The leukemogenic tyrosine kinase Bcr-Abl contains a highly conserved inhibitor-binding pocket (IBP), which serves as a binding site for imatinib mesylate. Mutations at the IBP may lead to resistance of the Abl kinase against imatinib mesylate. To examine the mechanisms of imatinib mesylate binding and resistance in more detail, we created several point mutations at amino acid positions 315 and 380 of Abl, blocking the access to the IBP and rendering Bcr-Abl imatinib mesylate–resistant. Moreover, introduction of a mutation destabilizing the inactive conformation of Abl (Asp276Ser/Glu279Ser) also led to imatinib mesylate resistance, suggesting that the inhibitor required inactivation of the kinase prior to binding. These Bcr-Abl mutants were then used to evaluate the binding mode and specificity of 2 compounds, PP1 and CGP76030, originally characterized as Src kinase inhibitors. Both compounds inhibited Bcr-Abl in a concentration-dependent manner by overlapping binding modes. However, in contrast to imatinib mesylate, PP1 and CGP76030 blocked cell growth and survival in cells expressing various inhibitor-resistant Abl mutants. Studies on the potential signaling mechanisms demonstrated that in cells expressing inhibitor-resistant Bcr-Abl mutants, PP1 and CGP76030 inhibited the activity of Src family tyrosine kinases and Akt but not signal transducer and activator of transcription–5 (STAT5) and JUN kinase (Jnk). The results suggest that the use of Src kinase inhibitors is a potential strategy to prevent or overcome clonal evolution of imatinib mesylate resistance in Bcr-Abl+ leukemia.
Introduction
The c-Abl is a nonreceptor tyrosine kinase that contributes to several leukemogenic fusion proteins, including Bcr-Abl. The latter was found to be the hallmark of chronic myeloid leukemia (CML), a clonal malignancy of a hematopoietic stem cell. In addition, Bcr-Abl is detectable in a subset (25% to 30%) of patients with acute lymphoblastic leukemias (ALLs). Fusion of c-Abl to Bcr is caused by a reciprocal translocation between chromosomes 9 and 22 and has been shown to result in constitutive activation of the Abl kinase domain.1
Imatinib mesylate (STI571) is an adenosine triphosphate (ATP)–competitive inhibitor of Abl. Several clinical trials revealed that imatinib mesylate induces hematologic remissions in up to 98% of patients, depending on the disease phenotype and stage.2-5 The lack of dose-limiting toxicity obviously reflects the specificity of imatinib mesylate. In addition to Abl, only Arg, as well as c-Kit and platelet-derived growth factor (PDGF) receptor (PDGF-R) kinases, have been reported to be inhibited by imatinib mesylate.6 7
A recently published crystal structure of Abl in complex with an analog of imatinib mesylate suggested that the high specificity of imatinib mesylate results from binding into an inhibitor-binding pocket (IBP), which is located in close proximity to the ATP-binding site.8 In Abl, 6 amino acids, alanine (Ala) at residue 269, lysine (Lys) at residue 271, valine (Val) at residue 299, threonine (Thr) at residue 315, alanine (Ala) at residue 380, and phenylalanine (Phe) at residue 382 (Figure 1A), define the entrance of the IBP. Two of these positions, residues 315 and 380, show high variability among different protein kinases. Position 315 is occupied by either threonine (Thr), valine (Val), leucine (Leu), isoleucine (Ile), methionine (Met), glutamine (Gln) or phenylalanine (Phe), and position 380 by either alanine (Ala) cysteine (Cys), serine (Ser) or threonine (Thr). Interestingly, in most imatinib mesylate–insensitive kinases, the side chains of the amino acids at these positions are bulkier than in Abl.
Effect of amino acid substitutions at positions 315 and 380 on inhibition of Bcr-Abl by imatinib mesylate.
(A) Stick representations of imatinib mesylate in complex with Abl or mutants of Abl at position 315 to methionine and 380 to threonine. Mutants were introduced into the Abl structure by means of Swiss Prot Structure Viewer software and are based on previously published coordinates of the Abl/imatinib mesylate analog complex.8(B) Cos7 cells were transfected with either pApuro, Bcr-Ablwt, or the indicated mutants of Bcr-Abl at positions 315 and 380. Cells were either left untreated or incubated with 10 μM imatinib mesylate at 4 hours prior to lysis. Phosphorylation of Bcr-Abl was assessed by means of anti–phospho-Abl antibodies. Expression of Bcr-Abl was controlled by means of anti-Abl antibody. (C) Survival and proliferation of 32D cells expressing Bcr-Ablwt or the various mutants of Bcr-Abl at positions 315 and 380. First, 5 × 104 cells were seeded per 6 wells and treated with DMSO, 1 or 2.5 μM imatinib mesylate. Survival and proliferation were determined 48 hours later as described in “Materials and methods.” Data are shown as means ± SD.
Effect of amino acid substitutions at positions 315 and 380 on inhibition of Bcr-Abl by imatinib mesylate.
(A) Stick representations of imatinib mesylate in complex with Abl or mutants of Abl at position 315 to methionine and 380 to threonine. Mutants were introduced into the Abl structure by means of Swiss Prot Structure Viewer software and are based on previously published coordinates of the Abl/imatinib mesylate analog complex.8(B) Cos7 cells were transfected with either pApuro, Bcr-Ablwt, or the indicated mutants of Bcr-Abl at positions 315 and 380. Cells were either left untreated or incubated with 10 μM imatinib mesylate at 4 hours prior to lysis. Phosphorylation of Bcr-Abl was assessed by means of anti–phospho-Abl antibodies. Expression of Bcr-Abl was controlled by means of anti-Abl antibody. (C) Survival and proliferation of 32D cells expressing Bcr-Ablwt or the various mutants of Bcr-Abl at positions 315 and 380. First, 5 × 104 cells were seeded per 6 wells and treated with DMSO, 1 or 2.5 μM imatinib mesylate. Survival and proliferation were determined 48 hours later as described in “Materials and methods.” Data are shown as means ± SD.
It was suggested that a third gatekeeping amino acid position, Phe382, acted as a determinant of specificity. In imatinib mesylate–inhibited Abl, Phe382 adopts an orientation that markedly increases the access to the IBP. This orientation is different from the configuration found in the inactive structures of kinases such as Src, hematopoietic cell kinase (Hck), fibroblast growth factor receptor tyrosine kinase (FGF-RK), or EphB2-RK.9-12 This specific orientation of Phe382 in inactive Abl seemed to depend closely on the conformation of the activation loop, which in many kinases structurally varies considerably between the active and inactive states. Therefore, it was hypothesized that the activation loop (A-loop) of Abl needs to adopt the inactive conformation prior to imatinib mesylate binding.
Emerging data about the resistance of patients to imatinib mesylate has somewhat reduced the initial enthusiasm about imatinib mesylate.2,13 Gorre and colleagues13have reported Bcr-Abl gene amplification and a point mutation at position 315 to isoleucine to be potential causes of such resistances, the latter observation corroborating the importance of this position for imatinib mesylate binding. Other mutations, such as Tyr253His, Tyr253Phe, Glu255Lys, or Glu255Val, have been reported. Their mechanism of reducing imatinib mesylate sensitivity, however, remains not fully understood, as most residues are not directly involved in imatinib mesylate binding.14-16 Moreover, these residues are not conserved among imatinib mesylate–sensitive kinases, and some of the residues, such as phenylalanine at position 253 or lysine at position 255, are actually found at homologous positions in PDGF-R and c-Kit.
In addition, the up-regulation of expression of signaling molecules such as Bruton tyrosine kinase (Btk) or Src family kinases has been observed in cells from imatinib mesylate–resistant patients.17,18 It was hypothesized that such signaling molecules might at least in part substitute for Abl kinase activity to induce important signaling pathways. Interestingly, we were previously able to show that Bcr-Abl activates Src family kinases in an Abl-kinase–independent manner and that active Src kinases are able to induce phosphorylation of Bcr-Abl and its interaction with growth factor receptor-bound protein 2 (Grb2).19,20Moreover, imatinib mesylate does not inhibit Src family kinases directly,21 further corroborating the possible importance of Src kinases in the development of clinical imatinib mesylate resistance.
This study was undertaken to investigate the use of dual-specific Src/Abl kinase inhibitors to treat Bcr-Abl+ leukemias and to set up an experimental system that would enable us to dissect the biologic properties of such inhibitors. We show here that dual-specific Src/Abl inhibitors target Src kinases to induce growth arrest and apoptosis in Bcr-Abl+ cells independently of Abl inhibition. The results suggest the use of dual-specific Src/Abl-inhibitors as a potential strategy to prevent or overcome imatinib mesylate resistance in Bcr-Abl+ leukemias.
Materials and methods
Antibodies
Rabbit polyclonal antibodies (abs) to Lyn(44), Fyn (Fyn3), Src (N16), STAT5 (C-17), Akt (N19), and Jnk (FL) as well as antiphosphotyrosine antibody (PY99) were obtained from Santa Cruz Biotechnology (CA). The monoclonal α-Abl ab Ab3 was purchased from Oncogene Sciences (Uniondale, NJ). Phospho-specific polyclonal antibodies to Abl (pTyr245), STAT5 (pTyr694), Jnk (pThr183/pTyr185), Akt (pThr308, pSer473) and Src kinases (pTyr416) were obtained from Cell Signaling (Beverly, MA). All secondary abs for Western blot were provided by Amersham (Braunschweig, Germany).
Tyrosine kinase inhibitors
The pyrazolo-pyrimidine derivative PP1 was bought from Alexis Biochemicals (San Diego, CA) and dissolved in dimethyl sulfoxide (DMSO) to a concentration of 25 mM. Imatinib mesylate and CGP76030 were from Novartis Pharmaceuticals (Basel, Switzerland).
Plasmids and site-directed mutagenesis
To introduce point mutations into Bcr-Abl, aKpnI/Eco47III subfragment of Bcr-Abl containing the sequence coding for Bcr-Abl's kinase domain was cloned into pUCΔNdeI/XbaI, engineered by site-directed mutagenesis to contain a Eco47III site in the polylinker. Cloning of pUCΔNdeI/XbaI was described elsewhere.20 After introduction of point mutations, this fragment was first recloned into pcDNA3abl. Thereafter, the 5′ part of abl up to the KpnI site was replaced by Bcr coding sequences, and the respective part of Abl by aKpnI-fragment from pcDNA3bcr-abl. All mutations were confirmed by sequencing. For expression in COS7 and 32D cells, cDNAs were cloned into the EcoRI sites of pApuro or pMSCV (murine stem cell virus).
Cell lines
Parental 32D cells as well as 32D cells expressing Bcr-Abl and mutants thereof (32Dp210) were grown in Iscove modified Dulbecco media (IMDM) supplemented with 10% fetal bovine serum (FBS). COS7 cells were cultured in Dulbecco modified eagle medium (DMEM) containing 4.5 g/L glucose) and supplemented with 10% FBS. K562, LAMA, U937, HL60, and OCI-AML5 cells were grown in RPMI supplemented with 10% FBS. All media and FBS were purchased from Gibco Life Technologies (Karlsruhe, Germany).
Transfection of cells
COS7 cells were transfected with the use of Effecten transfection reagent according to the guidelines of the manufacturer (Quiagen, Hilden, Germany).
Generation of retroviral stocks
For generation of retroviral stocks, all DNAs were purified by means of endotoxin-free DNA preparation kits (Quiagen). With the use of a conventional calcium precipitation, 293T cells were transfected with 10 μg retroviral vector DNA(pMSCV) and 5 μg MSCV-ecopac, an ecotropic packaging construct, in a 6-cm dish. Medium was changed at 24 hours, and virus-containing medium was harvested at 48 hours after transfection. Virus titer for neomycin resistance was determined by transduction of Rat1 fibroblast cells.
Retroviral infection of 32D cells
First, 32D cl3 cells were grown in RPMI media containing 10% FBS and 10% WEHI supernatant. Then, 5 × 104 cells per 24 wells were transduced with equal multiplicities of infection (MOIs) of retroviral stocks in a total volume of 500 μL in the presence of polybrene for 4 to 5 hours. Thereafter, 1.5 mL medium was added, and cells were incubated at 37°C. At 48 hours after infection, G418 was added at a concentration of 1 mg/mL. Finally, 4 days after infection, cells were selected for interleukin 3 (IL-3) independence. At day 6, dead cells were removed by density centrifugation; viable cells were counted and plated at 5 × 104 cells per milliter in a TC25 flask in the presence of either vehicle (DMSO) or various kinase inhibitors (imatinib mesylate, PP1, CGP76030). The number of viable cells and the rate of cell death were determined 48 hours later.
Cell lysis
COS7 cells were lysed as described recently.20 For lysis of 32D cells, cells were harvested and washed twice in cold phosphate-buffered saline (PBS). For experiments evaluating the activity profile of PP1 or imatinib mesylate, cells were incubated with the indicated concentrations of inhibitor or with DMSO at a density of 5 × 106 cells per milliliter for 1.5 to 2 hours. Then, 107 cells were lysed in 100 μL lysis buffer containing 1% Nonidet P-40 (NP-40), 20 mM Tris (tris(hydroxymethyl)aminomethane) (pH 8.0), 50 mM NaCl, and 10 mM EDTA (ethylenediaminetetraacetic acid), 1 mM phenylmethylsulfonylfluoride, 10 μg/mL aprotinin, 10μg/mL leupeptin, and 2 mM sodium orthovanadate. After resuspension in lysis buffer, cells were incubated for 25 minutes on ice. Finally, insoluble material was removed by centrifugation at 15 000 g. Clarified lysates were checked for protein concentrations by means of a BioRad (BioRad Laboratories, Muenchen, Germany) protein assay.
Gel electrophoresis and immunoblotting
Gel electrophoresis and immunoblotting were performed as described with some minor modifications.20 Proteins were routinely blotted on nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany). Membranes were blocked in 5% milk powder for 1 hour. Primary and secondary antibodies were diluted 1:500 to 1:5000 in Tris-buffered saline (TBS) containing 1% milk powder. Phospho-specific antibodies were used according to the manufacturer's guidelines. Proteins were detected by means of the ECL or ECL Plus detection system as recommended by the manufacturer (Amersham).
Detection of apoptosis by flow cytometry
For assessing apoptosis induced by the various kinase inhibitors, cells were incubated with the indicated concentrations of PP1 or imatinib mesylate at a density of 5 × 104/mL. Apoptosis was routinely assessed by measuring the binding of fluorescein isothiocyanate (FITC)–conjugated annexin V to the membranes of cells undergoing apoptosis. About 5 × 104 cells were taken at the indicated time points and washed once in PBS. Thereafter, cells were resuspended in 195 μL annexin V binding buffer, and 5μL annexin V–FITC (Bender MedSystems Diagnostics, Vienna, Austria) was added. Cells were mixed and incubated at room temperature for 10 to 20 minutes. Afterward, cells were pelleted again, washed once, and resuspended in 190 μL annexin V binding buffer. Then, 10 μL of a 20-μg/mL propidium iodide stock solution was added, and the ratio of annexin V+ to annexin V− cells was determined by fluorescence-activated cell sorting (FACS) analysis by means of a Coulter EPICS XL 4-color cytometer (Beckman Coulter, Unterschleissheim, Germany).
Generation of computer-animated structural data
Figures containing structural data were prepared by means of WebLab ViewerPro software (Cambridge, United Kingdom) and are based on the previously published coordinates of the Abl/imatinib mesylate crystal structure complex.8 Amino acid exchanges were introduced into this structure by means of Swiss Prot Structure Viewer software (Glaxo SmithKline, Geneva, Switzerland).
Results
Mutation of positions Thr315 and Ala380 of Abl to bulky amino acids induces resistance to imatinib mesylate
Computer models of the Abl/imatinib mesylate complex with mutations Thr315Met or Ala380Thr suggested that bulkier amino acid side chains at these positions restrict the access for imatinib mesylate to an IBP, located adjacent to the ATP-binding site (Figure1A). Thus, amino acid positions 315 and 380 might act as molecular gatekeepers controlling the access for imatinib mesylate to the IBP in individual protein kinases. To prove this genetically, mutations to valine, leucine, isoleucine, methionine, glutamine, or phenylalanine at position 315 and to cysteine or threonine at position 380 were generated; these represent most of the amino acids found at these positions in other kinases. When expressed in Cos7 cells, all mutants at position 315 showed a similar, or slightly enhanced, spontaneous kinase activity as compared with wild-type Bcr-Abl (Bcr-Ablwt) (Figure 1B, and data not shown). In contrast, activity of mutants Ala380Cys and Ala380Thr was decreased by approximately 50% (Figure 1B and data not shown). More important, imatinib mesylate inhibited substrate and autophosphorylation (pTyr245-Abl) of Bcr-Ablwt in a concentration-dependent manner (data not shown), but failed to inhibit mutants Thr315Val, Thr315Leu, Thr315Ile, Thr315Met, Thr315Gln, and Thr315Phe as well as Ala380Thr at concentrations up to 10 μM (Figure 1B).
We next introduced Bcr-Ablwt and the various mutants into the murine IL-3–dependent cell line 32D. Concentration kinetics showed that exposure of 32DBcr-Ablwt cells to about 1 to 2.5 μM imatinib mesylate was both necessary and sufficient to induce a complete stop of cell proliferation and to cause apoptosis in more than 90% of cells within 2 days (data not shown). These effects correlated to a complete down-regulation of Bcr-Abl autophosphorylation as well as inhibition of STAT5 and Jnk phosphorylation/activation (data not shown). In contrast, when imatinib mesylate–resistant Bcr-Abl mutants were expressed in 32D cells, both the blocking of cell proliferation and the induction of apoptosis caused by 1 and 2.5 μM imatinib mesylate were completely abolished (Figure 1C). Comparable results were obtained for all mutants except Ala380Cys, which did not confer any resistance to imatinib mesylate in 32D cells (Figure 1C). Resistance of cells expressing mutant Ala380Thr was incomplete, with about 20% to 25% apoptotic cells and a considerable reduction of cell proliferation at 2.5 μM imatinib mesylate (Figure1C). Taken together, the results confirmed the importance of positions 315 and 380 as molecular gatekeepers controlling the access to the IBP, thereby determining the specificity of imatinib mesylate for Abl.
Destabilization of the inactive conformation of the A-loop decreases the sensitivity of Abl to imatinib mesylate
Adoption of the inactive conformation of the A-loop was suggested to be indispensable for imatinib mesylate binding.8 To prove this hypothesis, we studied the intramolecular interactions of the A-loop of imatinib mesylate–inhibited Abl with other regions of the enzyme. We hypothesized that the disruption of such interactions would destabilize the A-loop, thereby hyperactivating Abl and rendering it less sensitive to imatinib mesylate. This led us to the identification of an H-bond system between Arg386, located on the A-loop, and Asp276/Glu279, located on the β3-αC-loop (Figure 2A). Importantly, this motive was found to be highly conserved within imatinib mesylate–sensitive kinases (not shown), and none of these amino acids contribute directly to imatinib mesylate binding.8
Genetic characterization of a possible role of Phe382 (F382) as a functional gatekeeper controlling imatinib mesylate binding.
(A) Schematic representation of the interaction between the A-loop and the β3-αC-loop mediated by H-bonds between Arg386 (R386) and Asp276/Glu279 (D276/E279). (B) Cos7 cells were transfected with 1 μg pApuro, Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser. Activity of the various Bcr-Abl mutants was assessed by means of phospho-Abl and phosphotyrosine antibodies. (C) Phospho-Abl and Abl immunoblots of lysates from Cos7 cells transfected with either pApuro, Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser. Cells were either left untreated or incubated with the indicated concentrations of imatinib mesylate. (D) Proliferation (top panel) and survival (bottom panel) of 32D cells expressing either Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser grown in the presence of the indicated concentrations of imatinib mesylate. Data are shown as means ± SD.
Genetic characterization of a possible role of Phe382 (F382) as a functional gatekeeper controlling imatinib mesylate binding.
(A) Schematic representation of the interaction between the A-loop and the β3-αC-loop mediated by H-bonds between Arg386 (R386) and Asp276/Glu279 (D276/E279). (B) Cos7 cells were transfected with 1 μg pApuro, Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser. Activity of the various Bcr-Abl mutants was assessed by means of phospho-Abl and phosphotyrosine antibodies. (C) Phospho-Abl and Abl immunoblots of lysates from Cos7 cells transfected with either pApuro, Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser. Cells were either left untreated or incubated with the indicated concentrations of imatinib mesylate. (D) Proliferation (top panel) and survival (bottom panel) of 32D cells expressing either Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser grown in the presence of the indicated concentrations of imatinib mesylate. Data are shown as means ± SD.
To test the importance of this interaction for kinase regulation and imatinib mesylate binding, both residues were mutated to serine. When expressed in Cos7 cells, the resultant mutant, Bcr-AblAsp276Ser/Glu279Ser, displayed an increased kinase activity (Figure 2B) and a markedly decreased sensitivity toward imatinib mesylate inhibition (Figure 2C). Moreover, if expressed in 32D cells, the inhibitory concentration 50% (IC50) for inhibition of cell proliferation by imatinib mesylate was increased about 10-fold, from 0.069 μM in 32DBcr-Ablwt cells to 0.71 μM in 32DBcr-AblAsp276Ser/Glu276Ser cells (Figure 2D, top panel), and the rate of apoptotic cell death at 2 μM imatinib mesylate was reduced from greater than 90% to about 30% (Figure 2D, bottom panel).
The residue Arg386 was shown to interact with conserved autophosphorylated tyrosines located on the A-loop in structures of active kinases, such as lymphoid T-cell protein tyrosine kinase (Lck) or insulin receptor kinase (IRK).22 23 This suggested that dephosphorylation of Tyr393, the major autophosphorylation site of Abl, is another prerequisite for imatinib mesylate binding. We hypothesized that a mutant of this autophosphorylation site to phenylalanine would not be dephosphorylated prior to imatinib mesylate binding and would therefore possess increased sensitivity to imatinib mesylate. Surprisingly, however, the mutation of the respective autophosphorylation site in Bcr-Abl to phenylalanine (Tyr393Phe) had little effect on the sensitivity of Bcr-Abl to imatinib mesylate in Cos7 and almost no effect in 32D cells (Figure 2C-D), suggesting that dephosphorylation of Tyr393 by endogenous phosphatases is not a rate-limiting factor for imatinib mesylate binding.
PP1 and CGP76030 inhibit Bcr-Abl and induce growth arrest and apoptosis in Bcr-Abl+ cells
The rapid development of direct imatinib mesylate resistance during therapy of patients with advanced Bcr-Abl+ leukemias suggests that inhibition of additional signaling molecules might be needed to treat primary Bcr-Abl+ B-lineage ALL (B-ALL) or CML in blast crisis. We recently showed that Bcr-Abl activates—by an Abl kinase independent mechanism—Src family kinases, which are not directly inhibited by imatinib mesylate.19,20 The lack of inhibition of Src kinases by imatinib mesylate might be due to differences of Abl and Src kinases in the conformation of the A-loop and the orientation of Phe382, only.8 Still, Bcr-Abl and Src kinases share high homology in terms of the amino acids forming the imatinib mesylate–binding pocket, and as a consequence, many Src family kinase inhibitors do indeed show cross-reactivity on Abl kinase. We used the Bcr-Abl mutants to validate the binding mode and biologic action of 2 such dual-specific Src/Abl kinase inhibitors, PP1 and CGP76030 (Figure 3A-B).24 25
As shown in Figure 3C-D, both compounds, PP1 and CGP76030, inhibited Bcr-Abl in a concentration-dependent manner. Approximately 25 to 50 μM PP1 or 5 to 10 μM CGP76030 were necessary to inhibit substrate phosphorylation by, and autophosphorylation of, Bcr-Abl in Cos7 and 32DBcr-Ablwt cells (Figure 3C-D, and data not shown). Interestingly, in contrast to the close correlation of inhibition of substrate and autophosphorylation of Bcr-Abl caused by imatinib mesylate (data not shown), Bcr-Abl inhibited by PP1 and CGP76030 retained considerable levels of autophosphorylation at concentrations that completely abolished substrate phosphorylation (Figure 3C-D and data not shown). Both compounds induced growth arrest and apoptosis in 32DBcr-Ablwt cells; 50 μM PP1 (Figure 3E, left panel) and 5 μM CGP76030 (Figure3F, left panel) were equivalent in potency to 2.5 μM imatinib mesylate. This effect correlated well with the concentrations needed for inhibition of Bcr-Abl and Src kinases in 32DBcr-Ablwt cells, as assessed by immunoblots with phospho-specific antibodies (Figure 7C and data not shown). However, in marked contrast to imatinib mesylate, both inhibitors also reduced survival and proliferation of parental 32D cells, probably reflecting the inhibition of Src kinases in IL-3–dependent signaling, which is well known.26 The action of both compounds on parental 32D cells correlated with their inhibition of Src kinase and Akt kinase activity (data not shown). However, since PP1 and CGP76030 also acted on parental 32D cells, it was not possible to link the biologic effects of PP1 and CGP76030 to the inhibition of either Bcr-Abl or Src kinases in these cells.
PP1 and CGP76030 inhibition of Bcr-Abl.
PP1 and CGP76030 inhibit Bcr-Abl in a concentration-dependent manner to induce apoptosis in 32DBcr-Ablwt cells. (A-B) Structural formula of the kinase inhibitors CGP76030 and PP1. (C-D) Cos7 cells were transfected with either pApuro or Bcr-Ablwt. Cells were either left untreated or incubated with the indicated concentrations of CGP76030 and PP1 4 hours prior to lysis. Phosphorylation of Bcr-Abl and cellular proteins was assessed with the use of anti–phospho-Abl or antiphosphotyrosine antibodies. Expression of Bcr-Abl was controlled by means of anti-Abl antibody. (E-F) Survival (solid line) and proliferation (dashed line) of 32DBcr-Ablwt cells in the presence of the indicated concentrations of PP1 (panel E) or CGP76030 (panel F).
PP1 and CGP76030 inhibition of Bcr-Abl.
PP1 and CGP76030 inhibit Bcr-Abl in a concentration-dependent manner to induce apoptosis in 32DBcr-Ablwt cells. (A-B) Structural formula of the kinase inhibitors CGP76030 and PP1. (C-D) Cos7 cells were transfected with either pApuro or Bcr-Ablwt. Cells were either left untreated or incubated with the indicated concentrations of CGP76030 and PP1 4 hours prior to lysis. Phosphorylation of Bcr-Abl and cellular proteins was assessed with the use of anti–phospho-Abl or antiphosphotyrosine antibodies. Expression of Bcr-Abl was controlled by means of anti-Abl antibody. (E-F) Survival (solid line) and proliferation (dashed line) of 32DBcr-Ablwt cells in the presence of the indicated concentrations of PP1 (panel E) or CGP76030 (panel F).
To further evaluate the potential of CGP76030 to reduce cell growth and survival of Bcr-Abl+ cells, 2 Bcr-Abl+ cell lines (K562 and LAMA84) and a number of Bcr-Abl− leukemic cell lines (U937, HL-60, OCI-AML5) were incubated with various concentrations of imatinib mesylate or CGP76030. Similarly to imatinib mesylate, CGP76030 inhibited cell growth and survival only in Bcr-Abl+ cell lines (Figure 4A-B), suggesting that this compound was not generally cytotoxic but rather acted by a specific mechanism. Similar results were obtained for PP1 (data not shown).
Imatinib mesylate, PP1, and CGP76030 share overlapping binding modes
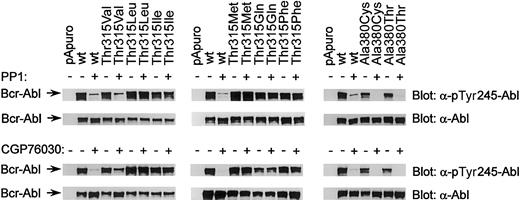
Using the mutants, we evaluated the binding of PP1 and CGP76030 to Abl in more detail. Similarly to imatinib mesylate, PP1 (100 μM) and CGP76030 (25 μM) failed to inhibit Bcr-Abl in cells expressing the mutants Thr315Leu, Thr315Ile, Thr315Met, Thr315Gln, and Thr315Phe (Figure 5). In marked contrast to imatinib mesylate, normal PP1 sensitivity was found for the mutant Thr315Val. Inhibition of Bcr-AblThr315Val by 25 μM CGP76030 was significantly reduced (Figure 5). Taken together, these results suggested a differential requirement for H-bonding between Thr315 and PP1 versus Thr315 and CGP76030 for inhibitor binding.
Effect of CGP76030 on growth and survival of Bcr-Abl+ and AML cell lines.
CGP76030 inhibits growth and survival of Bcr-Abl+ but not of AML cell lines. The indicated cell lines were seeded in triplicate into 6-well plates and treated with the indicated concentrations of imatinib mesylate or CGP76030. DMSO was used as a vehicle control. The ratio of annexin-V+ to annexin-V− cells (A) and cell counts (B) were determined 3 days later as described in “Materials and methods.”
Effect of CGP76030 on growth and survival of Bcr-Abl+ and AML cell lines.
CGP76030 inhibits growth and survival of Bcr-Abl+ but not of AML cell lines. The indicated cell lines were seeded in triplicate into 6-well plates and treated with the indicated concentrations of imatinib mesylate or CGP76030. DMSO was used as a vehicle control. The ratio of annexin-V+ to annexin-V− cells (A) and cell counts (B) were determined 3 days later as described in “Materials and methods.”
Binding mode of the dual-specific Src/Abl kinase inhibitors PP1 and CGP76030.
Phospho-Abl and Abl immunoblots of lysates from Cos7 cells transfected with pApuro, Bcr-Ablwt, or the indicated mutants of Bcr-Abl at positions 315 and 380. Cells were either left untreated or incubated with 25 μM CGP76030 or 100 μM PP1.
Binding mode of the dual-specific Src/Abl kinase inhibitors PP1 and CGP76030.
Phospho-Abl and Abl immunoblots of lysates from Cos7 cells transfected with pApuro, Bcr-Ablwt, or the indicated mutants of Bcr-Abl at positions 315 and 380. Cells were either left untreated or incubated with 25 μM CGP76030 or 100 μM PP1.
While position 315 was of critical importance for PP1 and CGP76030 binding, position 380 was not. Both mutants (Ala380Cys and Ala380Thr) displayed a sensitivity to PP1 and CGP76030 similar to that of Bcr-Ablwt (Figure 5). This observation is in accordance with the ability of PP1 and CGP76030, but not imatinib mesylate, to inhibit epidermal growth factor receptor (EGF-R), a kinase that has a threonine at the position homologous to position 380 in Abl. Finally, PP1 and CGP76030 inhibited Bcr-AblAsp276Ser/Glu279Ser expressed in Cos7 cells with kinetics similar to that of Bcr-Ablwt (Figure 6A-B). Taken together, the results suggest that PP1, CGP76030, and imatinib mesylate bind into the same IBP adjacent to the ATP-binding site of Abl and have largely overlapping binding modes. However, PP1 and CGP76030 seem to dock into Abl in a manner that is independent of the conformation of the A-loop. Obviously, both inhibitors fill the entrance of the IBP of Abl less perfectly than imatinib mesylate. This conclusion is consistent with their ability to additionally inhibit Src kinases and other targets, but needs to be confirmed by crystal structure analysis.
Mutation of Thr315 of Bcr-Abl to bulkier amino acids does not completely disrupt the inhibitory effects of PP1 and CGP76030 on cell growth and survival
Importantly, comparison of the binding modes of imatinib mesylate, PP1, and CGP76030 with the use of the various mutants of the gatekeeping amino acid positions identified Bcr-Abl mutants (Thr315Leu, Thr315Ile, Thr315Met, Thr315Gln, Thr315Phe), which were equally resistant to all 3 inhibitors. We next wished to make use of these mutants to dissect Abl- and Src-dependent effects of such inhibitors. Therefore, 32D cells expressing the various mutants of the gatekeeping positions 315 and 380 were incubated with imatinib mesylate, PP1, or CGP76030. Expression of the imatinib mesylate–resistant Bcr-Abl mutants Thr315Val, Thr315Leu, Thr315Ile, Thr315Met, Thr315Gln, Thr315Phe, and Ala380Thr caused a complete or partial (Ala380Thr) resistance of 32D cells to the inhibitory effects of imatinib mesylate with regard to cell proliferation and survival (Figure 7A-B). In marked contrast, PP1 and CGP76030 considerably blocked the proliferation and inhibited the survival of 32D cells expressing the PP1- and CGP76030-resistant Bcr-Abl mutants Thr315Leu, Thr315Ile, Thr315Met, Thr315Gln, and Thr315Phe (Figure 7A-B). Interestingly, while both compounds inhibited cell proliferation to a similar degree, CGP76030 was more potent in inducing apoptosis than PP1. As expected from the above experiments in Cos7 cells, 32D cells expressing the Bcr-Abl mutants Thr315Val, Ala380Cys, and Ala380Thr showed rates of cell death and inhibition of proliferation similar to cells expressing Bcr-Ablwt (Figure 7A-B). Finally no significant influence of the mutation Asp276Ser/Glu279Ser was seen on the inhibition of cell proliferation and survival by PP1 and CGP76030 in 32D cells (Figure 6C-D), an observation that might be due to both the partially Abl-unrelated action of this compound and the unaltered sensitivity of the Asp276Ser/Glu279Ser mutant for PP1 and CGP76030 as observed in Cos7 cells.
We next determined the effect of PP1 and CGP76030 on Bcr-Abl activity in cells expressing either Bcr-Ablwt or an inhibitor-resistant mutant, such as Thr315Gln. As shown in Figure 7C, all 3 inhibitors greatly reduced the activity of Bcr-Ablwt as assessed by using phospho-specific antibodies. In contrast, no influence of the various inhibitors on the activity of Bcr-Abl was seen in 32DBcr-AblThr315Gln cells. Similar results were obtained from cells expressing other inhibitor-resistant mutants such as Thr315Leu, Thr315Ile, Thr315Met, or Thr315Phe. In summary, these results suggest that PP1 and CGP76030 are able to induce growth arrest and apoptosis independently of Abl inhibition.
Effect of the A-loop conformation on inhibition of Abl by PP1 and CGP76030.
The conformation of the A-loop does not influence inhibition of Abl by PP1 and CGP76030. (A-B) Phospho-Abl and Abl immunoblots of lysates from Cos7 cells transfected with pApuro, Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser. Cells were either left untreated or incubated with the indicated concentrations of PP1 (panel A) or CGP76030 (panel B). (C-D) Proliferation (left panels) and survival (right panels) of 32DBcr-Ablwt or 32DBcr-AblAsp276Ser/Glu279Ser cells grown in the presence of the indicated concentrations of PP1 (panel C) or CGP76030 (panel D).
Effect of the A-loop conformation on inhibition of Abl by PP1 and CGP76030.
The conformation of the A-loop does not influence inhibition of Abl by PP1 and CGP76030. (A-B) Phospho-Abl and Abl immunoblots of lysates from Cos7 cells transfected with pApuro, Bcr-Ablwt, Bcr-AblTyr393Phe, or Bcr-AblAsp276Ser/Glu279Ser. Cells were either left untreated or incubated with the indicated concentrations of PP1 (panel A) or CGP76030 (panel B). (C-D) Proliferation (left panels) and survival (right panels) of 32DBcr-Ablwt or 32DBcr-AblAsp276Ser/Glu279Ser cells grown in the presence of the indicated concentrations of PP1 (panel C) or CGP76030 (panel D).
Incomplete biologic resistance of 32DBcr-Abl cells expressing mutants resistant to CGP76030 and PP1.
Incomplete biologic resistance of 32DBcr-Abl cells expressing mutants resistant to CGP76030 and PP1 correlates with inhibition of Src kinases and Akt. (A-B) Proliferation (panel A) and survival (panel B) of 32D cells expressing Bcr-Ablwt or the various mutants of Bcr-Abl at positions 315 and 380 in response to imatinib mesylate, PP1, or CGP76030. First, 5 × 104 cells were seeded per 6 wells and treated with DMSO, 2.5 μM imatinib mesylate, 50 μM PP1, and 5 μM CGP76030. Survival and proliferation were determined 48 hours later by FACS analysis and trypan blue exclusion. (C-D) Phospho-Abl, phospho-Src, panphosphotyrosine, phospho-STAT5, phospho-Jnk, and phospho-Akt Western blots of 32DBcr-Ablwtor 32DBcr-AblThr315Gln cells treated with the indicated concentrations of imatinib mesylate, PP1, and CGP76030 1.5 hours prior to lysis. Equal expression of the various signaling molecules in the presence of the various inhibitors was assessed with the use of rabbit polyclonal antibodies to Abl, Src, Fyn, Lyn, STAT5, Jnk, and Akt.
Incomplete biologic resistance of 32DBcr-Abl cells expressing mutants resistant to CGP76030 and PP1.
Incomplete biologic resistance of 32DBcr-Abl cells expressing mutants resistant to CGP76030 and PP1 correlates with inhibition of Src kinases and Akt. (A-B) Proliferation (panel A) and survival (panel B) of 32D cells expressing Bcr-Ablwt or the various mutants of Bcr-Abl at positions 315 and 380 in response to imatinib mesylate, PP1, or CGP76030. First, 5 × 104 cells were seeded per 6 wells and treated with DMSO, 2.5 μM imatinib mesylate, 50 μM PP1, and 5 μM CGP76030. Survival and proliferation were determined 48 hours later by FACS analysis and trypan blue exclusion. (C-D) Phospho-Abl, phospho-Src, panphosphotyrosine, phospho-STAT5, phospho-Jnk, and phospho-Akt Western blots of 32DBcr-Ablwtor 32DBcr-AblThr315Gln cells treated with the indicated concentrations of imatinib mesylate, PP1, and CGP76030 1.5 hours prior to lysis. Equal expression of the various signaling molecules in the presence of the various inhibitors was assessed with the use of rabbit polyclonal antibodies to Abl, Src, Fyn, Lyn, STAT5, Jnk, and Akt.
Differential effects of PP1, CGP76030, and imatinib mesylate on the activity of Src kinases
We next investigated whether the Abl-independent biologic effects of PP1 and CGP76030 correlated with their ability to additionally inhibit Src kinases. In the absence of inhibitors, a high level of Src kinase autophosphorylation was seen with the use of a phospho-specific pan-Src antibody (Figure 7C). Four major autophosphorylated bands were detected (not resolved in Figure 7C). In 32DBcr-Ablwt cells, PP1 and CGP76030, but not imatinib mesylate, completely blocked the autophosphorylation of Src kinases, corroborating the concept of Abl-kinase–independent activation of Src kinases.20 Inhibition of Src kinase autophosphorylation by PP1 and CGP76030 occurred independently of whether Bcr-Ablwt or an inhibitor-resistant mutant was expressed (Figure 7C). Moreover, in 32DBcr-AblThr315Gln cells, PP1 and CGP76030 still led to a subtle but reproducible reduction of the tyrosine phosphorylation of cytosolic phosphoproteins (Figure 7C), whereas imatinib mesylate showed no effect.
We also wished to study the influence of the various inhibitors on the activity of potential downstream signaling intermediates. All 3 inhibitors led to a disruption of the phosphorylation and/or activity of signal transducer and activator of transcription 5 (STAT5), Jnk, and Akt in cells expressing Bcr-Ablwt. Inhibition of STAT5 and Jnk was not seen in cells expressing the inhibitor-resistant mutant Thr315Gln, suggesting that these signaling pathways are directly downstream of Abl kinase activity (Figure 7D). Similarly, inhibition of Akt by imatinib mesylate was reversed by expression of Thr315Gln (Figure 7D). Surprisingly, however, activity of Akt was still reduced by PP1 and CGP76030 in cells expressing Thr315Gln (Figure 7D). These data were reproduced with the use of other inhibitor-resistant mutants. This suggested that, in addition to Bcr-Abl, Src kinases contributed to the regulation of Akt in Bcr-Abl+ cell lines.
In summary, these findings suggest that the differential biologic effects of PP1/CGP76030 versus imatinib mesylate on 32DBcr-Ablwt and 32DBcr-AblThr315 mutant cells are explained—at least in part—by their ability to inactivate Lyn and other Src kinases. However, we wish to emphasize at this point that PP1 and CGP76030 might inhibit additional, yet unknown, kinases that explain their biologic properties in our experiments.
Discussion
Based on a recently published crystal structure analysis,8 work presented in this manuscript characterizes the functional role of 2 amino acid positions, 315 and 380, as gatekeepers that control the access for imatinib mesylate to an IBP. Moreover, we could confirm by genetic experiments the hypothesis that imatinib mesylate would bind into inactive rather than active Abl, a feature that contributes considerably to the high specificity of imatinib mesylate.8 A mutation was introduced into Abl that was proposed to destabilize the A-loop and to shift the equilibrium of the different states of the A-loop toward the active conformation. This mutation was based on the disruption of an H-bond system between Arg386, located on the A-loop, and Asp276 as well as Glu279, both located within the β3-αC-loop. This interaction is of special interest because in the structures of active IRK and Lck, the arginine residues homologous to Arg386 are H-bond donors for the phosphate groups of autophosphorylated tyrosines located within the A-loop, thereby contributing to the stabilization of the active conformation.22 23 In our study, disruption of the bonding system generated a Bcr-Abl mutant that was considerably less sensitive to imatinib mesylate when compared with Bcr-Ablwt.
Also, this work proposes a more general use of inhibitor-resistant mutants to validate kinase inhibitors. In our experiments, we used inhibitor-resistant mutants to validate the binding mode and action of 2 small-molecule Abl kinase inhibitors, PP1 and CGP76030, that inhibit Src kinases in addition to Abl.24,25,27 We could show that both inhibitors block growth and survival of Bcr-Abl+ 32D cells to a degree similar to imatinib mesylate. Moreover, both compounds killed Bcr-Abl+ cell lines derived from CML patients, but not several AML cell lines. However, both compounds also affected growth and survival of parental 32D cells, possibly reflecting the published role of Src kinases in the IL-3–dependent growth and survival of several leukemic cells lines.26
A comparison of the binding of PP1, CGP76030, and imatinib mesylate to the various Bcr-Abl mutants showed that these inhibitors had largely overlapping binding modes in that all of them were sensitive to mutations at position 315. However, binding of PP1 and CGP76030 was not influenced by the mutation of Asp276/Glu279 to serine. This suggested that both compounds bound and inhibited Bcr-Abl independently of the A-loop conformation. Clearly, a crystal structure analysis will be needed to prove this interesting concept. However, our hypothesis is in agreement with a recently published crystal structure of a different Abl kinase inhibitor, PD173955, in complex with the kinase domain of Abl. In this study, the authors were able to show that PD173955 binds to a conformation of Abl similar to the active conformation of active tyrosine kinases, especially with regard to the structure of the A-loop.28
Finally, mutations influencing position 380 did not interfere with inhibition of Bcr-Abl by PP1 and CGP76030. This observation might reflect the larger molecular distance between position 380 and PP1/CGP76030 versus position 380 and imatinib mesylate,27 29 but again, crystal structure analysis will be necesarry to elucidate the nature of this finding.
The reduced complexity of binding concluded from testing both compounds with inhibitor-resistant mutants explains the somewhat broder target profile of PP1 and CGP76030.24,25 Therapeutically, however, the reduced specificity of PP1 and CGP76030 may be an advantage, as the additional targets of these compounds, particularly Src kinases, fit into a therapeutic concept. Despite the success of imatinib mesylate in clinical trials, patients with blast crisis or Bcr-Abl+ B-ALL rapidly relapse on imatinib mesylate.2 A recent publication suggested that specific point mutations interfering with imatinib mesylate binding might contribute to imatinib mesylate resistance: a mutation of Bcr-Abl at position 315 to isoleucine was identified in 3 patients with clinical imatinib mesylate resistance.13 Additional mutations have been found at other positions, but the importance of these mutations for clinical resistance needs to be determined.14-16 In our study, we confirmed that a mutation of Thr315 to isoleucine, but also to other amino acids, resulted in a complete resistance of 32D cells to the inhibitory effects of imatinib mesylate. In contrast, PP1 and CGP76030 still significantly reduced cell growth and survival of cell lines expressing inhibitor-resistant Bcr-AblThr315 mutants, suggesting that both compounds inhibited additional kinase targets. This might help to avoid selection for inhibitor-resistant point mutations, as observed for imatinib mesylate. We are fully aware that our data did not formally prove the inhibition of Src kinases as the sole or major explanation for the Abl-unrelated biologic effects observed. However, the biochemical experiments showed a strong correlation of the effects of PP1 and CGP76030 on the growth of cells expressing inhibitor-resistant Bcr-Abl mutants with their ability to inactivate Src kinases.
Interestingly, data on a different Abl inhibitor, PD180970, that also inhibits Src kinases was published recently.30 In this study, the authors concluded that most of the effect of this compound depended on Abl inhibition. However, only indirect evidence was shown to prove this hypothesis, and it would be interesting to see how PD180970 acts, compared with PP1 or CGP76030, on cells engineered to express inhibitor-resistant Bcr-Abl.
Importantly, inhibition of Src kinases in cells expressing Thr315 mutants was associated with a loss of Akt activity. It was only recently shown that Src kinases influence Akt activity by phosphorylation of tyrosines in the Akt A-loop and by regulating 3-phosphoinositide–dependent protein kinase–1 (PDK1), a serine/threonine kinase upstream of Akt.31,32 Importantly, neither PP1 nor CGP76030 is a direct inhibitor of Akt or the upstream kinase PDK1 (M.W, D.F., and M.H.; unpublished observations, 2001). This suggests that by direct phosphorylation of PDK1 and, above all, Akt, Src kinases might contribute to the activation of this pathway, which is crucial in regulating cell growth and survival of Bcr-Abl+ cells.33 Future experiments will clarify this question.
In summary, the results presented here point to a potential role of dual-specific inhibitors of Abl and Src kinases, such as PP1 or CGP76030, for the treatment of advanced or imatinib mesylate–resistant Philadelphia chromosome–positive (Ph+) leukemias. Owing to their less complex binding mode and the additional inhibition of Src (and maybe other) kinases, they might avoid selection of inhibitor-resistant cells, as is observed with imatinib mesylate. Importantly, CGP76030 has already been used in a rat model for osteoporosis without causing serious toxicity.25 No influence on bone marrow cellularity or on peripheral white blood cell (WBC) counts was seen (M.W. and M.H; unpublished observations). It now remains to be determined whether these or similar Src kinase inhibitors are sufficiently specific and safe for clinical use.
We thank S. Reis for technical assistance, C. Kurzeder and C. Schuster for advice with FACS analysis, and R. van Etten and S. Li (Center for Blood Research, Boston, MA) for providing reagents.
Prepublished online as Blood First Edition Paper, September 5, 2002; DOI 10.1182/blood-2002-01-0288.
Supported by grants Ha 1680/2-3 and Ha 1680/2-4 from the Deutsche Forschungsgemeinschaft (M.H.) and by grants from the Novartis Foundation for Therapeutic Research (M.H. and M.W.).
Several of the authors (D.F., P.W.M., E.B.) are employed by a company (Novartis) whose (potential) product was studied in the present work.
M.W. and N.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Michael Hallek, Medizinische Klinik III, Universität München, Grosshadern, Marchioninistrasse 15, D-81377 München, Germany; e-mail:mhallek@med3.med.uni-muenchen.de.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal