The Scianna blood group encompasses the high-frequency antigens Sc1 and Sc3 and the low-frequency antigen Sc2. Another low-frequency antigen Rd (Radin) was suggested to belong to the Scianna blood group. The molecular basis of the Scianna blood group was unknown. The erythrocyte membrane-associated protein (ERMAP) shared the genomic location, protein product size, and localization to the red blood cell (RBC) membrane surface with Scianna. TheERMAP gene was sequenced in probands with known Scianna and Radin phenotypes. In a Sc:-1,-2 proband, only anERMAP allele with a 2-bp deletion in exon 3 causing a frameshift could be detected. A Sc:-1,2 proband was homozygous for theERMAP(Gly57Arg) allele. An Rd+proband was heterozygous for the ERMAP(Pro60Ala) allele. Polymerase chain reaction with sequence-specific priming (PCR-SSP) systems was developed to detect the Sc2 and Rd alleles of theERMAP gene. The 2 alleles occurred with about 1% and less than 1% frequency in the population, which was compatible with the frequency of the Sc2 and Rd antigens known in whites. Two Sc2+ and one Rd+ samples that were found by genotyping were confirmed by serology. The antigens of the Scianna blood group include Rd and are expressed by the human ERMAP protein. Sc2 is caused by an ERMAP(Gly57Arg) allele and Rd by an ERMAP(Pro60Ala) allele. Scianna is the last of the previously characterized protein-based blood group systems whose molecular basis was discerned. Hence, the phenotype prediction by genotyping became possible for all human blood group systems encoded by proteins.
Introduction
The molecular characterization of polymorphism underlying blood group antigens has made enormous advances during the past 15 years.1 Currently, 26 blood group systems are recognized.2 The antigens of 22 blood group systems are determined by epitopes on proteins. They may function as transport or adhesion proteins, ectoenzymes, or cytokine receptors present on the red blood cell (RBC) surface.1 Most recently, theDOK1 gene determining the Dombrock blood group system (ISBT 014) had been characterized,3 leaving Scianna (ISBT 013) the last protein-based blood group system whose molecular basis remained obscure.
The Scianna blood group system comprises 2 antithetical antigens, the high-frequency antigen Sc1 (ISBT 013.001, formerly Sm) and the low-frequency antigen Sc2 (ISBT 013.002, formerly Bua). Lewis et al4 recognized the relationship of the antigens and coined the name Scianna.5 The antigen Sc3 (ISBT 013.003) is expressed on all RBCs except those of the very rare Sc:-1,-2 phenotype, 3 clusters of which were observed in circumscribed populations.5
The Scianna antigens were shown to reside on a Scianna glycoprotein of about 60 to 68 kD.6 Scianna antigens were inherited linked to the RH antigens but were shown to be distinct.7 The latest data indicated a genomic position of 1p34 to 1p36.8A second low-frequency antigen, Radin (Rd; 700.015) shared this position9,10 and was found on a protein of similar size.11 Both anti-Sc212 and anti-Rd13 are clinically relevant, having caused hemolytic disease of the newborn. The molecular basis and function of the Scianna blood group system and its glycoprotein remained unknown.
We demonstrated that the Scianna antigens are expressed by the red cell adhesion protein erythrocyte membrane-associated protein (ERMAP). The polymorphism underlying the Sc2 and Rd antigens was determined. Polymerase chain reaction with sequence-specific priming (PCR-SSP) methods that allow the genotypic prediction of these antigens was developed.
Materials and methods
Rare Scianna phenotypes
Sc:2 and Rd+ blood samples were obtained from the Serum, Cells and Rare Fluid (SCARF) exchange program. Sc:-1,2 and Sc:-1,-2 samples were from the reference collection at the International Blood Group Reference Laboratory. The Sc:-1,-2 pedigree came from Saudi Arabia. Control blood samples were derived from South-Western German blood donors.
Nucleotide sequencing
Nucleotide sequencing was performed by using Prism dye terminator cycle-sequencing kit with AmpliTaq FS DNA polymerase and Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Weiterstadt, Germany) with a DNA sequencing unit (ABI 3100; Applied Biosystems). Nucleotide sequencing of genomic DNA stretches representative for all 11 ERMAP exons and parts of the promoter was accomplished by using primers (Table1) and amplification procedures (Table2) that obviated the need for subcloning steps. PCR was performed by using Expand high-fidelity PCR system or Expand long-range PCR system (Boehringer Mannheim, Mannheim, Germany). Thermocycling conditions consisted of an initial denaturation of 2 minutes at 92°C, followed by 45 cycles of 20-second denaturation at 92°C, 30-second annealing at 60°C, and 10-minute extension at 68°C, with an incremental elongation of the extension time of 20 seconds for each cycle after cycle 10.
Primers used
| Name . | Nucleotide sequence . | Genomic region . | Position* . | Strandedness . |
|---|---|---|---|---|
| e18c | CATACAGCCGGAGGAAGACG | Exon 2 | 73 to 54 | Antisense |
| e18t | CATACAGCCGGAGGAAGACA | Exon 2 | 73 to 54 | Antisense |
| e26h | gatcagaaactcctacCTGACACTTG | Exon 2 | 16† to 85 | Antisense |
| e26y | gatcagaaactcctacCTGACACTTA | Exon 2 | 16† to 85 | Antisense |
| e80a | GCAGCCACCTCACCTCCTTTGC | Exon 3 | 199 to 178 | Antisense |
| edra2b | TCACCTCCTTGGGTACCGTACT | Exon 3 | 190 to 169 | Antisense |
| edra2x | TCACCTCCTTGGGTACCGTACC | Exon 3 | 190 to 169 | Antisense |
| ei5r | CCTCCGTCACATGTTCATAGAGAAG | Exon 5 | 580 to 556 | Antisense |
| ei11a | GTGTGTGGGGATTTTCCTGGAC | Exon 11 | 1068 to 1089 | Sense |
| ei11b | GCCAGGCGACTGAAGTCCAG | Exon 11 | 1493 to 1474 | Antisense |
| ei11c | GGGACTTGTTGGTCACATTGTAG | Exon 11 | 1135 to 1113 | Antisense |
| en1a | gtaaggagaaaggggattatttctctac | Intron 1 | 65 to 92 | Antisense |
| en1b | cacccttgaaaatgttcttgtgcgtaag | Intron 1 | 88 to 115 | Antisense |
| en10r | cataaggaggaagtgctctgagatatg | Intron 10 | 52 to 26 | Antisense |
| en3r | ggcaagtaaaaactacactgcagatg | Intron 3 | 120 to 95 | Antisense |
| en4r | caccagcctgagctcccttac | Intron 4 | 32 to 12 | Antisense |
| en5r | gtggattcttgaaattcccatatatccc | Intron 5 | 342 to 315 | Antisense |
| en6r | ggatatatgtgtgcatgtgtgtacag | Intron 6 | 126 to 101 | Antisense |
| en7r | gtccccatggcttgaattac | Intron 7 | 125 to 106 | Antisense |
| ev1a | gagtcaagcgattacttcagctcac | Promoter | −2106 to −2082 | Sense |
| ev1b | gcctcctctttgctctgttc | 5UTR | −278 to −259 | Sense |
| ev2a | gcctcggctcctgggttatac | Intron 1 | −77 to −57 | Sense |
| ev2b | tctgtcacgccaagcccatcag | Intron 1 | −54 to −33 | Sense |
| ev2c | agcccgcttgtgctgtctcc | Intron 1 | −34 to −14 | Sense |
| ev3a | ctcatcccttcccaagctttc | Intron 2 | −65 to −45 | Sense |
| ev3b | ctcccagttggccttgtctc | Intron 2 | −33 to −14 | Sense |
| ev3c | cattatgtcttttggggttctgttc | Intron 2 | −156 to −132 | Sense |
| ev4a | ccatctgaggcatgccagaag | Intron 3 | −68 to −48 | Sense |
| ev4b | gacatctccctgaggtcag | Intron 3 | −41 to −23 | Sense |
| ev5a | atgcccgcttacctgtagtgac | Intron 4 | −125 to −104 | Sense |
| ev6a | gggaggtggaggtgaagatag | Intron 5 | −117 to −97 | Sense |
| ev7a | gagtgatgaggctgccacag | Intron 6 | −80 to −61 | Sense |
| ev8a | cattatgggagggcctctacatag | Intron 7 | −65 to −42 | Sense |
| ev8b | gggtatagctattgagtatctgtctg | Intron 7 | −115 to −90 | Sense |
| ev11a | ctaaaatgggctaatgacagtgatgg | Intron 10 | −116 to −91 | Sense |
| ev11b | acagtgatggcagacctaagattc | Intron 10 | −100 to −77 | Sense |
| en11a | TCACTGTGATAGTTAATCTTATGTATCCAC | Exon 11 | 3122 to 3093 | Antisense |
| Name . | Nucleotide sequence . | Genomic region . | Position* . | Strandedness . |
|---|---|---|---|---|
| e18c | CATACAGCCGGAGGAAGACG | Exon 2 | 73 to 54 | Antisense |
| e18t | CATACAGCCGGAGGAAGACA | Exon 2 | 73 to 54 | Antisense |
| e26h | gatcagaaactcctacCTGACACTTG | Exon 2 | 16† to 85 | Antisense |
| e26y | gatcagaaactcctacCTGACACTTA | Exon 2 | 16† to 85 | Antisense |
| e80a | GCAGCCACCTCACCTCCTTTGC | Exon 3 | 199 to 178 | Antisense |
| edra2b | TCACCTCCTTGGGTACCGTACT | Exon 3 | 190 to 169 | Antisense |
| edra2x | TCACCTCCTTGGGTACCGTACC | Exon 3 | 190 to 169 | Antisense |
| ei5r | CCTCCGTCACATGTTCATAGAGAAG | Exon 5 | 580 to 556 | Antisense |
| ei11a | GTGTGTGGGGATTTTCCTGGAC | Exon 11 | 1068 to 1089 | Sense |
| ei11b | GCCAGGCGACTGAAGTCCAG | Exon 11 | 1493 to 1474 | Antisense |
| ei11c | GGGACTTGTTGGTCACATTGTAG | Exon 11 | 1135 to 1113 | Antisense |
| en1a | gtaaggagaaaggggattatttctctac | Intron 1 | 65 to 92 | Antisense |
| en1b | cacccttgaaaatgttcttgtgcgtaag | Intron 1 | 88 to 115 | Antisense |
| en10r | cataaggaggaagtgctctgagatatg | Intron 10 | 52 to 26 | Antisense |
| en3r | ggcaagtaaaaactacactgcagatg | Intron 3 | 120 to 95 | Antisense |
| en4r | caccagcctgagctcccttac | Intron 4 | 32 to 12 | Antisense |
| en5r | gtggattcttgaaattcccatatatccc | Intron 5 | 342 to 315 | Antisense |
| en6r | ggatatatgtgtgcatgtgtgtacag | Intron 6 | 126 to 101 | Antisense |
| en7r | gtccccatggcttgaattac | Intron 7 | 125 to 106 | Antisense |
| ev1a | gagtcaagcgattacttcagctcac | Promoter | −2106 to −2082 | Sense |
| ev1b | gcctcctctttgctctgttc | 5UTR | −278 to −259 | Sense |
| ev2a | gcctcggctcctgggttatac | Intron 1 | −77 to −57 | Sense |
| ev2b | tctgtcacgccaagcccatcag | Intron 1 | −54 to −33 | Sense |
| ev2c | agcccgcttgtgctgtctcc | Intron 1 | −34 to −14 | Sense |
| ev3a | ctcatcccttcccaagctttc | Intron 2 | −65 to −45 | Sense |
| ev3b | ctcccagttggccttgtctc | Intron 2 | −33 to −14 | Sense |
| ev3c | cattatgtcttttggggttctgttc | Intron 2 | −156 to −132 | Sense |
| ev4a | ccatctgaggcatgccagaag | Intron 3 | −68 to −48 | Sense |
| ev4b | gacatctccctgaggtcag | Intron 3 | −41 to −23 | Sense |
| ev5a | atgcccgcttacctgtagtgac | Intron 4 | −125 to −104 | Sense |
| ev6a | gggaggtggaggtgaagatag | Intron 5 | −117 to −97 | Sense |
| ev7a | gagtgatgaggctgccacag | Intron 6 | −80 to −61 | Sense |
| ev8a | cattatgggagggcctctacatag | Intron 7 | −65 to −42 | Sense |
| ev8b | gggtatagctattgagtatctgtctg | Intron 7 | −115 to −90 | Sense |
| ev11a | ctaaaatgggctaatgacagtgatgg | Intron 10 | −116 to −91 | Sense |
| ev11b | acagtgatggcagacctaagattc | Intron 10 | −100 to −77 | Sense |
| en11a | TCACTGTGATAGTTAATCTTATGTATCCAC | Exon 11 | 3122 to 3093 | Antisense |
5UTR indicates 5 untranslated region of exon 1.
The positions of the synthetic oligonucleotides are indicated relative to their distances from the first nucleotide position of the start codon ATG in the cDNA for all primers in the promoter and in the exons including the 3 untranslated region of exon 10, or relative to their adjacent exon/intron boundaries for all other primers.
Intron 2
Sequencing method for the 11 ERMAP exons from genomic DNA
| ERMAP . | PCR primers* . | Sequencing primers*,† . | ||
|---|---|---|---|---|
| Sense . | Antisense . | Sense . | Antisense . | |
| Exon 1 | ev1a | en1b | ev1b | en1b |
| Exon 2 | ev2a | ei5r | ev2b, ev2c | — |
| Exon 3 | ev2a | ei5r | ev3c, ev3a | en3r |
| Exon 4 | ev2a | ei5r | ev4a, ev4b | — |
| Exon 5 | ev4a | en10r | ev5a | — |
| Exon 6 | ev4a | en10r | ev6a | — |
| Exon 7 | ev4a | en10r | ev7a | — |
| Exon 8 | ev4a | en10r | ev8a | — |
| Exon 9 | ev4a | en10r | ev8a | — |
| Exon 10 | ev4a | en10r | ev8a | — |
| Exon 11 | ev11a | en11a | ev11b, ei11a | ei11b, ei11c |
| ERMAP . | PCR primers* . | Sequencing primers*,† . | ||
|---|---|---|---|---|
| Sense . | Antisense . | Sense . | Antisense . | |
| Exon 1 | ev1a | en1b | ev1b | en1b |
| Exon 2 | ev2a | ei5r | ev2b, ev2c | — |
| Exon 3 | ev2a | ei5r | ev3c, ev3a | en3r |
| Exon 4 | ev2a | ei5r | ev4a, ev4b | — |
| Exon 5 | ev4a | en10r | ev5a | — |
| Exon 6 | ev4a | en10r | ev6a | — |
| Exon 7 | ev4a | en10r | ev7a | — |
| Exon 8 | ev4a | en10r | ev8a | — |
| Exon 9 | ev4a | en10r | ev8a | — |
| Exon 10 | ev4a | en10r | ev8a | — |
| Exon 11 | ev11a | en11a | ev11b, ei11a | ei11b, ei11c |
— indicates not applicable.
Primer sequences are given in Table 1.
For each sample, one or more sequencing primers were used, if applicable.
PCR-SSP
PCR-SSP methods were devised to detect the C>T polymorphism at nucleotide 54 (counted relative to the A of the start codon in the cDNA), the C>T polymorphism at position 76, the G>A polymorphism at position 169 causing the Sc1/Sc2 polymorphism, and the G at position 178 in Rd (Table 3). The reactions were triggered to work under similar PCR conditions as a PCR-SSP system previously developed for RHDtyping.14,15 Thermocycling conditions consisted of an initial denaturation of 2 minutes at 94°C, followed by 10 cycles of 10-second denaturation at 94°C and 1-minute annealing/extension at 65°C and finally of 25 cycles of 30-second denaturation at 94°C, 1 minute annealing at 61°C, and 30-second extension at 72°C.15 Amplifications were carried out with Taq (Qiagen, Hilden, Germany) in a final volume of 10 μL. HGHcontrol primers were used at a concentration of 0.75 μM to amplify the 434-bp control PCR fragment derived from the human growth hormone gene.
PCR-SSP for ERMAP alleles
| Polymorphism detected . | Detection primers . | Size of specific product, bp . | ||
|---|---|---|---|---|
| Specific . | Nonspecific . | Concentration, μM . | ||
| 54C | e18c | ev2b | 2 | 134 |
| 54T | e18t | ev2b | 2 | 134 |
| 76C | e26h | ev2b | 3 | 167 |
| 76T | e26y | ev2b | 3 | 167 |
| 169A (Sc2) | edra2b | ev3b | 4 | 147 |
| 169G (Sc1) | edra2x | ev3b | 4 | 147 |
| 178G (Rd) | e80a | ev3b | 2 | 156 |
| Polymorphism detected . | Detection primers . | Size of specific product, bp . | ||
|---|---|---|---|---|
| Specific . | Nonspecific . | Concentration, μM . | ||
| 54C | e18c | ev2b | 2 | 134 |
| 54T | e18t | ev2b | 2 | 134 |
| 76C | e26h | ev2b | 3 | 167 |
| 76T | e26y | ev2b | 3 | 167 |
| 169A (Sc2) | edra2b | ev3b | 4 | 147 |
| 169G (Sc1) | edra2x | ev3b | 4 | 147 |
| 178G (Rd) | e80a | ev3b | 2 | 156 |
PCR-RFLP
The PCR product also used for exon 3 sequencing (Table 2) was digested for 3 hours at 37°C with PvuII andSmaI (Pharmacia, Freiburg, Germany). In this assay (restriction fragment length polymorphism [RFLP]),PvuII was added in addition to SmaI to ensure that the resulting fragments were in a size range that allowed easy distinction of fragments that were or were not cut bySmaI.
Population screen for ERMAP alleles
The prevalence of the 54C>T and 76C>T polymorphism was determined in 111 blood samples from blood donors in Baden-Württemberg (South-Western Germany) by PCR-SSP. Maximum likelihood estimates of allele frequencies were calculated by using the counting method.16 The presence of ERMAPalleles representative of Sc2 and Rd was checked by PCR-SSP in 305 samples.
Modeling
The extent of the signal peptide was determined by using Signal P V2.0 (http://www.cbs.dtu.dk/services/SignalP-2.0/) web server.17,18 A 3-dimensional model of the immunoglobulin domain of ERMAP was modeled at the 3D-JIGSAW server (http://www.bmm.icnet.uk/∼3djigsaw/)19,20 by using the heavy chain of the Fab fragment of the murine monoclonal antibody 3A221 as template. Stereoscopic views were rendered by using Deep View Swiss-PdbViewer (http://www.expasy.ch/spdbv/).
Results
A Sc:-1-2 proband is probably homozygous for a nonfunctionalERMAP allele
We determined the nucleotide sequence of the 11 ERMAPexons in a proband that lacked the Sc1, Sc2, and Sc3 antigens (EMBL/GenBank/DDBJ accession nos. AJ505027 to AJ505034). In exon 3, theERMAP allele harbored a 2-bp deletion that was possibly caused by a slippage error of the DNA polymerase in the GAGAGA repetitive sequence. This frameshift caused a truncated protein of 113 amino acids (Figure 1). Additional nucleotide substitutions 54C>T and 76C>T were detected in exon 2, the latter of which caused a His→Thr amino acid substitution at codon 26 (not shown). No evidence for a second ERMAP allele was observed, suggesting homozygosity for the ERMAP(54C>T, 76C>T, 307del2) allele. As an unlikely alternative, theERMAP(54C>T, 76C>T, 307del2) could be in trans to a second nonfunctional allele that possessed gross structural changes, preventing amplification of exons 2 and 3 by PCR.
A 2-bp deletion in the ERMAP allele of the Sc:-1,-2 proband.
The nucleotide sequences (nt) and derived amino acid sequences (aa) of a part of the ERMAP exon 3 are shown for the proband with the Sc:-1,-2 phenotype (A) and a control with the prevalentERMAP allele encoding Sc:1,-2 (B). The control harbors a triple GA repeat at nucleotides 303 to 308, one of which is deleted in the Sc:-1,-2 proband (asterisks). Because of the induced frameshift, a nonsense amino acid sequence is translated beyond codon 102 and a stop codon occurs at codon 114, resulting in a nonfunctional protein of 113 amino acids. All nucleotides are in capitals, indicating an exon sequence.
A 2-bp deletion in the ERMAP allele of the Sc:-1,-2 proband.
The nucleotide sequences (nt) and derived amino acid sequences (aa) of a part of the ERMAP exon 3 are shown for the proband with the Sc:-1,-2 phenotype (A) and a control with the prevalentERMAP allele encoding Sc:1,-2 (B). The control harbors a triple GA repeat at nucleotides 303 to 308, one of which is deleted in the Sc:-1,-2 proband (asterisks). Because of the induced frameshift, a nonsense amino acid sequence is translated beyond codon 102 and a stop codon occurs at codon 114, resulting in a nonfunctional protein of 113 amino acids. All nucleotides are in capitals, indicating an exon sequence.
Sc2 antigen is due to an ERMAP(Gly57Arg) allele
Sc:2 samples carry an ERMAP(Gly57Arg) allele.
The ERMAP gene of a Sc:-1,2 proband was sequenced to determine the molecular basis of the Sc2 antigen and to corroborate that Scianna and ERMAP are identical (EMBL/GenBank/DDBJ accession nos. AJ505035 to AJ505042). A substitution of guanosine (G) by adenosine (A) was detected at the predicted cDNA position 169 (relative to the A of the start codon) that indicated the homozygous presence of an ERMAP allele with a Gly→Arg substitution in codon 57. The nucleotide substitution destroyed a SmaI site and allowed an independent confirmation by PCR-RFLP (data not shown). The homozygous presence of the ERMAP(Gly57Arg) allele was demonstrated by nucleotide sequencing of ERMAP exon 3 in 2 additional unrelated Sc:-1,2 probands. Two Sc:1,2 samples were heterozygous for the ERMAP(Gly57Arg) allele. PCR-SSP methods for the detection of the ERMAP(Gly57Arg) and the wild-typeERMAP alleles were developed (Figure2).
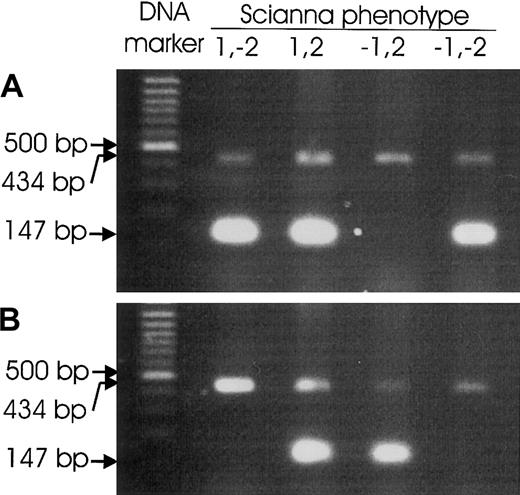
Scianna genotyping by PCR-SSP.
PCRs were devised to amplify specific PCR products of 147-bp size from the wild-type ERMAP allele representative of Sc1 (A) or from the ERMAP(Gly57Arg) allele carrying the Sc2 antigen (B), respectively. Representative results for Sc:1,-2; Sc:1,2; Sc:-1,2; and Sc:-1,-2 samples are shown. Control PCR products of 434-bp size occurred in all reactions.
Scianna genotyping by PCR-SSP.
PCRs were devised to amplify specific PCR products of 147-bp size from the wild-type ERMAP allele representative of Sc1 (A) or from the ERMAP(Gly57Arg) allele carrying the Sc2 antigen (B), respectively. Representative results for Sc:1,-2; Sc:1,2; Sc:-1,2; and Sc:-1,-2 samples are shown. Control PCR products of 434-bp size occurred in all reactions.
ERMAP(Gly57Arg) is rare in the population.
DNA from 305 blood donors was checked for the presence of anERMAP(Gly57Arg) allele by PCR-SSP. Five samples carried this allele in heterozygous form, as a wild-type allele could be detected by PCR-SSP, too. A haplotype frequency of 0.008 was deduced for ERMAP(Gly57Arg).
ERMAP(Gly57Arg)-positive samples are Sc:2.
Red cells of 2 ERMAP(Gly57Arg) heterozygous donors detected by PCR-SSP were tested for the Sc1 and Sc2 antigens by serology. Both donors were found to be Sc:1,2.
Rd antigen is due to an ERMAP(Pro60Ala) allele
An Rd+ sample carries an ERMAP(Pro60Ala) allele.
The ERMAP gene of an Rd+ proband was sequenced to assess if Rd belongs to the Scianna blood group system (EMBL/GenBank/DDBJ accession nos. AJ505043 to AJ505050). A C>G heterozygosity was detected at the predicted cDNA position 178 that indicated the heterozygous presence of an ERMAP allele with a Pro→Ala substitution at codon 60. A PCR-SSP for the detection of the ERMAP(Pro60Ala) allele was developed (Figure3).
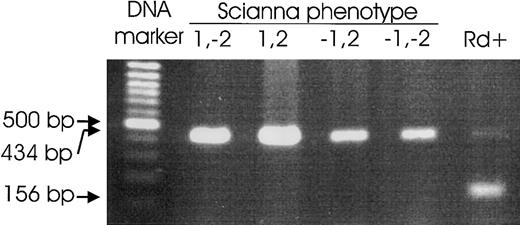
Radin genotyping by PCR-SSP.
A PCR was devised to amplify a specific PCR product of 156-bp size from the ERMAP(Pro60Ala) allele carrying the Rd antigen. Representative results for Sc:1,-2; Sc:1,2; Sc:-1,2; and Sc:-1,-2 samples and an Rd+ sample are shown. The 434-bp control product derives from the HGH gene, but this product may be lacking because of competition, if a specific product is present.
Radin genotyping by PCR-SSP.
A PCR was devised to amplify a specific PCR product of 156-bp size from the ERMAP(Pro60Ala) allele carrying the Rd antigen. Representative results for Sc:1,-2; Sc:1,2; Sc:-1,2; and Sc:-1,-2 samples and an Rd+ sample are shown. The 434-bp control product derives from the HGH gene, but this product may be lacking because of competition, if a specific product is present.
ERMAP(Pro60Ala) is very rare in the population.
Among the 305 blood donors previously tested forERMAP(Gly57Arg), no donor was positive for theERMAP(Pro60Ala) allele. However, among a different set of 20 random donors, an ERMAP(Pro60Ala)-positive sample was inadvertently found.
The second ERMAP(Pro60Ala)-positive sample is Rd+.
Red cells of the ERMAP(Pro60Ala)-positive donor detected by PCR-SSP were tested for the Sc1, Sc2, and Rd antigens by serology and were found to be Sc:1,-2; Rd+.
Population frequencies of the ERMAP(His26Tyr) allele and the silent 54C>T polymorphism
The 54C>T and 76C>T polymorphisms initially observed in the rare Sc:-1,-2 phenotype were also encountered in control samples. Therefore, 111 blood donors were systematically checked for the presence of anERMAP(His26Tyr) allele or a 54C>T polymorphism (Table4). Maximum likelihood estimates of the allele frequencies were derived. ERMAP(His26Tyr) occurred with a frequency of 0.33. Most of these alleles (allele frequency 0.28) were predicted to carry a 54C>T substitution, too, whereas anERMAP(His26Try) with the wild-type C at position 54 occurred less frequently (allele frequency 0.05). In contrast, the ERMAPalleles encoding a His at position 26 carried a C at position 54 (allele frequency 0.67) and represented the wild-type allele.22 23
Linkage of 54T and 76T polymorphism in 111 samples investigated
| Position 54 . | Position 76 . | Total . | ||
|---|---|---|---|---|
| C . | C/T . | T . | ||
| C | 51 | 7 | 0 | 58 |
| C>T | 0 | 40 | 3 | 43 |
| T | 0 | 0 | 10 | 10 |
| Total | 51 | 47 | 13 | 111 |
| Position 54 . | Position 76 . | Total . | ||
|---|---|---|---|---|
| C . | C/T . | T . | ||
| C | 51 | 7 | 0 | 58 |
| C>T | 0 | 40 | 3 | 43 |
| T | 0 | 0 | 10 | 10 |
| Total | 51 | 47 | 13 | 111 |
Maximum likelihood estimates of allele frequencies were 0.67 forERMAP(54C,76C), 0.28 forERMAP(54T,76T), 0.05 for ERMAP(54C,76T), and 0 for ERMAP(54T,76C).
Modeling of the ERMAP immunoglobulin domain
On the basis of the known similarity of ERMAP to immunoglobulin, we modeled the tertiary structure of the amino acids 46 to 131 representing part of the extracellular domain ofERMAP. The Pro substituted in the Rd allele was exposed on the protein's surface. The amino acid causing the Sc1/Sc2 polymorphism was located in a pocket of the protein's surface, thereby rendering it less accessible to antibodies. However, the amino acids involved in the Sc2 polymorphism were predicted to alter the 3-dimensional structure of this extracellular domain considerably, which may explain the immunogenicity of the ERMAP polymorphism (Figure4).
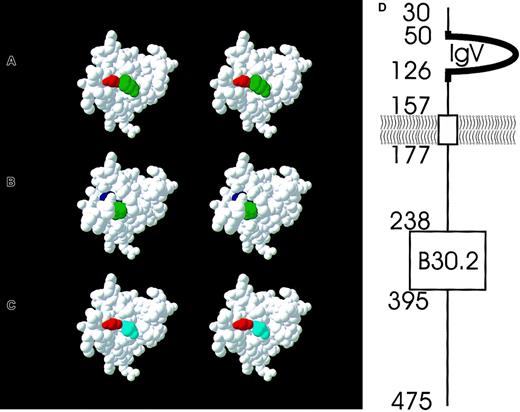
Stereo view of a 3D model of the ERMAP immunoglobulin domain and a schematic representation of the protein topology.
The 2 amino acid positions that are involved in the Sc1/Sc2 and Rd polymorphism are colored. In the wild-type ERMAP expressing Sc1 but lacking Rd antigens, there is a Gly (red) at position 57 and a Pro (green) at position 60 (A). The Sc2 antigen is caused by an Arg (blue) at position 57 (B), the Rd antigen by an Ala (light blue) at position 60 (C). The topology of the ERMAP protein as predicted by Su et al23 is shown (D). The mature protein starts at amino acid position 30. The aminotermial protein segment up to amino acid 157 is located extracellularly. The immunoglobulin V domain (amino acids 50-126) and a few adjacent amino acids (heavy line, amino acids 46-131) is modeled as shown in panels A to C. Almost two thirds of the protein (amino acids 177-475) are predicted to be located intracellularly and encompass a B30.2 domain.
Stereo view of a 3D model of the ERMAP immunoglobulin domain and a schematic representation of the protein topology.
The 2 amino acid positions that are involved in the Sc1/Sc2 and Rd polymorphism are colored. In the wild-type ERMAP expressing Sc1 but lacking Rd antigens, there is a Gly (red) at position 57 and a Pro (green) at position 60 (A). The Sc2 antigen is caused by an Arg (blue) at position 57 (B), the Rd antigen by an Ala (light blue) at position 60 (C). The topology of the ERMAP protein as predicted by Su et al23 is shown (D). The mature protein starts at amino acid position 30. The aminotermial protein segment up to amino acid 157 is located extracellularly. The immunoglobulin V domain (amino acids 50-126) and a few adjacent amino acids (heavy line, amino acids 46-131) is modeled as shown in panels A to C. Almost two thirds of the protein (amino acids 177-475) are predicted to be located intracellularly and encompass a B30.2 domain.
Discussion
The antigens of the Scianna blood group system are carried by the ERMAP protein. We formally proved that the low-frequency Rd antigen belongs to the Scianna blood group system, because it is expressed by a variant ERMAP protein. A third amino acid polymorphism His26Tyr was frequently observed but located in the signal peptide.
The human ERMAP gene was a good candidate gene for the Scianna blood group locus: Its genomic position23 was almost in the midst of the position 1p34 to 1p36 reported for the Scianna locus.8 ERMAP was expressed on RBCs22,23 and shared homology to chicken blood group antigens.23 The molecular weight of 60 to 68 kD determined for the glycosylated Scianna protein11 was compatible with 446 amino acids predicted for the mature ERMAP protein.
The identity of the ERMAP gene with the gene underlying the Scianna blood group system was based on 2 independent lines of evidence: (1) The expression of the Sc antigens was lacking in a proband who carried no functional ERMAP allele. (2) The expression of the Sc1 and Sc2 antigens correlated with the presence of a wild-type ERMAP allele and the otherwise rareERMAP(Gly57Arg) allele, respectively. Each observation alone could have been explained by a linkage to a neighboring gene; their coincidence, however, must be considered convincing evidence of the identity of ERMAP and Scianna. Alternative explanations for the accrued data are unlikely: The assumption that Scianna antigens depend on the interaction of several membrane proteins, like Wrb that is defined by an interaction of the anion exchanger AE1 and glycophorin A,24,25 could hardly be reconciled with the earlier demonstration11 that Scianna antigens reside on one glycoprotein as defined by immunoblotting. The most parsimonious explanation for the observed coincidence of mutations in the ERMAP immunoglobulin domain with antigens of the Scianna glycoproteins is the identity of ERMAP and Scianna. However, a formal proof would have to rely on the expression of ERMAP recombinant proteins carrying the proposed Sc/Rd mutations in eukaryotic cells followed by cell surface analysis with antibodies specific for Sc and Rd antigens. Such studies could also address the question of whether the ERMAP protein is expressed in a complex with other membrane proteins, like the Rh proteins, the in vitro expression of which is regulated by the RhAG protein.26
The low-frequency Rd antigen was often suspected to belong to the Scianna blood group,11 but a final verification was lacking. We showed Rd to be associated with the distinct rareERMAP(Pro60Ala) allele, rendering Rd part of the Scianna blood group system. Sc:-1,-2 RBCs have been reported to lack several distinguishable high-incidence antigens.27 It will be straightforward to check for the association of high- and low-frequency antigens with ERMAP alleles, possibly extending the number of antigens known to belong to this blood group system.
ERMAP is the fourth blood group protein belonging to the immunoglobulin superfamily, along with LU (ISBT 005),28 LW (ISBT 016),29 and OK (ISBT 024).30 As expected, the amino acids involved in the Sc1/Sc2 and Rd blood group polymorphisms occurred in an extracellular protein segment. The Gly at position 57 representative of the Sc1 antigen was rather inaccessible at the protein's surface, as shown by modeling of the immunoglobulin domain of ERMAP. However, the amino acid substitution in Sc2 was predicted to cause considerable displacement of the neighboring amino acids, which may explain its immunogenicity.
A third amino acid polymorphism His26Tyr was frequent in our population. Because the 29 aminoterminal amino acids of ERMAP probably represented its signal peptide, this polymorphism was not expected to be part of the mature ERMAP protein. Accordingly, a correlation with a blood group antigen was neither expected nor established. Although the His26Tyr polymorphism was also observed in the ERMAP allele of the Sc:-1,-2,-3 proband, there was no indication that it prevented Sc1 expression: The His26Tyr substitution and the frameshift destroying the functional peptide sequence occurred in the same PCR product encompassing ERMAP exons 2 and 3, excluding the possibility that these 2 aberrations from the normal nucleotide sequence related to 2 different nonfunctional alleles. Furthermore, the frequent occurrence of a nonfunctional His26Tyr allele in the donor population would be incompatible with the known extreme rarity of Sc:-1,-2,-3 individuals.
Scianna was the last previously defined blood group system encoded by a protein whose molecular basis was resolved. Hence, the identification of the gene underlying Scianna rendered genotyping possible for all protein-encoded blood groups. The ERMAP protein is believed to represent an adhesion protein and possesses a C1q recognition sequence.22,23 It is characterized by the presence of an extracellular immunoglobulin domain and an intracellular B30.2 domain. The immunoglobulin domain is most similar to myelin/oligodendrocyte glycoprotein (MOG), a component of the myelin sheath on the surface of oligodendrocytes and Schwann cells and is a common autoantigen in patients with multiple sclerosis.23 B30.2 domains occur in combination with quite dissimilar N-terminal structures, like immunoglobulin domains, RING finger domains, and leucine zippers,31 and are found in transmembrane, intracellular, and secreted proteins.31 Because the familial Mediterranean fever (MIM 249100) and Opitz G/BBB syndrome (MIM 300000) are associated with mutations in the B30.2 domains of pyrin and of MID1, respectively, they are assumed to be functionally important.31 The concatenation of immunoglobulin and B30.2 domains, as observed in ERMAP, resembles most closely butyrophilin, a major protein of the milk droplet. Additional functional motifs detected in ERMAP include several phosphorylation consensus sequences for tyrosine kinase, casein kinase, and protein kinase C, a motif resembling a consensus site for SH3 domain binding, a dileucine motif that can mediate internalization of membrane receptors, and a possible leucine zipper.22Although the function of ERMAP has not been fully established, it is known to be expressed throughout erythropoesis.22 Because the probands of the known kindreds with Sc:-1,-2 phenotype appear to be healthy,5 its function may likely be compensated by other proteins. Even the lack of functional important proteins as the water transporter in Co(a-b-) individuals32 may become symptomatic only in very special circumstances.33 It may be worthwhile to check if the polymorphisms observed inERMAP influence its binding characteristics and function.
We thank John J. Moulds (Ortho Diagnostics, Raritan, NJ) and Joann M. Moulds (Hahnemann University, Philadelphia, PA) for rare RBC and serum samples from SCARF, and Cornelie B. Lonicer (Munich, Germany) for providing an anti-Sc2 serum. We acknowledge the expert technical assistance of Marianne Lotsch, Hedwig Bohnacker, Anita Hacker, and Nicole Stampfl in Ulm and Jackie Banks in Bristol.
Prepublished online as Blood First Edition Paper, August 22, 2002; DOI 10.1182/blood-2002-07-2064.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Willy A. Flegel, Abteilung Transfusionsmedizin, Universitätsklinikum Ulm, and DRK-Blutspendedienst Baden-Württemberg-Hessen, Institut Ulm, Helmholtzstrasse 10, D-89081 Ulm, Germany; e-mail:waf@ucsd.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal