML cells
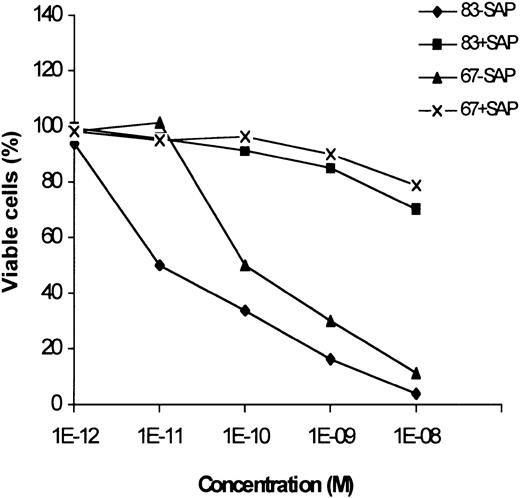
The effect of 2 immunotoxins, obtained by chemically conjugating anti–CTLA-4 scFvs 67 and 83 to the saporin type-1 RIP, on the induction of apoptosis of leukemic cells was evaluated by the annexin V/propidium iodide staining. Both the immunotoxins at 10−8M concentration caused apoptosis of more than 90% of neoplastic cells from an M2 AML sample with a high percentage of neoplastic elements (Figure 6). Saporin mixed with scFvs was weakly toxic only at 10−8 M concentration. Since anti–CTLA-4 immunotoxins have been previously shown17 to be only slightly toxic for bone marrow precursors at 10−8M concentration, these results suggest their possible in vivo application for killing leukemic cells.
Effect of anti–CTLA-4 immunotoxins on apoptosis induction of AML cells.
Percentage of viable neoplastic cells from an adult AML leukemia treated for 72 hours with scalar doses of 2 immunotoxins (83-Sap and 67-Sap), or a mixture of saporin and scFv (83+Sap and 67+Sap). Results are expressed as means of 3 different experiments. SD never exceeded 10%.
Effect of anti–CTLA-4 immunotoxins on apoptosis induction of AML cells.
Percentage of viable neoplastic cells from an adult AML leukemia treated for 72 hours with scalar doses of 2 immunotoxins (83-Sap and 67-Sap), or a mixture of saporin and scFv (83+Sap and 67+Sap). Results are expressed as means of 3 different experiments. SD never exceeded 10%.
Discussion
There is much information on the expression of CTLA-4 receptor in human cells of the T-lymphoid lineage, but very little information is available about other types of cells. In this study, we analyzed the CTLA-4 distribution pattern in a variety of human normal and transformed hematopoietic and neoplastic cells of lymphoid or myeloid origin.
To this purpose, cell reactivity was tested with a panel of human anti–CTLA-4 scFv antibodies and with the commercial murine BN13 mAb in commonly used techniques, such as immunofluorescence and flow cytometry, immunohistochemistry and Western blotting.
Immunofluorescence staining with either the scFvs or BN13 mAb indicated that CTLA-4, although regularly present in the cytoplasm, was never expressed at the cell surface of resting normal PBMCs, T cells, B cells, granulocytes, or CD34+ stem cells, but its expression was induced upon activation. On the contrary, CTLA-4 was constitutively weakly expressed on monocytes. The 4 different CTLA-4 epitopes (40, 55, 67, and 83) recognized by the scFvs were similarly expressed in the various cell types except for 1 epitope (3) that consistently appeared more weakly expressed on the cell surface but not in the cytoplasm. All together, these results correlate with previous findings that most CTLA-4 molecules accumulate in the intracellular compartment, from which they traffic to the cell surface and are rapidly internalized.26 27
CTLA-4 expression was observed to various degrees on cell lines of T-lymphoid (Jurkat, CEM, Molt-4), B-lymphoid (Raji, HOM-2, Daudi), myeloid (HL-60, KG1a), and erythroid (K562) origin, with an expression pattern resembling the one observed in their normal activated cell counterparts.
Our RT-PCR analysis confirmed previous studies showing that nonstimulated human PBMCs constitutively expressed membrane full-length CTLA-4 transcript24 but also expressed an additional spliced variant named CTLA-4delTM.25 As expected, this variant, which lacks both transmembrane and intracellular domains, was not observed in PMA/PHA–stimulated PBMCs.
None of the cell lines tested, whether resting or stimulated, expressed both CTLA-4 full-length and extracellular domain RNA by conventional RT-PCR, except activated Jurkat cells that were positive for the full-length transcript. Only an RT-PCR/nested PCR combination was able to detect a CTLA-4 transcript (extracellular domain) in all the cell lines analyzed. This finding may be explained by a low amount of CTLA-4 RNA molecules, probably due to a very high RNA translation rate or to low RNA stability. Such evidence appears to be in striking contrast to the high CTLA-4 protein levels observed in cell lines, but it actually confirms previous findings28 that the signals regulating CTLA-4 RNA transcription rate and protein expression may be different (M.P.P., P.L.T., G.L.P., et al, manuscript in preparation).
The finding of CTLA-4 expression in the Jurkat cell line, in resting as well as activated conditions, was surprising since previous studies never found such a cell line positive, at either the protein or the mRNA level.29,30 This discrepancy may be attributed to the different specificity and characteristics of the anti–CTLA-4 antibodies used in the studies. In fact, previous reports used a xeno-polyclonal serum recognizing mainly the monomeric form of CTLA-4 in immunoprecipitation experiments, in contrast to our human scFv fragments recognizing the homodimerized CTLA-4 (Pistillo et al15 and data not shown). In addition, previous analysis of CTLA-4 mRNA in Jurkat cell line was performed by Northern blot,29 31 whereas we used the more sensitive nested-PCR technique.
Neoplastic cells from most AMLs or CMLs and B-CLLs were positive for all CTLA-4 epitopes at either the surface or the cytoplasm, whereas neoplastic cells from T-CLL LGL were completely negative. Absence of surface CTLA-4 expression in T-CLL cells is in concordance with the T-resting–like phenotype of this leukemia, although resting T cells differ in their cytoplasmic expression of CTLA-4. In general, the highly frequent expression of CTLA-4 on various subtypes of myeloid and lymphoid leukemias are likely to reflect their differentiation and maturation stages, as confirmed by the phenotypic pattern of expression observed in cell lines belonging to the same lineage.
Since incubation with the anti–CTLA-4 scFvs described above did not affect proliferation or survival of either normal or neoplastic hematopoietic cells, we evaluated the possibility of using them as vehicles for apoptosis-inducing toxins. Therefore, we investigated whether immunotoxins containing 2 different scFvs, against CTLA-4 and the type-1 RIP saporin, were able to kill CTLA-4–expressing leukemic cells in vitro. It was already reported that RIP-containing immunotoxins are able to induce apoptosis in target cells,32,33 with cytotoxic mechanisms comprehending protein synthesis inhibition and possibly direct damages to DNA or other types of RNA.34 We demonstrated that they were highly efficient in inducing apoptosis of neoplastic cells from an AML sample, supporting the importance of CTLA-4 as a new target molecule for in vivo immunotherapeutic strategies of human leukemias.
Our results point out that the availability of a panel of anti–CTLA-4–specific scFvs will be very important for monitoring CTLA-4 expression in neoplastic cell populations from different leukemia subtypes and for studying the physiological function of this molecule in such neoplastic cells. In fact, the expression of CTLA-4 in leukemias might indicate that the tumor cells have the ability to interact with the CD80/CD86 ligands on antigen-presenting cells, and actually transduce an immunosuppressive signal that plays a role in initiating and maintaining the neoplastic process. Whether this signal would follow the same inhibitory pathway typical of activated T cells remains to be established.
We are grateful to D. Neri (Swiss Federal Institute of Technology, Zurich) for kindly providing the ETH-2 phage library used in this study.
Prepublished online as Blood First Edition Paper, August 15, 2002; DOI 10.1182/blood-2002-06-1668.
Supported in part by funds from Progetto Finalizzato Consiglio Nazionale Ricerche (PF CNR) Target Project on Biotechnology 2001, Ministero della Sanità Ricerca Finalizzata (RF), 1999-2000, and Progetto Strategico Oncologia N.74 from Ministero Istruzione Università e Ricerca, Rome, Italy.
M.P.P. and P.L.T. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Maria Pia Pistillo, Laboratory of Immunogenetics, National Cancer Research Institute, c/o Advanced Biotechnology Center, Largo R Benzi 10, 16132 Genova, Italy; e-mail:mariapia.pistillo@istge.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal