Abstract
Fanconi anemia (FA) is a condition that induces susceptibility to bone marrow failure, myelodysplastic syndrome (MDS), and leukemia. We report on a high incidence of expanding clonal aberrations with partial trisomies and tetrasomies of chromosome 3q in bone marrow cells of 18 of 53 FA patients analyzed, detected by conventional and molecular cytogenetics. To determine the clinical relevance of these findings, we compared the cytogenetic data, the morphologic features of the bone marrow, and the clinical course of these patients with those of 35 FA patients without clonal aberrations of 3q. The 2 groups did not differ significantly with respect to age, sex, or complementation group. There was a significant survival advantage of patients without abnormalities of chromosome 3q. Even more pronounced was the risk assessment of patients with gains of 3q material with respect to the development of morphologic MDS and acute myeloid leukemia (AML). Thus, our data from 18 patients with 3q aberrations reveal that gains of 3q are strongly associated with a poor prognosis and represent an adverse risk factor in FA.
Introduction
Fanconi anemia (FA) is an autosomal recessive chromosomal instability disorder with a high risk for bone marrow failure, myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and solid tumors. Recent data demonstrate that FA cells have a reduced fidelity in processing DNA double-strand breaks,1-5 which leads to unbalanced chromosomal aberrations such as deletions, insertions, and translocations. This specific intrinsic susceptibility might, together with extrinsic factors, influence the course of the disease, resulting in the outgrowth of clones with chromosomal aberrations in bone marrow (BM) cells and, subsequently, in circulating peripheral blood cells. The most frequently reported acquired clonal aberrations in FA patients are trisomies of chromosome 1q and monosomies of chromosome 7; the latter is thought to be associated with a poor prognosis. The significance and the predictive value of such clonal alterations with respect to hematopoietic function and malignant progress are not fully understood.6-9
Study design
We analyzed 132 bone marrow samples from 53 FA patients who had a normal constitutional karyotype in their peripheral T lymphocytes. FA was proven in all patients by chromosomal breakage test after mitomycin C treatment. In addition to conventional cytogenetics, where up to 50 BM metaphases after synchronization and GTG banding were analyzed, we used comparative genomic hybridization (CGH) and fluorescence in situ hybridization (FISH). Our approach of investigating all BM samples by CGH allows the comprehensive analysis of the entire genome in just one experiment, providing information not only about the chromosomal assignment but also about the size of the chromosomal imbalances.10 11 All CGH results were validated by FISH with specific probes onto BM metaphases.
Results and discussion
Of 53 FA patients, 28 showed a normal karyotype in BM cells after conventional cytogenetics and CGH, whereas 25 patients had clonal aberrations. Of these, 18 (72%) revealed partial trisomies or tetrasomies for the long arm of chromosome 3, indicating an extremely high incidence of 3q aberrations in our FA patients (Figure1A). The 3q aberrations of almost all our FA patients were mosaics with subtle aberrations due to unbalanced translocations of distal 3q to various other chromosomes. An example is given in Figure 1B, demonstrating that the identification of additional material in BM metaphase spreads is extremely difficult by conventional cytogenetics alone.
Chromosomal assignment and size of gains of chromosome 3 material (vertical lines) in bone marrow cells of 18 FA patients detected by CGH and validated by FISH analyses.
(A) The critical trisomic or tetrasomic region 3q26q29 shared by all FA patients is shadowed in gray. (B) Cytogenetic and molecular cytogenetic analysis of patient 11. (i) Partial karyotype of chromosomes 3 and 10 after GTG banding of a bone marrow cell. The arrow points to the translocated material of chromosome 3 onto chromosome 10. (ii) Whole chromosome paints for chromosome 3 (green) and chromosome 10 (red) on a BM metaphase plate for the validation of CGH results. (iii) Averaged CGH copy number karyotype of patient 11 indicating the gain of chromosome 3q25qter material (green bar right of chromosome 3). Ratio deviations in heterochromatic parts of chromosomes and in the X and Y chromosome are generally excluded from evaluation.
Chromosomal assignment and size of gains of chromosome 3 material (vertical lines) in bone marrow cells of 18 FA patients detected by CGH and validated by FISH analyses.
(A) The critical trisomic or tetrasomic region 3q26q29 shared by all FA patients is shadowed in gray. (B) Cytogenetic and molecular cytogenetic analysis of patient 11. (i) Partial karyotype of chromosomes 3 and 10 after GTG banding of a bone marrow cell. The arrow points to the translocated material of chromosome 3 onto chromosome 10. (ii) Whole chromosome paints for chromosome 3 (green) and chromosome 10 (red) on a BM metaphase plate for the validation of CGH results. (iii) Averaged CGH copy number karyotype of patient 11 indicating the gain of chromosome 3q25qter material (green bar right of chromosome 3). Ratio deviations in heterochromatic parts of chromosomes and in the X and Y chromosome are generally excluded from evaluation.
In the literature, only 3 other FA patients with additional material of chromosome 3q in BM cells have been described. All had trisomies or tetrasomies for almost the entire long arm from 3q13 to 3qter,7,12 13 permitting identification by conventional cytogenetics alone, as in our patients 10, 13, and 14 (Figure 1A). In consideration of the incidence of 3q gains in our FA patient group compared with the number of published cases, we assume that other studies might have failed to detect the more subtle partial trisomies and tetrasomies of 3q.
In 16 of 18 FA patients with 3q gains, serial bone marrow analyses were performed (mean, 5.6 analyses per patient). By conventional cytogenetics, we observed in all of them a considerable increase of the clone with additional 3q material over time. Furthermore, 3 patients developed a tetrasomic 3q clone that derived from the trisomic by a subsequent duplication. In these 3 patients the tetrasomic clone replaced the trisomic clone over time. Thus, our data strongly suggest that gains of 3q confer either a higher proliferative advantage or an increased survival to bone marrow cells. No transient appearance of 3q gains, as described for other clonal aberrations in FA patients, was noticed.
Of 18 patients, 8 had an additional monosomy 7. In 2 of these patients the clone with the 3q gain and the monosomy 7 was already present at the time of the first BM analysis, whereas in the other 6 patients the monosomy 7 developed in the 3q aberrant clone as a secondary event. Even though the number of patients affected with both a gain of 3q and monosomy 7 was small, our data imply that gains of 3q might increase the risk for subsequently developing a monosomy 7.
In order to determine the percentage of aberrant cells in nondividing BM cells and peripheral blood mononuclear cells (PBMCs), we performed interphase FISH analyses with a 1010-kb yeast artifical chromosome (YAC) mapped to 3q27q28. Our first results demonstrated that the FA patients with 3q aberrations exhibited up to 70% of the BM cells and PBMCs with more than 2 signals in the interphase FISH, as compared with a maximum of 2% to 3% in healthy control subjects. Since the karyotype was normal in all T and B lymphocytes investigated, these results suggest that almost all circulating granulocytes are affected by the aberration in these patients. In 3 FA patients the 3q aberration was detected only by screening PBMCs by interphase FISH prior to the conventional cytogenetic analysis of BM cells. This proves the high sensitivity and specificity of interphase FISH to screen for aberrant clones in PBMC.
Four genes involved in MDS and/or AML have been identified in the critical region 3q shared by our FA patients: the myelodysplasia–myeloid leukemia factor 1 (MLF1), the myelodysplasia syndrome–associated sequence 1 (MDS1), the murine myeloid leukemia–associated gene EVI1, and the Epstein-Barr–associated protein EAP. It has been demonstrated in non-FA patients with MDS or AML that balanced structural aberrations between these genes and between the nucleophosmin gene (NPM) and the acute myeloid leukemia 1 gene (AML1) generate the expression of fusion proteins that seem to be implicated in MDS/AML progression.14-18 In contrast to these balanced aberrations, all FA patients whose results are reported here had an unbalanced status, with additional material of 3q and different breakpoints in 3q. This indicates that mechanisms other than the expression of fusion transcripts must be responsible for the outgrowth of these clonal aberrations.
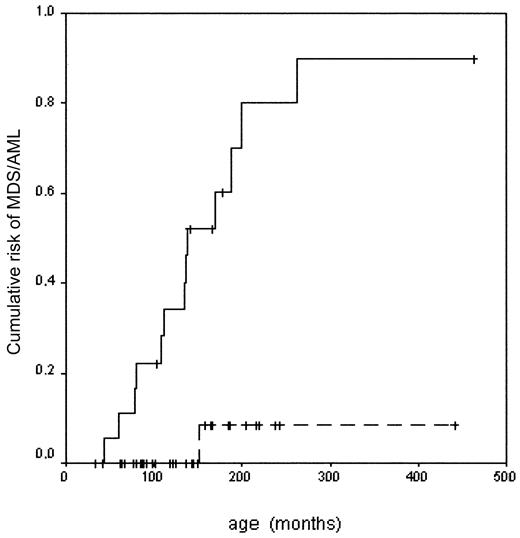
In order to determine the clinical relevance of these findings, we compared the cytogenetic data, the morphologic features of the bone marrow, and the clinical course of 18 FA patients with chromosome 3 aberrations with those of 35 FA patients without clonal aberrations of 3q. The 2 groups did not differ significantly with respect to age, sex, complementation group, or genotypic reversion. There was only a slight trend toward male sex and FANCC in the group with 3q aberrations (Table1). Despite the fact that more hematopoietic stem cell transplantations have been performed in this group, there was a significant survival advantage for patients without abnormalities of chromosome 3q. Even more pronounced was the risk assessment of patients with gains of 3q material with respect to the development of morphologic MDS and AML (Table 1; Figure2). Thus, our data from 18 patients with 3q aberrations reveal that gains of 3q are strongly associated with a poor prognosis and represent an adverse risk factor in FA.
Characteristics and outcomes of FA patients with and without trisomies or tetrasomies of chromosomal segment 3q26-3q29
| . | Total . | With chromosome 3 aberration . | Without chromosome 3 aberration . |
|---|---|---|---|
| n | 53 | 18 | 35 |
| Age, median (range), mo | 141 (34-463) | 149 (95-463) | 125 (34-442) |
| Sex, no. | |||
| Male | 28 | 11 | 17 |
| Female | 25 | 7 | 18 |
| Complementation group, no. | |||
| FANCA | 28 | 10 | 18 |
| FANCC | 4 | 4 | 0 |
| FANCG | 8 | 3 | 5 |
| Unknown | 13 | 1 | 12 |
| Genotypic reversion* | 3 | 1 | 2 |
| Outcome, no. | |||
| MDS†,‡ | 9 | 9 | 0 |
| AML1-153 | 5 | 4 | 1 |
| MDS+AML† | 14 | 13 | 1 |
| Alive1-155 | 44 | 11 | 33 |
| HSCT | 20 | 12 | 8 |
| . | Total . | With chromosome 3 aberration . | Without chromosome 3 aberration . |
|---|---|---|---|
| n | 53 | 18 | 35 |
| Age, median (range), mo | 141 (34-463) | 149 (95-463) | 125 (34-442) |
| Sex, no. | |||
| Male | 28 | 11 | 17 |
| Female | 25 | 7 | 18 |
| Complementation group, no. | |||
| FANCA | 28 | 10 | 18 |
| FANCC | 4 | 4 | 0 |
| FANCG | 8 | 3 | 5 |
| Unknown | 13 | 1 | 12 |
| Genotypic reversion* | 3 | 1 | 2 |
| Outcome, no. | |||
| MDS†,‡ | 9 | 9 | 0 |
| AML1-153 | 5 | 4 | 1 |
| MDS+AML† | 14 | 13 | 1 |
| Alive1-155 | 44 | 11 | 33 |
| HSCT | 20 | 12 | 8 |
FANCA, FANCC, and FANCG indicate Fanconi anemia complementation groups A, C, and G, respectively; MDS, myelodysplastic syndrome; AML, acute myeloid leukemia; and HSCT, hematopoietic stem cell transplantation.
Spontaneous reversion of the cellular FA phenotype in lymphocytes by back mutation or gene conversion in compound heterozygotes.
P < .001, Fisher exact test (2-sided).
MDS was defined according to the French-American-British (FAB) classification, and as described elsewhere.6Eight of 9 MDS patients presented 3% to 26% blasts in the marrow.
AML was defined by more than 30% blasts in the marrow.
P = .005, Fisher exact test (2-sided).
Risk of developing MDS or AML in Fanconi anemia with or without chromosome 3 aberration.
With chromosome 3 aberration (—): n = 18, MDS/AML 13, risk 0.90 ± 0.09. Without chromosome 3 aberration (- - -): n = 35, MDS/AML 1, risk 0.08 ± 0.08. Log rank: P < .001.
Risk of developing MDS or AML in Fanconi anemia with or without chromosome 3 aberration.
With chromosome 3 aberration (—): n = 18, MDS/AML 13, risk 0.90 ± 0.09. Without chromosome 3 aberration (- - -): n = 35, MDS/AML 1, risk 0.08 ± 0.08. Log rank: P < .001.
Considering the high MDS/AML risk and the significantly higher mortality in the group of FA patients with 3q gains, we recommend the systematic assessment of all individual FA patients by molecular cytogenetics in order to detect these aberrations as early as possible. In case of 3q gains, the decision for a more aggressive clinical intervention, that is, early unrelated donor transplantation, has to be seriously considered.
Further data on the role of genes located on the described chromosomal region 3q26q29 are needed to elucidate the pathomechanisms of MDS and leukemia in FA and non-FA patients.
We are indebted to all the Fanconi anemia patients, their parents, and their clinicians who supported this study. We are grateful to Hans Joenje, Free University Amsterdam; Detlev Schindler, University Würzburg; and Helmut Hanenberg, University Düsseldorf, for data on the complementation analyses. We thank Sylke Niehage, Marianne Plieth, Marlies Schwanke, and Britta Teubner for excellent technical assistance.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/blood-2002-10-3243.
Supported by grants from the Deutsche Fanconi Anämie Hilfe e.V. and the Charité Research Fund (No.2000-627), Humboldt-University, Berlin, Germany.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Heidemarie Neitzel, Institute of Human Genetics, Charité, Humboldt-University, Augustenburger Platz 1, 13353 Berlin, Germany; e-mail:heidemarie.neitzel@charite.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal