Abstract
Mantle cell lymphoma (MCL) is believed to originate from a naive B cell. However, we recently demonstrated that a subset of MCL displayed mutated VH genes. We also reported restricted use of certain VH genes. To assess the prognostic impact of these new findings, we performed VH gene analysis of 110 patients, revealing that 18 (16%) patients had mutated and 92 (84%) patients had unmutated VH genes. Because the mutation rate was low in the mutated group (2.2%-6.7%), further investigation of the germline VH gene in T cells from 5 patients with mutated VH genes was carried out; results showed that the unrearranged VH gene was identical to the published sequence. These data confirm that the base pair substitutions within the rearranged VH genes represent hypermutations, and indicate germinal center exposure. However, VH gene mutation status did not correlate with prognosis because there was no difference in clinical outcome between the unmutated and mutated groups. The most frequently used VH genes were VH3-21 (21 patients) and VH4-34 (19 patients). A novel finding was that VH3-21+ MCL almost exclusively expressed λ light chains and displayed highly restricted use of the Vλ3-19 gene. VH3-21+patients had longer median survival than the remaining patients (53 vs 34 months; P = .03), but they tended to be younger at diagnosis. The combined use of VH3-21/Vλ3-19 suggests a possible role for antigen(s) in the pathogenesis of these tumors and indicates that VH3-21+ patients constitute a new MCL entity.
Introduction
Mantle cell lymphoma (MCL) is generally an aggressive and incurable malignant lymphoma with a median survival time of 3 to 4 years.1,2 Most patients are diagnosed with widespread disease with disseminated lymphadenopathy and frequent involvement of the bone marrow (BM), spleen, and gastrointestinal (GI) tract.1,2 Tumor cells typically express CD5, CD19, and CD20 but lack CD23 expression.3 Almost all MCLs exhibit the (11;14) translocation, leading to rearrangement of theBCL1 gene and overexpression of cyclin D1.4,5 The up-regulation of this cell cycle regulatory protein contributes to the pathogenesis of MCL, but alone it is insufficient for tumor progression.6 Despite this characteristic diagnostic marker, the disease entity includes a heterogeneous group of patients with various clinical courses. Few known markers can further define prognostic subgroups within the disease, with the exceptions of the morphologically distinct blastoid variant of MCL (MCL-BV) and the high proliferation index that have been reported to identify patients with aggressive disease.1 7Thus, the identification of more predictors of outcome will be the first step in revealing biologic differences in the disease, which may ultimately be exploited to enable improved treatment strategies.
Analysis of somatic hypermutation of the immunoglobulin variable heavy chain (IgVH) genes has provided valuable insight into the origin of B-cell lymphomas. This process occurs during a T-cell–dependent immune response in the germinal center (GC) of secondary lymphoid follicles.8 Therefore, a mutated VH gene in a B-cell lymphoma indicates GC or post-GC B-cell origin, whereas an unmutated VH gene indicates derivation from a pre-GC B cell that has proliferated independently of T-cell help. In MCL, the postulated cell of origin is a B cell deriving from the mantle zone of a lymphoid follicle that has not entered the GC and, therefore, lacks hypermutation.9 This fact has been questioned because MCL patients with mutated VH genes have been reported by our group and others.10-13 Recently, we investigated 51 MCL patients and showed that approximately 20% of them displayed hypermutated VH genes (more than 2% sequence deviation from the corresponding germline gene was defined as mutated); the remaining had unmutated VHgenes.13 This led us to suggest that MCL may consist of 2 subsets—one larger unmutated group and one smaller mutated group. Similarly, studies of chronic lymphocytic leukemia (CLL) have convincingly revealed that this disease comprises 2 subsets but the proportion of mutated and unmutated cases is approximately equal.14-16 Although VH gene mutation status has been shown to be an independent prognostic marker in CLL, with mutated cases having better clinical outcomes, our pilot survival analysis (of 48 MCL patients) did not reveal a significant association between VH gene mutation status and overall survival in MCL.13 However, the 2% mutation cut-off level empirically chosen for CLL17 may not necessarily be correct for MCL, especially because the true biologic border that distinguishes between mutated and unmutated lymphomas is unknown.
VH gene use has been of great interest in B-cell malignancies—a biased use of individual VH gene family members has been shown in several lymphoma and leukemia entities.18-26 It has been hypothesized that B-cell receptors encoded by certain VH genes may predispose those B cells to malignant transformation as they may have a greater tendency or capacity to proliferate more frequently due to the recognition of many self- or nonself-antigens.19,21,22,27,28 We have recently shown restricted VH gene use in MCL, demonstrating an overuse of 3 different VH genes: VH3-21, VH4-34, and VH5-51.13 The 2 former VH genes have a known, strong association with autoimmunity.29-33 Additionally, we demonstrated that the VH3-21 gene is overrepresented in CLL with mutated VH genes26 and is associated with a significantly worse overall survival than other mutated cases.
In the present study, we investigated the prognostic impact of somatic hypermutation and restricted VH gene use in a large cohort of MCL patients. We could confirm our recent finding that MCL comprises a mutated (16%) and an unmutated (84%) group, but no significant difference in survival was found between the 2 subsets even when choosing different mutation cut-off levels (1%-5%). Analysis of the corresponding unrearranged germline gene was performed in T cells from 5 mutated cases and showed 100% homology to the published sequence, verifying that the incorporated mutations in the rearranged VH genes corresponded to hypermutations. A novel and interesting finding was that VH3-21+ MCL patients showed almost exclusive use of the Vλ light chain gene, Vλ3-19.
Patients, materials, and methods
Patients and materials
Tumor samples from 110 MCL patients diagnosed between 1982 and 2002 were identified from the registries of the Departments of Pathology at Uppsala University Hospital, Umeå University Hospital, Lund University Hospital, Karolinska Hospital, and Huddinge University Hospital, Sweden, where tumors were registered as either centrocytic lymphoma or MCL. Approval for this study (Ups 01-182) was obtained from the ethical committee at Uppsala University; informed consent was not required since the analyses were performed on retrospective patient material. The only denominator that the patients chosen from the 5 centers shared was the availability of frozen tumor material from which DNA could be successfully extracted and amplified, though we initially included 10 patients from whom paraffin-embedded tissue was used. Besides that, there was no obvious bias in our patient selection, which is also reflected by the typical clinical characteristics and prognoses found in these MCL patients. Tumor material was obtained from lymph nodes (n = 79), BM (n = 9), spleen (n = 8), peripheral blood (PB) (n = 5), GI tract (n = 3), tonsil (n = 3), and other sources (n = 3). MCL diagnosis was based on morphologic and immunophenotypic criteria according to the World Health Organization (WHO) classification34; cyclin D1 overexpression was confirmed by immunohistochemistry in 103 patients, and t(11;14) was identified by fluorescence in situ hybridization (FISH) analysis in the other 7 patients. All MCL patients were diagnosed at the individual institutions and were centrally reviewed by an experienced hematopathologist (C.S.). Medical records were reviewed for all but one patient. Staging was performed according to Ann Arbor staging methods, including computed tomography (CT) of the abdomen/thorax or ultrasound of the abdomen, chest x-ray examination, and BM biopsy/aspiration. Age, sex, stage, nodal/extranodal disease, BM/spleen involvement, blood lymphocyte count, serum lactate dehydrogenase (s-LDH) level, treatment, and disease course were recorded, and the international prognostic index (IPI) was calculated for each age group.35 Survival information was obtained from medical records and local Swedish cancer registries.
PCR amplification and nucleotide sequence analysis
DNA was prepared from frozen or paraffin-embedded tumor material using standard protocols as previously described13 or with the QIAamp DNA Mini Kit (Qiagen, Valencia, CA), according to manufacturer's instructions. VH and V light-chain (VL) gene family-specific polymerase chain reaction (PCR) amplification was performed as outlined previously.13,36 Monoclonality of the VH and VL PCR products was distinguished from a polyclonal product by single-strand conformation polymorphism analysis, as detailed earlier.13
In most samples (n = 78), clonal products from the VHgene PCR were sequenced directly using the BigDye Terminator Cycle Sequencing Reaction Kit version 2.0 (Perkin-Elmer, ABI, Foster City, CA). Cloning of the VH gene PCR products was performed in 32 patients as previously described,37 and 3 to 10 colonies from each PCR product were sequenced. To investigate the presence of ongoing mutations in patients with mutated VHgenes, further subcloning of the amplified rearrangements was performed in 5 patients using the Zero Blunt TOPO PCR Cloning kit, which includes the proofreading enzyme Pfu (Invitrogen, Paisley, United Kingdom). At least 10 colonies were subsequently sequenced from each PCR product. This cloning kit was also used for the analysis of germline VH gene sequences amplified from T-cell DNA. VLgene PCR products of VH3-21+ patients were cloned using the previously described method37 or the Zero Blunt TOPO PCR Cloning kit. All sequences were analyzed using an automated DNA sequencer (ABI 377 or ABI 3700; Applied Biosystems, Foster City, CA).
Sequencing of the unrearranged germline gene from T cells of 5 patients
T cells were isolated from frozen tumor samples from 5 patients with mutated VH genes (from PB in 3 patients, from lymph node or spleen in the other 2 patients) using a CD4 Positive Isolation Kit (Dynal ASA, Oslo, Norway). Briefly, magnetic beads coated with mouse anti-CD4 IgM monoclonal antibody were incubated with the sample, which allowed the CD4+ T cells in the sample to bind to the magnetic beads. Subsequent washing with the use of a magnet enabled isolation of the bound cells.
Primers were designed to amplify the unrearranged VHgermline gene for VH3-23, VH3-72, VH3-74, VH4-34, and VH4-59 using the Primer3 software program.38 Forward primers hybridized to a region upstream of framework region (FR)1, and the reverse primer hybridized to a region downstream of FR3, hence excluding a rearranged germline VH gene from being amplified. Primer sequences (5' to 3') were: VH3-23 forward, AGTTTGGGCTGAGCTGGCTTTTTC; VH3-23 reverse, AATCTGCATTTGGTGCGTGTGAG; VH3-72 forward, CCCCAGAATTCCCAGGTGTTTTC; VH3-72 reverse, AAGGCTCCAGAGTCCTGCAAAAA; VH3-74 forward, TTGCTGATCAGGACTGCACACAG; VH3-74 reverse, ACTTGATGAATCAGCCCCAGATG; VH4-34 forward, ATGGACCTCCTGCACAAGAACAT; VH4-34 reverse, CAGACCACTGAGCTGTTGAGGAA; VH4-59 forward, CACCTCTCCATACAAAGGCACCAC; VH4-59 reverse, GGCGCTGAGCAGCACCTG. The PCR reaction was carried out using the same conditions as for VH gene family-specific PCR except that annealing temperatures of 65°C for VH3-23, VH3-72, VH4-59 and 64°C for VH3-74 and VH4-34 were used. PCR products were cloned, and at least 10 colonies were sequenced per sample.
Analyses of immunoglobulin sequences
The obtained sequences were aligned to immunoglobulin sequence databases—Ig BLAST (National Center for Biotechnology Information, USA), V-BASE (MRC Centre for Protein Engineering, Cambridge, United Kingdom), and International ImMunoGeneTics (IMGT).39VH gene sequences containing mutations resulting in less than 98% homology to the germline gene were defined as mutated. The presence of mutations in hotspot regions was evaluated by analyzing the number of mutations that occurred in the germline sequence motif RGYW/WRCY.40 Ongoing somatic hypermutation was defined as the presence of a replacement mutation in at least 2 of 10 sequences analyzed.
Statistical analyses
Survival was calculated from the date of diagnosis until last follow-up or death. Kaplan-Meier survival analysis and log-rank analysis were performed to study the prognostic impact of various parameters in our MCL cohort, and P was calculated as 2-sided. Cox proportional hazards analysis was used to study the interrelationship between univariate significant parameters. Fisher exact test with 1-tailed P and McNemar χ2 test were used to calculate the significance of differences in VH gene use between MCL and healthy B cells. The Mann-Whitney U test was applied to calculate the significance of differences in distribution of age at diagnosis between groups. Statistica 6.0 software (StatSoft, Tulsa, OK) was used for all calculations.
Results
Clinical data
We investigated 110 patients; 90 were men and 20 women. Eighty-four patients were diagnosed with classical MCL and 26 with MCL-BV. Median age at diagnosis was 68 years (range, 34-85 years). At follow-up, 56 patients had died and 53 were alive. Median follow-up time (n = 109) was 23 months (range, 0-162 months), and median survival time was 38 months (range, 2-162 months). Adequate staging was performed for 102 patients, revealing that 9 were in stage I-II and 93 were in stage III-IV. One hundred patients had nodal disease; only 6 did not have any nodal involvement (information from 4 patients lacking). Of these 6 patients without nodal involvement, 3 had BM/blood involvement as the sole site. Seventy-four patients had BM involvement, 24 had splenic involvement, 39 patients had extranodal involvement, and 32 had leukemic disease (of which only 8 had lymphocyte counts exceeding 20 × 109/L). The distribution of IPI scores (n = 89) was as follows: 0 to 2, 44%; 3 to 5, 56%. The median s-LDH value (n = 93) was 6.6 μkatal (μkat)/L (range, 3.8-29 μkat/L; normal upper limit, 6.7 μkat/L). Treatment was recorded for 106 patients; 4 patients were untreated, 2 received radiotherapy alone, and the rest received 1 or more chemotherapy regimens or antibody treatment. Most patients received CHOP (cyclophosphamide, doxorubicin, vincristine, prednisone)/CNOP (cyclophosphamide, mitoxantrone, vincristine, prednisone) treatment (n = 78), but chlorambucil/COP (cyclophosphamide, vincristine, prednisone) (n = 18), rituximab (n = 36), and nucleoside analogues (n = 40) were also used. Nineteen patients underwent high-dose chemotherapy with autologous stem cell transplantation (ASCT). Major clinical factors are summarized in Table 1.
Clinical features of MCL patients, including distributions between subsets
| Parameter . | All MCL n = 110 . | VH gene mutation status . | VH gene use . | ||
|---|---|---|---|---|---|
| Mutated MCL n = 18 . | Unmutated MCL n = 92 . | VH3-21+MCL n = 21 . | MCL without VH3-21 n = 89 . | ||
| Blastoid morphology | 26 of 110 | 3 of 18 | 23 of 92 | 8 of 21 | 18 of 89 |
| Stage III–IV | 93 of 102 | 14 of 18 | 79 of 84 | 20 of 20 | 73 of 82 |
| IPI score 3-5 | 50 of 89 | 7 of 14 | 43 of 75 | 11 of 19 | 39 of 70 |
| Nodal involvement | 100 of 106 | 18 of 18 | 82 of 88 | 20 of 20 | 80 of 86 |
| Leukemic disease | 32 of 102 | 5 of 16 | 27 of 86 | 5 of 20 | 27 of 82 |
| Splenomegaly | 46 of 108 | 6 of 18 | 40 of 90 | 11 of 21 | 35 of 87 |
| ASCT* | 19 of 106 | 3 of 18 | 16 of 88 | 8 of 21† | 11 of 85† |
| Parameter . | All MCL n = 110 . | VH gene mutation status . | VH gene use . | ||
|---|---|---|---|---|---|
| Mutated MCL n = 18 . | Unmutated MCL n = 92 . | VH3-21+MCL n = 21 . | MCL without VH3-21 n = 89 . | ||
| Blastoid morphology | 26 of 110 | 3 of 18 | 23 of 92 | 8 of 21 | 18 of 89 |
| Stage III–IV | 93 of 102 | 14 of 18 | 79 of 84 | 20 of 20 | 73 of 82 |
| IPI score 3-5 | 50 of 89 | 7 of 14 | 43 of 75 | 11 of 19 | 39 of 70 |
| Nodal involvement | 100 of 106 | 18 of 18 | 82 of 88 | 20 of 20 | 80 of 86 |
| Leukemic disease | 32 of 102 | 5 of 16 | 27 of 86 | 5 of 20 | 27 of 82 |
| Splenomegaly | 46 of 108 | 6 of 18 | 40 of 90 | 11 of 21 | 35 of 87 |
| ASCT* | 19 of 106 | 3 of 18 | 16 of 88 | 8 of 21† | 11 of 85† |
High-dose chemotherapy and ASCT.
ASCT at age 65 or younger: 8 of 12 VH3-21+patients versus 10 of 33 of the remaining patients with MCL.
IgH rearrangements
One hundred seventeen IgH gene rearrangements were amplified and sequenced from 110 patients, including 7 patients with double rearrangements. Sequence results of the first 51 patients have been published.13 Of the 117 sequences, most had in-frame rearrangements, whereas 9 sequences had a stop codon or an out-of-frame rearrangement. Four of these nonfunctional sequences were detected in patients who had a double rearrangement though the other rearranged allele was in-frame. Of the other 5 nonfunctional sequences, 2 were VH3-21+ rearrangements. Percentages of VH gene family use of the 117 sequences were as follows: VH1, 14%; VH2, 3%; VH3, 50%; VH4, 25%; VH5, 7%; VH6, 2%. The most frequently used VH genes were VH3-21 (18%; n = 21), VH4-34 (16%; n = 19), VH3-23 (8%; n = 9), VH5-51 (7%; n = 8), and VH1-8 (6%; n = 7). In Table2, we compared the percentage use of these VH genes with the use reported in CD5+ B cells from the PB of 2 healthy elderly donors41 and in PB B cells from 5 healthy elderly donors.42 This comparison revealed a higher frequency of the VH3-21 gene and the VH4-34 gene in MCL than in controls (P < .001). Use of the 2 most frequent VHgenes is referred to in “Discussion” in terms of percentage of 110 patients—19% for VH3-21 and 17% for VH4-34. The most commonly used diversity (D) genes were D6-13 (12%), D2-2 (10%), and D3-3 (9%). Percentages of use of the joining (JH) genes were JH1, 0%; JH2, 3%; JH3, 8%; JH4, 34%; JH5, 10%, and JH6, 33%. The D and JH genes could not be assigned for 8% and 12% of the sequences, respectively.
Most common VH genes in MCL
| VH gene . | Use in MCL, % . | Use in CD5+ B cells, % . | Use in PB B cells, % . |
|---|---|---|---|
| VH3-21 | 18 | 3 | 2 |
| VH4-34 | 16 | 10 | 10 |
| VH3-23 | 8 | 7 | 7 |
| VH5-51 | 7 | 2 | 4 |
| VH1-8 | 6 | 3 | 4 |
| VH gene . | Use in MCL, % . | Use in CD5+ B cells, % . | Use in PB B cells, % . |
|---|---|---|---|
| VH3-21 | 18 | 3 | 2 |
| VH4-34 | 16 | 10 | 10 |
| VH3-23 | 8 | 7 | 7 |
| VH5-51 | 7 | 2 | 4 |
| VH1-8 | 6 | 3 | 4 |
Somatic hypermutation status
Of the 110 patients, 18 (16%) had somatic hypermutations in their VH genes using the 2% cut-off level (Table3), and 92 (84%) were unmutated. Three patients with a double rearrangement had 1 mutated and 1 unmutated sequence; hence, they were regarded as MCL patients with mutated VH genes (patients 41, 60, and 87). The number of mutations ranged from 5 to 15, corresponding to 97.8% to 93.3% homology to the germline gene, respectively. Five mutated VH gene rearrangements were subcloned to investigate the presence of ongoing somatic hypermutation, but none of the mutated sequences showed evidence of this. Mutated VH genes were also evaluated for the percentage of mutations that occurred in the known hypermutation hotspots, the RGYW/WRCY motifs. Of the total number of mutations (n = 121) in the 18 patients, 50% (n = 60) were in these known hotspots. In addition to the 18 MCL patients with more than 2% mutations, another 17 patients contained 1% to 2% mutations in their VH genes, corresponding to 3 or 4 mutations, which may also represent somatic hypermutation. None of the VH3-21+ MCL patients had mutated VHgenes (Table 4).
MCL patients with mutated VH genes
| MCL patient . | Morphology . | VHgene . | No.mutations . | Homology, % . | D . | JH . | Frame . | Ongoing mutation . | Germline analyzed . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MCL-BV | VH3-23 | 7 | 96.9 | D2-15 | JH1-6 | IF | — | — |
| 10 | MCL | VH3-23 | 8 | 95.3 | D1-7+D5-12 | JH4 | IF | — | — |
| 11 | MCL-BV | VH4-34 | 5 | 97.7 | D1-26 | JH4 | IF | — | — |
| 20 | MCL | VH4-34 | 5 | 97.7 | D6-19 rev | JH6 | IF | — | — |
| 22 | MCL | VH3-33 | 5 | 97.8 | D1-26 | JH4 | IF | — | — |
| 37 | MCL | VH4-59 | 6 | 97.3 | D6-13 | JH4 | IF | — | Yes |
| 413-150 | MCL | VH4-30 | 7 | 96.9 | ND | JH3 | IF | — | — |
| 45 | MCL | VH3-72 | 5 | 97.8 | D1-20/D1-7 | JH4 | IF | — | Yes |
| 46 | MCL | VH4-34 | 6 | 97.3 | ND | JH6 | IF | No | Yes |
| 51 | MCL | VH3-23 | 6 | 97.3 | D4-23 | JH4 | IF | No | Yes |
| 59 | MCL | VH3-48 | 5 | 97.8 | ND | JH6 | IF | — | — |
| 603-150 | MCL | VH4-31 | 5 | 97.8 | D4-17 | JH4 | SC | — | — |
| 65 | MCL | VH1-8 | 7 | 96.8 | ND | JH1-6 | IF | — | — |
| 70 | MCL | VH3-30 | 6 | 97.3 | D6-19 | JH4 | IF | No | — |
| 75 | MCL-BV | VH3-30 | 15 | 93.3 | D6-13 | JH4 | IF | No | — |
| 873-150 | MCL | VH4-34 | 10 | 95.4 | D6-19 | JH2 | IF | — | — |
| 89 | MCL | VH3-74 | 8 | 96.4 | D7-27 | JH5 | IF | No | Yes |
| 99 | MCL-BV | VH3-74 | 5 | 97.8 | D6-13 | JH3 | IF | — | — |
| MCL patient . | Morphology . | VHgene . | No.mutations . | Homology, % . | D . | JH . | Frame . | Ongoing mutation . | Germline analyzed . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | MCL-BV | VH3-23 | 7 | 96.9 | D2-15 | JH1-6 | IF | — | — |
| 10 | MCL | VH3-23 | 8 | 95.3 | D1-7+D5-12 | JH4 | IF | — | — |
| 11 | MCL-BV | VH4-34 | 5 | 97.7 | D1-26 | JH4 | IF | — | — |
| 20 | MCL | VH4-34 | 5 | 97.7 | D6-19 rev | JH6 | IF | — | — |
| 22 | MCL | VH3-33 | 5 | 97.8 | D1-26 | JH4 | IF | — | — |
| 37 | MCL | VH4-59 | 6 | 97.3 | D6-13 | JH4 | IF | — | Yes |
| 413-150 | MCL | VH4-30 | 7 | 96.9 | ND | JH3 | IF | — | — |
| 45 | MCL | VH3-72 | 5 | 97.8 | D1-20/D1-7 | JH4 | IF | — | Yes |
| 46 | MCL | VH4-34 | 6 | 97.3 | ND | JH6 | IF | No | Yes |
| 51 | MCL | VH3-23 | 6 | 97.3 | D4-23 | JH4 | IF | No | Yes |
| 59 | MCL | VH3-48 | 5 | 97.8 | ND | JH6 | IF | — | — |
| 603-150 | MCL | VH4-31 | 5 | 97.8 | D4-17 | JH4 | SC | — | — |
| 65 | MCL | VH1-8 | 7 | 96.8 | ND | JH1-6 | IF | — | — |
| 70 | MCL | VH3-30 | 6 | 97.3 | D6-19 | JH4 | IF | No | — |
| 75 | MCL-BV | VH3-30 | 15 | 93.3 | D6-13 | JH4 | IF | No | — |
| 873-150 | MCL | VH4-34 | 10 | 95.4 | D6-19 | JH2 | IF | — | — |
| 89 | MCL | VH3-74 | 8 | 96.4 | D7-27 | JH5 | IF | No | Yes |
| 99 | MCL-BV | VH3-74 | 5 | 97.8 | D6-13 | JH3 | IF | — | — |
Ongoing mutation was investigated by subcloning in the cases indicated. Unrearranged germline genes were analyzed from T cells of the 5 patients noted.
A second rearrangement was also detected.
MCL-BV indicates blastoid variant; IF, in-frame rearrangement; SC, rearrangement with stop codon; ND, not able to define; and —, not analyzed.
MCL patients using the VH3-21 gene
| MCL patient . | Morphology . | Light-chain expression . | VHgene homology, % . | N-D-N . | D . | JH . | VH gene frame . | Vλ . | Jλ . |
|---|---|---|---|---|---|---|---|---|---|
| 5 | MCL | λ | 100 | AGGAGG-GTATTACGATTTTTGGAGTGGTTATTAT-TCTTGG | D3-3 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 144-150 | MCL | λ | 99.6 | GATCCGAC-CAGCAGCTGG-GACGTAACACCCAAAT | D6-13 | JH4 | IF | na | na |
| 23 | MCL | λ | 99.1 | GA-AGTAGTACCAGCTGTTATA-GACATA | D2-2 | JH6 | IF | OF‡ | OF‡ |
| 29 | MCL | λ | 100 | GAAATGTCAGG-CGATTTTTGGAGTGGTTAT-A | D3-3 | JH6 | IF | nt | nt |
| 32 | MCL | λ | 98.6 | GAA-TATAGCAGCAGCT-CCA | D6-13 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 33 | MCL | λ | 100 | GATTTCTCGGCCGAGTCT-TGGTTCGGGGAGTTATTAT-CTAG | D3-10 | JH6 | IF | Vλ3-19 | Jλ3 |
| 34 | MCL-BV | λ | 99.6 | GGACTCTGG-TGGAGTGGTTATTATA-G | D3-3 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 414-150 | MCL | λ | 99.6 | GATCTCACGGGCTTCGCCCTG | ND | JH6 | IF | Vλ3-19 | Jλ3 |
| 54 | MCL | λ | 100 | AAGAATCACGATTTTCT-TTTGGGG-CACTGGG | D3-16 | JH6 | IF | Vλ3-194-151 | Jλ2/3 |
| 58 | MCL-BV | κ | 100 | TAGGATCATAC-TAGCAGCT-TCT | D6-6 | JH5 | SC | na | na |
| 66 | MCL-BV | λ | 100 | GAAAGC-ACTATGATAGTAGTGGTTA-CCCAGGCGC | D3-22 | JH2 | IF | Vλ3-19 | Jλ2/3 |
| 68 | MCL | λ | 98.7 | GGGGAGAAT-TATTACGATATTTTCACTGGTTAT-CACACAAC | D3-9 | JH4 | IF | Vλ9-49 | Jλ2/3 |
| 72 | MCL-BV | λ | 100 | GCCCTTAATCACGGGGGGTGGGAA | ND | JH6 | IF | Vλ3-19 | Jλ1 |
| 74 | MCL-BV | λ | 98.7 | GATACGACATACCTGCTATGGGGGATTATGAT-GTATTACT | D3-22 | JH4 | IF | Vλ3-19 | Jλ2/3 |
| 86 | MCL-BV | λ | 99.1 | GATAGT-CAGTGGCTGG-AAAT | D6-19 | JH6 | IF | Vλ3-19 | Jλ3 |
| 91 | MCL | λ | 99.6 | GATCAGC-TACTATGGTTCGGGGAGTTATTA-CAAGCCCAGGG | D3-10 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 92 | MCL | λ | 99.6 | GATCCT-GTATTACGATATTTTGACTGG-CTAT | D3-3 | JH3 | SC | Vλ3-19 | Jλ3 |
| 97 | MCL | λ | 99.1 | GAGAATCCCCGCCCGG-TTGTGGTGGTGATTGCTA-CCCAT | D2-15 | JH3 | IF | Vλ3-194-151 | Jλ2/3 |
| 98 | MCL-BV | λ | 99.6 | GATA-ATAGTGGCTACGA-CCTACGAGG | D5-12 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 103 | MCL | λ | 99.6 | GAACCT-TACCAGCTGCTAT-TGGGA | D2-2 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 105 | MCL | λ | 100 | GAAGG-TTACGATTTTTGGAGTGGTTATT-CAA | D3-3 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| MCL patient . | Morphology . | Light-chain expression . | VHgene homology, % . | N-D-N . | D . | JH . | VH gene frame . | Vλ . | Jλ . |
|---|---|---|---|---|---|---|---|---|---|
| 5 | MCL | λ | 100 | AGGAGG-GTATTACGATTTTTGGAGTGGTTATTAT-TCTTGG | D3-3 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 144-150 | MCL | λ | 99.6 | GATCCGAC-CAGCAGCTGG-GACGTAACACCCAAAT | D6-13 | JH4 | IF | na | na |
| 23 | MCL | λ | 99.1 | GA-AGTAGTACCAGCTGTTATA-GACATA | D2-2 | JH6 | IF | OF‡ | OF‡ |
| 29 | MCL | λ | 100 | GAAATGTCAGG-CGATTTTTGGAGTGGTTAT-A | D3-3 | JH6 | IF | nt | nt |
| 32 | MCL | λ | 98.6 | GAA-TATAGCAGCAGCT-CCA | D6-13 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 33 | MCL | λ | 100 | GATTTCTCGGCCGAGTCT-TGGTTCGGGGAGTTATTAT-CTAG | D3-10 | JH6 | IF | Vλ3-19 | Jλ3 |
| 34 | MCL-BV | λ | 99.6 | GGACTCTGG-TGGAGTGGTTATTATA-G | D3-3 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 414-150 | MCL | λ | 99.6 | GATCTCACGGGCTTCGCCCTG | ND | JH6 | IF | Vλ3-19 | Jλ3 |
| 54 | MCL | λ | 100 | AAGAATCACGATTTTCT-TTTGGGG-CACTGGG | D3-16 | JH6 | IF | Vλ3-194-151 | Jλ2/3 |
| 58 | MCL-BV | κ | 100 | TAGGATCATAC-TAGCAGCT-TCT | D6-6 | JH5 | SC | na | na |
| 66 | MCL-BV | λ | 100 | GAAAGC-ACTATGATAGTAGTGGTTA-CCCAGGCGC | D3-22 | JH2 | IF | Vλ3-19 | Jλ2/3 |
| 68 | MCL | λ | 98.7 | GGGGAGAAT-TATTACGATATTTTCACTGGTTAT-CACACAAC | D3-9 | JH4 | IF | Vλ9-49 | Jλ2/3 |
| 72 | MCL-BV | λ | 100 | GCCCTTAATCACGGGGGGTGGGAA | ND | JH6 | IF | Vλ3-19 | Jλ1 |
| 74 | MCL-BV | λ | 98.7 | GATACGACATACCTGCTATGGGGGATTATGAT-GTATTACT | D3-22 | JH4 | IF | Vλ3-19 | Jλ2/3 |
| 86 | MCL-BV | λ | 99.1 | GATAGT-CAGTGGCTGG-AAAT | D6-19 | JH6 | IF | Vλ3-19 | Jλ3 |
| 91 | MCL | λ | 99.6 | GATCAGC-TACTATGGTTCGGGGAGTTATTA-CAAGCCCAGGG | D3-10 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 92 | MCL | λ | 99.6 | GATCCT-GTATTACGATATTTTGACTGG-CTAT | D3-3 | JH3 | SC | Vλ3-19 | Jλ3 |
| 97 | MCL | λ | 99.1 | GAGAATCCCCGCCCGG-TTGTGGTGGTGATTGCTA-CCCAT | D2-15 | JH3 | IF | Vλ3-194-151 | Jλ2/3 |
| 98 | MCL-BV | λ | 99.6 | GATA-ATAGTGGCTACGA-CCTACGAGG | D5-12 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 103 | MCL | λ | 99.6 | GAACCT-TACCAGCTGCTAT-TGGGA | D2-2 | JH6 | IF | Vλ3-19 | Jλ2/3 |
| 105 | MCL | λ | 100 | GAAGG-TTACGATTTTTGGAGTGGTTATT-CAA | D3-3 | JH6 | IF | Vλ3-19 | Jλ2/3 |
Second VH gene rearrangement was also detected
MCL-BV indicates blastoid variant; N, N-nucleotides.
Second out-of-frame rearrangement was also detected.
OF, 2 out-of-frame Vλ gene rearrangements detected (Vλ7-43/Jλ1 and Vλ2-23/Jλ2).
IF, in-frame rearrangement; SC, stop codon in rearrangement, but no other IgH rearrangement was detected; na, not amplifiable; nt, insufficient DNA remaining and therefore not tested; and ND, not able to define.
VH3-21+ MCL light-chain analysis
Immunophenotype data on the VH3-21+ MCL patients showed that all but one patient (patient 58) expressed λ light chains (Table 4). VL analysis was performed on all VH3-21+ MCL patients, and in-frame light-chain rearrangements were detected and sequenced for 17 patients. Sixteen patients used the Vλ3-19 gene, whereas 1 used Vλ9-49. For 2 Vλ3-19+ patients, a second rearrangement was also detected; both were out-of-frame (patient 54, Vλ9-49; patient 97, Vλ2-11). Two out-of-frame rearrangements, Vλ7-43 and Vλ2-23, were detected for patient 23. The most frequently used Jλ genes were Jλ2/3 (sequence analysis did not distinguish between these genes in most patients because they are homologous). Use of Vλ3-19 in combination with the Jλ2/3 gene occurred in 11 patients. None of the Vλ genes had more than 2% mutations.
Analysis of unrearranged germline VH gene
In 5 patients with mutated VH genes displaying 96% to 97% homology to the published germline gene, we performed further analysis of the germline DNA from T cells obtained by cell sorting. In all 5 patients, VH gene sequences from the T cells were 100% identical to the germline gene. For example, 1 of the 5 patients had a clonal VH3-23 rearrangement in the MCL sample, and VH gene analysis showed 6 mutations between FR1 and FR3, whereas the unrearranged VH3-23 germline gene from the T cells of the same patient did not have any mutations. This demonstrated that the mutations in the rearranged VH gene represented true hypermutations.
Survival analysis
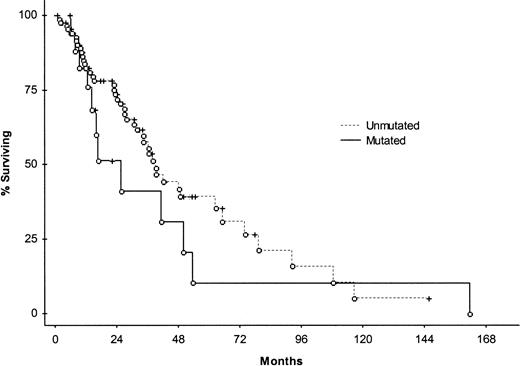
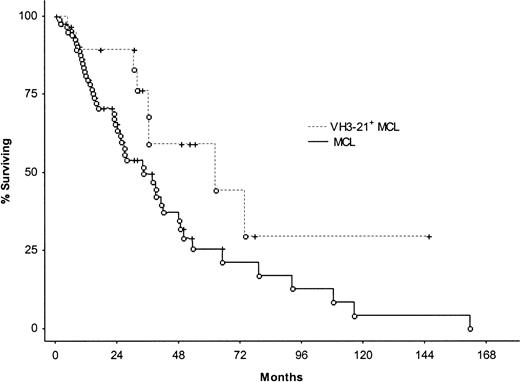
The impact of clinical and molecular parameters on survival in MCL was tested using the log-rank test (Table5). Patients 68 years and younger at diagnosis (n = 53) had significantly better survival times than those older than 68 years (n = 56); median survival times were 49 months (range, 2-162 months) and 23 months (range, 2-109 months), respectively. Other factors found to be significant predictors of survival were IPI and s-LDH level, whereas no significant differences with reference to overall survival were recorded between sex, classical MCL versus MCL-BV, stages I-II versus III-IV, nodal disease versus nonnodal disease, or the involvement of tonsil/spleen/GI tract versus noninvolvement. No association was found between mutation status and overall survival when cut-off levels of 1%, 1.5%, 2%, 2.5%, and 5% for defining a mutated case were applied. Figure1 shows the Kaplan-Meier curve comparing mutated and unmutated cases using the 2% cut-off level. Correlations were carried out between patients using specific VH genes and the remaining patients. Significantly longer overall survival was found in VH3-21+ patients compared with the rest of the material (log-rank test, P = .030); the VH3-21+ subset had a median survival time of 53 months (range, 5-146+ months) compared with 34 months (range, 2-162 months) for the remaining patients (Figure2). This subset had a lower median age at diagnosis (median, 62 years; range, 44-85 years) compared with the rest of the MCL patients (median, 70 years; range, 34-84 years), but the Mann-Whitney U test did not reveal any significant difference regarding the ages of the subsets (P = .15). Median follow-up time for the VH3-21+ patients was 33 months (range, 0-146 months), whereas the median follow-up time for the remaining MCL patients was 20 months (range, 0-162 months). Detailed clinical characteristics of the VH3-21+ patients are shown in Table6. There was no prognostic association with any of the other 4 frequently used VHgenes—VH4-34, VH3-23, VH5-51, and VH1-8.
Significance of different parameters on survival in 109 patients with MCL as determined by the log-rank test
| Parameter . | Median OS, mo . | Log-rank, P . |
|---|---|---|
| Age | < .001 | |
| 68 or younger, n = 53 | 49 | |
| Older than 68, n = 56 | 23 | |
| Sex | .88 | |
| Male, n = 89 | 39 | |
| Female, n = 20 | 35 | |
| Morphology | .72 | |
| Classical, n = 83 | 38 | |
| Blastoid variant, n = 26 | 36 | |
| Stage | .10 | |
| I-II, n = 9 | 47 | |
| III-IV, n = 92 | 37 | |
| IPI | .006 | |
| 0-2, n = 39 | 64 | |
| 3-5, n = 49 | 26 | |
| s-LDH | .004 | |
| 6.7 μkat/L or less, n = 46 | 65 | |
| More than 6.7 μkat/L, n = 46 | 25 | |
| Spleen involvement | .21 | |
| Yes, n = 45 | 36 | |
| No, n = 62 | 42 | |
| Tonsil involvement | .73 | |
| Yes, n = 17 | 38 | |
| No, n = 90 | 37 | |
| GI tract involvement | .93 | |
| Yes, n = 15 | 38 | |
| No, n = 92 | 35 | |
| VH gene mutation status | .28 | |
| Mutated, n = 18 | 18 | |
| Unmutated, n = 91 | 39 | |
| VH3-21 use | .030 | |
| Yes, n = 21 | 53 | |
| No, n = 88 | 34 |
| Parameter . | Median OS, mo . | Log-rank, P . |
|---|---|---|
| Age | < .001 | |
| 68 or younger, n = 53 | 49 | |
| Older than 68, n = 56 | 23 | |
| Sex | .88 | |
| Male, n = 89 | 39 | |
| Female, n = 20 | 35 | |
| Morphology | .72 | |
| Classical, n = 83 | 38 | |
| Blastoid variant, n = 26 | 36 | |
| Stage | .10 | |
| I-II, n = 9 | 47 | |
| III-IV, n = 92 | 37 | |
| IPI | .006 | |
| 0-2, n = 39 | 64 | |
| 3-5, n = 49 | 26 | |
| s-LDH | .004 | |
| 6.7 μkat/L or less, n = 46 | 65 | |
| More than 6.7 μkat/L, n = 46 | 25 | |
| Spleen involvement | .21 | |
| Yes, n = 45 | 36 | |
| No, n = 62 | 42 | |
| Tonsil involvement | .73 | |
| Yes, n = 17 | 38 | |
| No, n = 90 | 37 | |
| GI tract involvement | .93 | |
| Yes, n = 15 | 38 | |
| No, n = 92 | 35 | |
| VH gene mutation status | .28 | |
| Mutated, n = 18 | 18 | |
| Unmutated, n = 91 | 39 | |
| VH3-21 use | .030 | |
| Yes, n = 21 | 53 | |
| No, n = 88 | 34 |
OS indicates overall survival.
Median age at diagnosis was 68 years.
Kaplan-Meier survival curve comparing patients with mutated (n = 18) and unmutated (n = 91) disease.
No significant difference in median survival was found between the 2 subsets. Open circles represent patients who died, and crosses represent censored patients.
Kaplan-Meier survival curve comparing patients with mutated (n = 18) and unmutated (n = 91) disease.
No significant difference in median survival was found between the 2 subsets. Open circles represent patients who died, and crosses represent censored patients.
Kaplan-Meier survival curve comparing VH3-21+ MCL patients with the rest of the MCL patients.
Median survival for VH3-21+ MCL was 53 months compared with 34 months for the remaining patients. Log-rank test showed a significant difference at P = .03.
Kaplan-Meier survival curve comparing VH3-21+ MCL patients with the rest of the MCL patients.
Median survival for VH3-21+ MCL was 53 months compared with 34 months for the remaining patients. Log-rank test showed a significant difference at P = .03.
Clinical characteristics of VH3-21+MCL patients
| MCL patient . | Sex . | Age at diagnosis, y . | Stage . | Nodal disease . | Splenomegaly . | BM . | Absolute lymphocyte count . | Other extranodal sites involved . | s-LDH level . | IPI . | Treatment6-151 . | Survival, months . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | M | 60 | IVA | + | − | + | Normal | Tonsil | 7.8 | 4 | 2,3,5 | 786-152 |
| 14 | M | 66 | IVA | + | + | + | 6.9 | Gastric | Normal | 3 | 1,2,3,5 | 346-152 |
| 23 | M | 44 | IVA | + | − | − | Normal | Orbita, soft tissue | Normal | 2 | 2,4 | 1466-152 |
| 29 | M | 59 | IVA | + | + | + | 13 | – | 12.6 | 3 | 2 | 316-152 |
| 32 | M | 85 | IVA | + | ++ | + | Normal | – | 20.0 | 4 | 2 | 5 |
| 33 | M | 49 | IVA | + | − | + | Normal | – | 11.8 | 2 | 2,3,5,4 | 316-152 |
| 34 | F | 48 | IVB | + | ++ | + | Normal | Tonsil, liver | 11.6 | 4 | 1,2,3,4 | 37 |
| 41 | M | 80 | IVA | + | + | + | Normal | – | 13.1 | 4 | 2 | 66-152 |
| 54 | M | 79 | IVB | + | + | + | Normal | – | 9.6 | 4 | 2,3,1 | 526-152 |
| 58 | F | 70 | IIIA | + | ++ | − | Normal | – | 10.3 | 3 | 1,2,5 | 186-152 |
| 66 | M | 62 | IVB | + | − | + | 18 | Colon | Normal | 3 | 2,3,4,5 | 556-152 |
| 68 | F | 65 | IVA | + | ++ | + | 4.8 | Liver, pleura | 14.0 | 4 | 2,3,4,5 | 336-152 |
| 72 | M | 70 | IIIA | + | − | − | Normal | – | Normal | 2 | 0 | 8 |
| 74 | M | 49 | IVA | + | − | + | Normal | – | NA | NA | 2 | 626-152 |
| 86 | F | 79 | NA | NA | + | NA | NA | NA | NA | NA | 0 | 0 |
| 91 | F | 66 | IVA | + | − | + | Normal | – | Normal | 2 | 1,2,3 | 32 |
| 92 | M | 51 | IVA | + | − | + | Normal | – | Normal | 1 | 2,3,4,5 | 496-152 |
| 97 | M | 68 | IVB | + | ++ | + | 50 | Colon | Normal | 3 | 2 | 316-152 |
| 98 | F | 58 | IVB | + | ++ | + | Normal | – | 12.3 | 2 | 2,3,4,5 | 36 |
| 103 | M | 50 | IIIA | + | − | − | Normal | – | Normal | 1 | 2,4 | 74 |
| 105 | M | 60 | IVA | + | − | + | Normal | – | Normal | 2 | 2,3,5 | 66-152 |
| MCL patient . | Sex . | Age at diagnosis, y . | Stage . | Nodal disease . | Splenomegaly . | BM . | Absolute lymphocyte count . | Other extranodal sites involved . | s-LDH level . | IPI . | Treatment6-151 . | Survival, months . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | M | 60 | IVA | + | − | + | Normal | Tonsil | 7.8 | 4 | 2,3,5 | 786-152 |
| 14 | M | 66 | IVA | + | + | + | 6.9 | Gastric | Normal | 3 | 1,2,3,5 | 346-152 |
| 23 | M | 44 | IVA | + | − | − | Normal | Orbita, soft tissue | Normal | 2 | 2,4 | 1466-152 |
| 29 | M | 59 | IVA | + | + | + | 13 | – | 12.6 | 3 | 2 | 316-152 |
| 32 | M | 85 | IVA | + | ++ | + | Normal | – | 20.0 | 4 | 2 | 5 |
| 33 | M | 49 | IVA | + | − | + | Normal | – | 11.8 | 2 | 2,3,5,4 | 316-152 |
| 34 | F | 48 | IVB | + | ++ | + | Normal | Tonsil, liver | 11.6 | 4 | 1,2,3,4 | 37 |
| 41 | M | 80 | IVA | + | + | + | Normal | – | 13.1 | 4 | 2 | 66-152 |
| 54 | M | 79 | IVB | + | + | + | Normal | – | 9.6 | 4 | 2,3,1 | 526-152 |
| 58 | F | 70 | IIIA | + | ++ | − | Normal | – | 10.3 | 3 | 1,2,5 | 186-152 |
| 66 | M | 62 | IVB | + | − | + | 18 | Colon | Normal | 3 | 2,3,4,5 | 556-152 |
| 68 | F | 65 | IVA | + | ++ | + | 4.8 | Liver, pleura | 14.0 | 4 | 2,3,4,5 | 336-152 |
| 72 | M | 70 | IIIA | + | − | − | Normal | – | Normal | 2 | 0 | 8 |
| 74 | M | 49 | IVA | + | − | + | Normal | – | NA | NA | 2 | 626-152 |
| 86 | F | 79 | NA | NA | + | NA | NA | NA | NA | NA | 0 | 0 |
| 91 | F | 66 | IVA | + | − | + | Normal | – | Normal | 2 | 1,2,3 | 32 |
| 92 | M | 51 | IVA | + | − | + | Normal | – | Normal | 1 | 2,3,4,5 | 496-152 |
| 97 | M | 68 | IVB | + | ++ | + | 50 | Colon | Normal | 3 | 2 | 316-152 |
| 98 | F | 58 | IVB | + | ++ | + | Normal | – | 12.3 | 2 | 2,3,4,5 | 36 |
| 103 | M | 50 | IIIA | + | − | − | Normal | – | Normal | 1 | 2,4 | 74 |
| 105 | M | 60 | IVA | + | − | + | Normal | – | Normal | 2 | 2,3,5 | 66-152 |
Patient 86 died because of splenic rupture at diagnosis; therefore, information was limited.
BM indicates bone marrow involvement.
Normal s-LDH level was 6.7 μkat/L or less.
Leukemic involvement was defined by an absolute lymphocyte count greater than 4 × 109/L.
+ indicates presence; –, absence; and NA, data not available.
0 indicates no treatment; 1, chlorambucil/COP; 2, CHOP/CNOP; 3, rituximab; 4, ASCT; 5, purine/pyrimidine analogues.
Patients who are still alive.
All parameters were tested in a univariate fashion using Cox proportional hazards analysis to determine the effect on prognosis. Age (P < .001), IPI (P = .008), and s-LDH level (P = .003) were all significant. When these factors were included in multivariate analysis, only age and s-LDH level retained their importance (P = .003 and P = .038, respectively).
Discussion
For many years, it has been assumed that MCL harbors unmutated VH genes and therefore originates from naive B cells. However, in a recent report from our group investigating 51 MCL patients, we showed that approximately 20% of patients with MCL had somatic hypermutations (more than 2%) within their VHgenes.13 In this follow-up study, we extended our cohort to 110 patients, and we demonstrate that MCL comprises a mutated (16%; n = 18) and an unmutated (84%; n = 92) group. These data shed light on the postulated cell of origin of MCL because the presence of somatic hypermutations in a subset of MCL implies that these lymphomas may originate from a B cell that has been exposed to the GC environment. To ensure that the observed mutations truly represent somatic hypermutations and not single nucleotide polymorphisms, we analyzed the unrearranged VH gene in T cells from 5 selected patients whose corresponding rearranged clonal VHgene carried mutations in the range 2.2% to 3.6%. For each of these 5 patients, the unrearranged VH gene was 100% homologous to the published germline sequence, confirming that the mutations corresponded to hypermutations in these patients. Additionally, analysis of the amount of mutations that occurred in the known hypermutation hotspot regions (RGYW/WRCY motifs) showed that 50% of the total number of mutations (n = 121) were in these sequence motifs, further substantiating the data. We also investigated whether there were ongoing somatic hypermutations but did not find evidence of this in the 5 mutated cases analyzed. This indicates that the clonal B cells in MCL patients with mutated VH genes are not under prolonged antigenic stimulation in the GC and probably derive from a post-GC B cell. We believe that most MCLs derive from pre-GC B cells residing in the follicular mantle but that a small subset has been exposed to the GC environment.
An alternative explanation for mutated VH genes could be that the precursor lymphoma cells have undergone hypermutation outside the GC. Recently, it was reported that patients with X-linked hyper IgM syndrome, which is characterized by a mutation in the CD40Lgene and the inability to form GCs, can still have a subset of B cells (CD27+IgM+IgD+) with mutated VH genes.43 This is suggestive of the existence of a second diversification pathway to acquire hypermutations without the classical interaction between B and T cells. The rate of somatic hypermutations in these CD27+IgM+IgD+ B cells was generally low (approximately 1%-2%) in patients with hyper IgM syndrome. However, little is known about this separate route, and it remains to be investigated whether different B-cell subsets, such as MCL precursors, acquire hypermutations by this process.
In CLL, VH gene mutation status may be used to group patients into 2 prognostic subsets, in which patients with mutated VH genes have an overall survival time approximately twice as long as that of patients with unmutated VHgenes.14-16 In contrast, we could not find any significant differences in median survival times when comparing mutated and unmutated MCL at the 2% cut-off level. This mutation border, which has been empirically derived for CLL to avoid counting Taq polymerase errors and polymorphisms as hypermutations,14 may, however, not be appropriate for defining a mutated case in MCL. There is a large difference in the percentage of mutated cases in each disease (approximately 40%-50% in CLL vs 16% in MCL), and the mean mutation percentage of the mutated group is lower in MCL than in CLL14 (3% vs approximately 6%). In addition to the 18 patients with mutated VH genes, another 17 patients had VH genes with a mutation rate of 1% to 2%, which may also represent mutated MCL. Therefore, we performed survival analysis using other percentage cut-off levels for defining a mutated case (1%-5%), but similar results were obtained using different borders. Hence, in our cohort of MCL patients, VH gene mutation status was not prognostically useful. Interestingly, it has been reported in an abstract by Garand et al44 that patients with nonnodal MCL (patients without clinical lymphadenopathy) have mutations more frequently (19 of 34 patients) than patients with nodal engagement (5 of 31 patients). They stated that survival did not differ significantly between mutated and unmutated MCL but that there was a tendency toward improved survival in the mutated subset. They also showed that patients with nonnodal MCL had a less aggressive disease than nodal patients.44 The proportion of MCL patients without lymphadenopathy seems generally small. Only 6 patients in our cohort had nonnodal disease, all of which were unmutated. Further analysis of this subset would be of interest to determine whether mutation status has a greater clinical impact within this rare entity.
Biased use of individual VH genes has been demonstrated in several entities of B-cell lymphoma, as mentioned. Restricted use of the VH4-34 gene in MCL has been indicated through an immunohistochemical detection method.21 This gene has also been found overused in a small series of MCL patients.12In our earlier study, we confirmed the overrepresentation of the VH4-34 gene in MCL but could also show restricted use of VH3-21.13 In this report, the 2 VHgenes were still overused, with VH3-21 representing 19% of patients and VH4-34 representing 17% of patients. In addition, our finding of skewed VH3-21 gene use was recently supported by other groups, who have found increased use of VH3-21 in MCL.45-47 Preferential use of the VH4-34 gene has been found in several other B-cell malignancies, such as CLL, diffuse large B-cell lymphoma, and primary central nervous system (CNS) lymphoma,18,21,23,24 but this VH gene has also been found to be involved in autoimmunity, encoding antibodies to self-antigens31,32 and nonself-antigens.21,28 The current hypothesis in lymphoproliferative disease is that B cells using specific VH genes encode immunoglobulin molecules that may be stimulated by various antigens, resulting in an increased proliferation rate that makes them more susceptible to transformation.19,21,22,27 The VH3-21 gene has also been associated with autoimmunity—it has been shown to be involved in the production of autoantibodies in rheumatoid arthritis and Sjögren syndrome.29,30 We have recently reported a biased use of the VH3-21 gene in mutated CLL.26 Patients showed specific phenotypic and genotypic features, such as clonal λ light chain expression, restricted Vλ gene use (V2-14), and short, highly homologous CDR3s. Survival analysis also revealed that this subset of patients had worse overall survival than other mutated CLL.
In contrast to CLL, VH3-21+ MCL patients (n = 21) had unmutated VH genes and did not show any specific common features within CDR3. In parallel with CLL, they showed λ light-chain expression in all but one patient, and sequence analysis of 16 of 18 patients showed a strikingly skewed Vλ gene use of the Vλ3-19 gene (Table 4). This intriguing finding of restricted Vλ gene use in VH3-21+ MCL implicates a possible role for antigen(s) in the pathogenesis of this subset of MCL. VH3-21+ patients also had longer survival than the rest of the MCL patients, with a median overall survival time of 53 months versus 34 months (P = .03; Figure 2). Although these patients tended to be younger at diagnosis, they had no specific clinical characteristics that could account for the improved prognosis (Tables 1, 6). VH3-21+ patients were typical MCL patients with extensive nodal and extranodal disease, and could not be considered indolent because they had the usual spectrum of adverse prognostic factors. However, a large proportion of patients in this group underwent ASCT, which could be one reason for their prolonged survival (Table 1). This still does not explain, however, why these patients had a tendency to present with MCL earlier than the other patients. Indeed, our molecular data implicate that VH3-21+ patients constitute a new biologic entity considering the highly biased VH/Vλuse, but it remains to be evaluated in prospective studies whether they also represent a separate clinical subset with a different prognosis.
We thank Carin Backlin and Eva Pallin for their skillful technical assistance.
Prepublished online as Blood First Edition Paper, March 13, 2003; DOI 10.1182/blood-2002-11-3479.
Supported by grants from the Swedish Cancer Society, Lion's Cancer Research Foundation, Umeå University and Uppsala University, Gunnar Nilsson's Cancer Foundation, and the research foundation of the Department of Oncology at Uppsala University.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Richard Rosenquist, Department of Genetics and Pathology, Rudbeck Laboratory, Dag Hammarskjölds väg 20, Uppsala University, SE-751 85 Uppsala, Sweden; e-mail:richard.rosenquist@genpat.uu.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal