Abstract
T-cell receptor–B-variable (TCR-BV) gene usage and the CDR3 size distribution pattern were analyzed by reverse transcription–polymerase chain reaction (RT-PCR) in patients with B-cell chronic lymphocytic leukemia (B-CLL) to assess the T-cell repertoire. The use of TCR-BV families in CD4 and CD8 T cells stimulated with autologous activated leukemic cells was compared with that of freshly obtained blood T cells. Overexpression of individual TCR-BV families was found in freshly isolated CD4 and CD8 T cells. Polyclonal, oligoclonal, and monoclonal TCR-CDR3 patterns were seen within such overexpressed native CD4 and CD8 TCR-BV families. In nonoverexpressed TCR-BV families, monoclonal and oligoclonal populations were noted only within the CD8 subset. After in vitro stimulation of T cells with autologous leukemic B cells, analyses of the CDR3 length patterns showed that in expanded TCR-BV populations, polyclonal patterns frequently shifted toward a monoclonal/oligoclonal profile, whereas largely monoclonal patterns in native overexpressed TCR-BV subsets remained monoclonal. Seventy-five percent of CD8 expansions found in freshly obtained CD8 T cells further expanded on in vitro stimulation with autologous leukemic B cells. This suggests a memory status of such cells. In contrast, the unusually high frequency of CD4 T-cell expansions found in freshly isolated peripheral blood cells did not correlate positively to in vitro stimulation as only 1 of 9 expansions continued to expand. Our data suggest that leukemia cell–specific memory CD4 and CD8 T cells are present in vivo of patients with CLL and that several leukemia cell–associated antigens/epitopes are recognized by the patients' immune system, indicating that whole leukemia cells might be of preference for vaccine development.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) is a neoplastic clonal expansion of CD5+ B cells, expressing immunoglobulin M (IgM) molecules on the cell surface with a restricted immunoglobulin VH-gene usage.1,2 Leukemic cells may spontaneously induce a tumor-specific T-cell response in patients.3
Circulating mature T cells use the α/β heterodimeric T-cell receptor (TCR) for specific recognition of antigenic peptides in context with major histocompatibility complex (MHC) molecules.4,5 Genes encoding for the variable domains of the α and β polypeptide chains of the TCR are assembled by somatic recombination of one of each of the variable (V), diversity (D, only for β chain), and joining (J) exons. Three hypervariable or complementarity-determining regions (CDR1, CDR2, CDR3) of the TCR have been defined. The CDR3 loop is responsible for the specific interaction with the antigenic peptide.6 Positive T-cell recognition of a specific peptide induces a clonal expansion of T cells exhibiting a TCR-V gene product with an identical CDR3 region for each clone. Analyses of the CDR3 size distribution pattern might therefore define the degree of clonality of T cells in complex cell populations.7
Clonally expanded T cells with a restricted TCR–B-variable (TCR-BV) gene usage have been shown to recognize tumor cells in patients with malignant diseases.8-12 Clonal T-cell expansions determined as a preferential usage of a restricted TCR-BV gene have also been reported in CLL.13 14 However, the functional significance and specificity of such TCR-BV expansions are unclear.
Exploring the specificity and function of T cells in B-CLL may be of importance to better understand immune regulation of the leukemic clone and to form a basis for the development of immunotherapeutic strategies. In the present study, TCR-BV gene usage of CD4 and CD8 T cells activated in vitro with autologous leukemic B-CLL cells were analyzed and compared with that of native T cells through reverse transcription–polymerase chain reaction (RT-PCR) and the CDR3 length distribution pattern. The results further support that CLL patients may spontaneously mount leukemia cell-specific CD4 and CD8 T-cell responses recognizing multiple leukemia cell–associated antigens.
Patients, materials, and methods
Patients and controls
Three untreated patients with CLL (all women) whose mean age was 58 years (range, 38-68 years) were included in the study. Each patient was previously shown to have a specific T-cell response against the leukemic B cells.3 Diagnostic criteria were described earlier.15 The staging system according to Rai was applied.16 Clinical characteristics are shown in Table 1.
Clinical characteristics of patients
| Patient . | Age, y . | Sex . | Rai stage . | WBC count, 109/L . | CD19+, % . | CD3+, % . | CD3+CD4+, % CD3 . | CD3+CD8+, % CD3 . |
|---|---|---|---|---|---|---|---|---|
| CLL402 | 67 | F | III | 212.0 | 98 | 2 | 57 | 37 |
| CLL403 | 38 | F | 0 | 20.0 | 86 | 4 | 57 | 43 |
| CLL411 | 68 | F | I | 83.2 | 97 | 3 | 71 | 27 |
| Patient . | Age, y . | Sex . | Rai stage . | WBC count, 109/L . | CD19+, % . | CD3+, % . | CD3+CD4+, % CD3 . | CD3+CD8+, % CD3 . |
|---|---|---|---|---|---|---|---|---|
| CLL402 | 67 | F | III | 212.0 | 98 | 2 | 57 | 37 |
| CLL403 | 38 | F | 0 | 20.0 | 86 | 4 | 57 | 43 |
| CLL411 | 68 | F | I | 83.2 | 97 | 3 | 71 | 27 |
Nine healthy, age-matched controls whose average age was 60 years (range, 54-67 years) were also included. The study was approved by the Ethical Committee of the Karolinska Institute, and informed consent was obtained from the blood donors.
Purification of blood CD4+ and CD8+ T cells
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood by Ficoll-Hypaque (Amersham Pharmacia Biotech, Uppsala, Sweden) gradient centrifugation.17 T cells were enriched by nylon wool purification (Biotest, Breiech, Germany).18 CD4 and CD8 T cells were positively selected using anti-CD4 and anti-CD8 monoclonal antibodies (mAbs) coupled to magnetic beads according to the manufacturer's instructions (Dynal Biotech, Oslo, Norway).
Activation of T lymphocytes by leukemic B cells
Leukemic B cells were activated as described earlier.3 Briefly, 20 × 106 leukemic B cells in 20 mL were cultured with 2 × 106irradiated (100 Gy) mitomycin C (Sigma-Aldrich Sweden AB, Tyresö)–treated CDw32 cells, anti-CD40 mAb (G28-5 hybridoma producer cell line; American Type Culture Collection, Manassas, VA) (0.1 μg/mL) and recombinant human interleukin-4 (rhIL-4) (10 IU/mL) (Schering-Plough Research Institute, Kenilworth, NJ) in 260-mL cell culture flasks. Cells were cultured for 5 days at 37°C in humidified air with 5% CO2 in RPMI 1640 medium (Gibco, BRL, Paisley, United Kingdom) with 10% heat-inactivated pooled human AB serum, 1 mM L-glutamine, 100 IU/mL penicillin and 100 μg/mL streptomycin, and they were washed extensively, irradiated (50 Gy), and used as stimulator cells. T cells were then stimulated with the autologous activated tumor B cells for 5 days. A ratio of 10:1 T cells/B cells was used.3 After culturing, CD4 and CD8 T cells were then positively selected using anti-CD4 and anti-CD8 mAbs coupled to magnetic beads (Dynal Biotech). The purification procedure yielded more than 95% T cells of the respective subset as determined by flow cytometry.
RNA extraction and cDNA synthesis
Normalization of TCR B-chain–specific cDNA concentration
The TCR B-chain–specific cDNA concentration was normalized using TCR B-chain constant region (BC)–specific PCR by amplifying part of the TCR-BC gene.20 Briefly, a 5′ sense (5′ BC) and a 3′ antisense (3′ BC) primer, respectively,14 specific for the TCR-BC1 and TCR-BC2 genes, were used in a 30-cycle PCR with serial 2-fold dilutions of cDNA. The PCR products were then electrophoresed as described earlier.14 Based on scanned data, equal amounts of TCR B-chain–specific cDNA were used in the subsequent TCR-BV PCR amplifications.
Analysis of TCR-BV gene usage
PCR amplifications of BV-specific cDNA were performed using a panel of 22 TCR-BV–specific 5′ primers and a TCR-BC–specific 3′ primer as described earlier.14 To ensure the expected size of the amplified BV-BC PCR products, 8 μL of each product was electrophoresed on 1.5% ethidium bromide-stained agarose gels and photographed.
Southern blot analysis of TCR-BV and -BC PCR products
PCR products were subjected to electrophoresis on agarose gels. The gels were denatured using a solution containing 0.5 M NaOH and 1.5 M NaCl for 15 minutes and then were neutralized in 0.5 M Tris (tris[hydroxymethyl]aminomethane)–HCl and 1.5 M NaCl for 15 minutes followed by incubation for 20 minutes in 10 × SSC. The products were transferred to nylon membranes (Amersham Pharmacia Biotech). Membranes were air dried and baked for 2 hours at 80°C.
5′ end-labeling of BC oligonucleotide probe and hybridization to PCR products
A TCR-BC–specific oligonucleotide probe (BC reporter)14 recognizing BC1 and BC220 was designed, synthesized (Scandinavian Gene Synthesis AB, Köping, Sweden), and 5′-end labeled21 using T4 polynucleotide kinase enzyme (Amersham Pharmacia Biotech) and γ-32P–adenosine triphosphate (ATP) (Amersham Pharmacia Biotech). Hybridization and quantification of the relative use of BV genes were performed as described earlier.14 22
Analysis of CDR3 length polymorphism
CDR3 length polymorphism analysis was performed as described.7 Briefly, cDNA samples were amplified in 35 cycles of PCR using primers specific for the corresponding BV as 5′ primers and 3′ BC–fluorescein isothiocyanate (FITC) as the 3′ primer.14 The products were checked on 1.5% ethidium bromide–stained agarose gels. An aliquot was loaded onto 6% denaturing polyacrylamide-sequencing gels, and electrophoresis was run in the presence of fluorescence size markers (Amersham Pharmacia Biotech) in an ALF-DNA sequencing machine (Amersham Pharmacia Biotech). Data were collected by computer and analyzed by the Fragment Manager software program (Amersham Pharmacia Biotech). A dominant CDR3 peak (or peaks) was defined as monoclonal (high-intensity signal) when there was a dramatic reduction of other CDR3 signals within that particular TCR-BV family—for example, when the area under a single peak was 50% or more of the total area under the curve.7 23
Cloning and sequencing
PCR for TCR-BV gene products was performed as described.14 To avoid mutations during the PCR reaction, AmpliTaq Gold DNA polymerase (PE Biosystems, Stockholm, Sweden) was used for amplification. Briefly, the PCR product of the corresponding band for a BV family was cut and purified with the QIAquick gel extraction kit (Qiagen, Hilden, Germany). Cloning and sequencing of TCR-BV genes were performed as described earlier.3 Positive clones were randomly selected for plasmid preparation. Cycle-sequencing PCR was performed using BV-specific primers and Big Dye dideoxy terminator (PE Biosystems) in a GeneAmp PCR 2400 system (PE Biosystems) for 25 cycles.24Sequencing was carried out using an ABI Prism 310 sequencer (PE Biosystems). Results were analyzed through the SeqEd program (PE Biosystems). CDR3 regions were determined according to Rock et al.25 All TCR-BV CDR3 sequences were submitted to NCBI/GenBank.
Results
TCR-BV gene overexpression in unstimulated native T cells
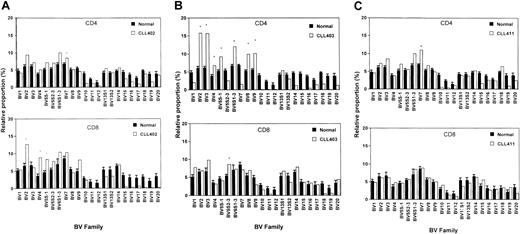
Any value for a given TCR-BV gene expression higher than mean + 3 SD of healthy age-matched controls is considered an overexpression.26 Overexpression of TCR-BV families within unstimulated native CD4 and CD8 T-cell populations of the individual patients are shown in Figure 1A-C. In total, 13 overexpressions were found.
TCR-BV families were overexpressed in the CD4 subset and 4 families in the CD8 subpopulation
To analyze whether overexpression was caused by polyclonal or monoclonal/oligoclonal expansion, the CDR3 length distribution pattern was determined. A monoclonal TCR-BV pattern represented by a single peak indicates recognition of a conventional antigen. In addition, an oligoclonal profile may indicate a conventional antigen-driven process. A polyclonal pattern is in line with a superantigen-derived expansion.7
In CLL patient 402, 4 TCR-BV families were expanded, one in CD4 and one in CD3 in the CD8 subsets (Figure 1A). The CDR3 pattern was monoclonal in 2 overexpressed CD8 TCR-BV families. An oligoclonal pattern was seen in the overexpressed CD4 TCR-BV population and in another CD8 BV-family. In the nonoverexpressed CD4 and CD8 TCR-BV counterparts, all families exhibited a polyclonal pattern (Tables2-3).
Relative TCR-BV gene segment use of 3 CLL patients and mean values of control donors (mean ± SEM) in native CD4 and CD8 T cells, respectively.
(A) CLL 402; (B) CLL403; (C) CLL411. *Statistically significant overexpression. A TCR-BV gene segment was regarded as overexpressed when the relative frequency was more than mean + 3 SD that of control donors (n = 9).
Relative TCR-BV gene segment use of 3 CLL patients and mean values of control donors (mean ± SEM) in native CD4 and CD8 T cells, respectively.
(A) CLL 402; (B) CLL403; (C) CLL411. *Statistically significant overexpression. A TCR-BV gene segment was regarded as overexpressed when the relative frequency was more than mean + 3 SD that of control donors (n = 9).
TCR CDR3 length polymorphism of native CD4 T cells in TCR-BV overexpressed and nonoverexpressed T-cell subsets
| Patient/BV . | CD4 T cells . | |||||
|---|---|---|---|---|---|---|
| Overexpressed . | Nonoverexpressed . | |||||
| Poly . | Oligo . | Mono . | Poly . | Oligo . | Mono . | |
| 402-BV7* | − | + | − | − | − | − |
| 402-BV2 | − | − | − | + | − | − |
| 402-BV4 | − | − | − | + | − | − |
| 402-BV5S1 | − | − | − | + | − | − |
| 403-BV2* | + | − | − | − | − | − |
| 403-BV3* | + | − | − | − | − | − |
| 403-BV4* | + | − | − | − | − | − |
| 403-BV5S1-3* | + | − | − | − | − | − |
| 403-BV5S2-3 | − | − | − | + | − | − |
| 403-BV6S1-3* | + | − | − | − | − | − |
| 403-BV8* | + | − | − | − | − | − |
| 403-BV9* | − | + | − | − | − | − |
| 411-BV3 | − | − | − | + | − | − |
| 411-BV7* | − | + | − | − | − | − |
| 411-BV13S1 | − | − | − | + | − | − |
| 411-BV14 | − | − | − | + | − | − |
| Patient/BV . | CD4 T cells . | |||||
|---|---|---|---|---|---|---|
| Overexpressed . | Nonoverexpressed . | |||||
| Poly . | Oligo . | Mono . | Poly . | Oligo . | Mono . | |
| 402-BV7* | − | + | − | − | − | − |
| 402-BV2 | − | − | − | + | − | − |
| 402-BV4 | − | − | − | + | − | − |
| 402-BV5S1 | − | − | − | + | − | − |
| 403-BV2* | + | − | − | − | − | − |
| 403-BV3* | + | − | − | − | − | − |
| 403-BV4* | + | − | − | − | − | − |
| 403-BV5S1-3* | + | − | − | − | − | − |
| 403-BV5S2-3 | − | − | − | + | − | − |
| 403-BV6S1-3* | + | − | − | − | − | − |
| 403-BV8* | + | − | − | − | − | − |
| 403-BV9* | − | + | − | − | − | − |
| 411-BV3 | − | − | − | + | − | − |
| 411-BV7* | − | + | − | − | − | − |
| 411-BV13S1 | − | − | − | + | − | − |
| 411-BV14 | − | − | − | + | − | − |
Poly indicates polyclonal; oligo, oligoclonal; mono, monoclonal; +, positive; −, negative.
indicate BVs that are overexpressed. A TCR-BV gene segment was regarded as overexpressed when the relative frequency was more than mean + 3 SD for that of control donors (n = 9).
TCR CDR3 length polymorphism of native CD8 T cells in TCR-BV overexpressed and nonoverexpressed T-cell subsets
| Patient/BV . | CD8 T cells . | |||||
|---|---|---|---|---|---|---|
| Overexpressed . | Non-overexpressed . | |||||
| Poly . | Oligo . | Mono . | Poly . | Oligo . | Mono . | |
| 402-BV7 | − | − | − | − | + | − |
| 402-BV23-150 | − | + | − | − | − | − |
| 402-BV43-150 | − | − | + | − | − | − |
| 402-BV5S13-150 | − | − | + | − | − | − |
| 403-BV2 | − | − | − | + | − | − |
| 403-BV3 | − | − | − | − | + | − |
| 403-BV4 | − | − | − | + | − | − |
| 403-BV5S1-3 | − | − | − | − | + | − |
| 403-BV5S2-33-150 | − | + | − | − | − | − |
| 403-BV6S1-3 | − | − | − | + | − | − |
| 403-BV8 | − | − | − | − | + | − |
| 403-BV9 | − | − | − | − | + | − |
| 411-BV3 | − | − | − | + | − | − |
| 411-BV7 | − | − | − | − | − | + |
| 411-BV13S1 | − | − | − | + | − | − |
| 411-BV14 | − | − | − | + | − | − |
| Patient/BV . | CD8 T cells . | |||||
|---|---|---|---|---|---|---|
| Overexpressed . | Non-overexpressed . | |||||
| Poly . | Oligo . | Mono . | Poly . | Oligo . | Mono . | |
| 402-BV7 | − | − | − | − | + | − |
| 402-BV23-150 | − | + | − | − | − | − |
| 402-BV43-150 | − | − | + | − | − | − |
| 402-BV5S13-150 | − | − | + | − | − | − |
| 403-BV2 | − | − | − | + | − | − |
| 403-BV3 | − | − | − | − | + | − |
| 403-BV4 | − | − | − | + | − | − |
| 403-BV5S1-3 | − | − | − | − | + | − |
| 403-BV5S2-33-150 | − | + | − | − | − | − |
| 403-BV6S1-3 | − | − | − | + | − | − |
| 403-BV8 | − | − | − | − | + | − |
| 403-BV9 | − | − | − | − | + | − |
| 411-BV3 | − | − | − | + | − | − |
| 411-BV7 | − | − | − | − | − | + |
| 411-BV13S1 | − | − | − | + | − | − |
| 411-BV14 | − | − | − | + | − | − |
Poly indicates polyclonal; oligo, oligoclonal; mono, monoclonal; +, positive; −, negative.
indicate BVs that are overexpressed. A TCR-BV gene segment was regarded as overexpressed when the relative frequency was more than mean + 3 SD for that of control donors (n = 9).
In CLL patient 403, 8 expanded TCR-BV families were found (7 in the CD4 subset and 1 in the CD8 fraction; Figure 1B). No monoclonal population was detected among these TCR-BV populations. Oligoclonal expansion was seen within one overexpressed CD4 TCR-BV family and one within a CD8 family.
In the remaining 6 overexpressed CD4 BV families, the pattern was polyclonal. In the 7 nonoverexpressed CD8 counterparts, 4 of the TCR-BV families showed an oligoclonal pattern and 3 showed a polyclonal pattern, whereas in the nonoverexpressed CD4 T cell counterparts all patterns were polyclonal (Tables 2-3).
In patient CLL 411, only one TCR-BV family was overexpressed. This expanded CD4 TCR-BV7 family showed an oligoclonal pattern, but in the nonoverexpressed CD8 counterpart a monoclonal population was found. Monoclonal and oligoclonal patterns are known to exist in nonoverexpressed CD8 TCR-BV families of healthy persons at a relatively high frequency.27 In patient CLL 411, 3 randomly selected nonoverexpressed families were also analyzed. A polyclonal CDR3 pattern was seen in the CD4 and the CD8 subsets (Tables 2-3).
To summarize, in the native blood of CLL patients, monoclonal TCR-BV populations were only found within the CD8 population, whereas oligoclonal subsets were seen within the CD4 and the CD8 T-cell fractions. No correlation was found between monoclonality or oligoclonality and TCR-BV expansions.
TCR-BV gene use of CD4+ and CD8+ T cells stimulated with autologous leukemic B cells
The relative TCR-BV gene use of CD4 and CD8 T cells after stimulation with autologous leukemic B cells was compared with that of native T cells (Figure 2A-C). The change in TCR-BV gene expression for each BV family is shown as a Δ value—that is, the percentage TCR-BV use after stimulation subtracted from the corresponding value before stimulation. An increase of 25% or more in the relative percentage of a TCR-BV family compared with baseline was thought to represent a positive response that could indicate specific TCR-BV gene usage induced by activation of the leukemic cells. In patient CLL 402, BV1, BV8, BV9, BV10, BV12, and BV17 within the CD4 population increased after stimulation with the autologous leukemic B cells. Within the CD8 T-cell fraction BV2, BV5S1, BV6S1-3, BV8, and BV10 subsets were augmented (Figure 2A).
Change in TCR-BV gene expression (Δ value) of CD4 and CD8 T cells after stimulation with autologous leukemic cells (percentage of a given TCR-BV family after stimulation subtracted from that of unstimulated cells [baseline]).
(A) CLL 402; (B) CLL403; (C) CLL411. A relative increase of more than 25% or a decrease (only for panel 2B) is indicated above the column by the percentage figure. *Subsets in which CDR3 length analyses were performed (Figures 3-5).
Change in TCR-BV gene expression (Δ value) of CD4 and CD8 T cells after stimulation with autologous leukemic cells (percentage of a given TCR-BV family after stimulation subtracted from that of unstimulated cells [baseline]).
(A) CLL 402; (B) CLL403; (C) CLL411. A relative increase of more than 25% or a decrease (only for panel 2B) is indicated above the column by the percentage figure. *Subsets in which CDR3 length analyses were performed (Figures 3-5).
In patient CLL403, BV1, BV5S2-3, BV7, BV10, BV11, BV12, BV13S2, BV14, BV15, BV16, BV17, BV18, BV19, and BV20 levels within the CD4 population and BV5S2-3, BV8, BV9, BV11, BV12, BV13S1, BV13S2, BV17, and BV18 levels within the CD8 population increased (Figure 2B).
In patient CLL 411, BV2, BV3, BV5S1, BV6S1-3, BV7, BV14, and BV19 levels in the CD4 fraction increased, and BV3, BV6S1-3, BV7, BV9, BV14, and BV17 levels in the CD8 population (Figure 2C).
TCR-BV CDR3 length analyses of leukemic cell-stimulated T cells
TCR-BV families, which increased (25% or more) after stimulation within CD4 or CD8 T-cell subsets, were further selected for CDR3 length polymorphism analyses. CDR3 length analyses could not be performed in all subfamilies because of the lack of material, as indicated in the legend to Figure 2.
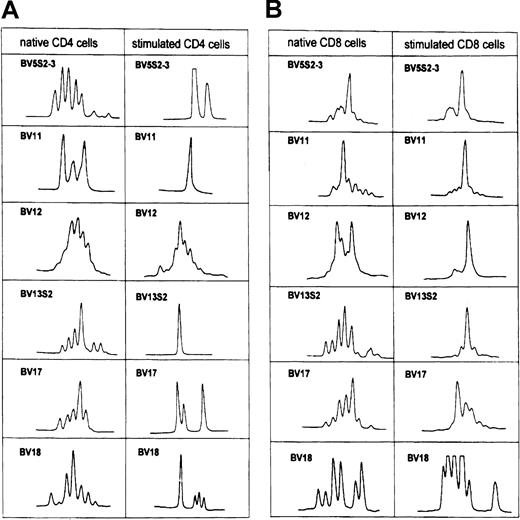
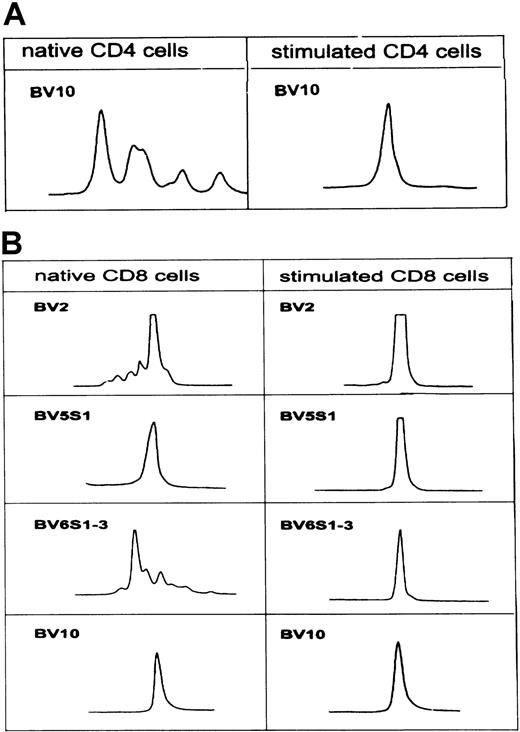
In patient CLL 402, CD4-expanded TCR-BV10 changed from an oligoclonal pattern to a monoclonal pattern in tumor cell–stimulated CD4 T cells. Among the analyzed CD8 TCR-BV families (BV2, BV5S1, BV6S1-3, BV10), the size distribution patterns were monoclonal and oligoclonal before stimulation, whereas a clear monoclonal pattern emerged in all families after stimulation (Figure 3A-B).
In patient CLL 403, the CDR3 length patterns of 6 TCR-BV families were analyzed (Figure 4A-B). The polyclonal pattern changed to monoclonal/oligoclonal profiles in CD4 and CD8 T cells. The BV 5S2-3, BV11, BV13S2, BV17, and BV 18 families within the CD4 T-cell fraction changed to a monoclonal/oligoclonal pattern, whereas BV12 remained polyclonal. In CD8 T cells a monoclonal profile of BV12 and BV13S2 families appeared.
TCR-BV CDR3 length pattern of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL403.
See Figure 2B.
TCR-BV CDR3 length pattern of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL403.
See Figure 2B.
BV 5S2-3 and BV11 within the CD8 T-cell subset remained nearly monoclonal after activation, whereas BV17 and BV18 remained polyclonal.
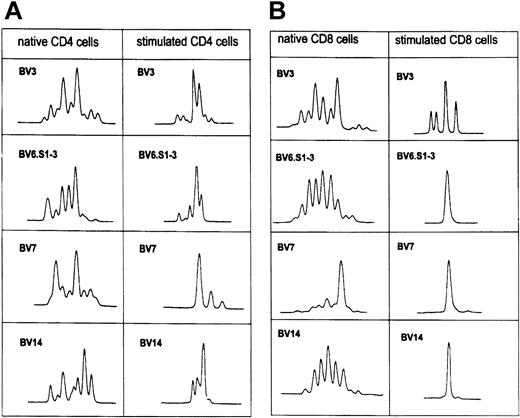
In patient CLL411, 4 TCR-BV families were analyzed. After stimulation a more restricted oligoclonal pattern (BV3, BV6 S1-3, BV7, BV14) was noted in all analyzed CD4 T cells, whereas 3 monoclonal T-cell populations (BV6 S1-3, BV7, BV14) appeared within the CD8 T-cell fraction. One TCR-BV–cell population (BV3) became more oligoclonal (Figure 5A-B).
TCR-BV CDR3 length pattern of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL411.
See Figure 2C.
TCR-BV CDR3 length pattern of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL411.
See Figure 2C.
TCR-BV gene segment use and TCR-CDR3 distribution pattern were also analyzed in T cells stimulated with phytohemagglutinin (PHA) or medium alone. No change in the frequency of TCR-BV use of CD4 and CD8 T cells or in the CDR3 length profile after in vitro activation was noted (data not shown).
TCR-BV CDR3 length pattern in nonexpanded T-cell populations
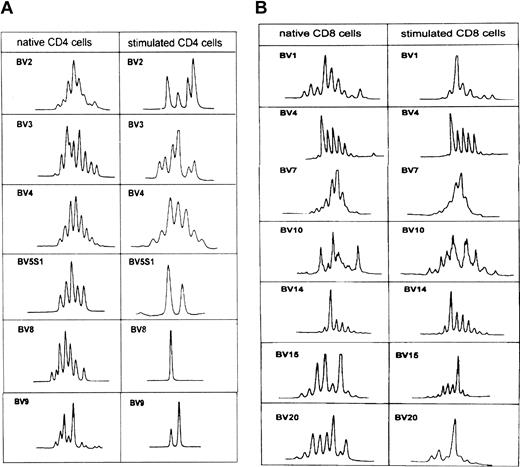
Monoclonal and oligoclonal patterns of the TCR-BV–CDR3 length distribution profile were seen in overexpressed native TCR-BV populations and in nonoverexpressed subsets (Table 2). To explore the possibility that the TCR-BV–CDR3 profile might also change in nonexpanded T-cell populations after stimulation with autologous leukemic B cells, the CDR3 length polymorphism was analyzed in TCR-BV families with a decreased expression.
These analyses were only performed in CD4 and CD8 subpopulations of patient CLL 403, as representative of the other patients. All nonexpanded TCR-BVs with a relative decrease of 25% or more were selected (Figure 6A-B). Polyclonal patterns of BV5S1, BV8, and BV9 of the CD4 subset changed to a monoclonal or an oligoclonal profile, whereas BV2, BV3, and BV4 remained polyclonal. BV1, BV15, and BV20 changed from a polyclonal pattern to a monoclonal or oligoclonal pattern in CD8 T cells, whereas BV4, BV7, BV10, and BV14 remained monoclonal (Figure 6A-B).
TCR-BV CDR3 length pattern of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL403.
See Figure 2B.
TCR-BV CDR3 length pattern of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL403.
See Figure 2B.
Cloning and sequencing
To further analyze and confirm monoclonality, sequence analyses were performed on the CDR3 of TCR-BV families stimulated with leukemic B cells.
In patient CLL 403, the nonoverexpressed native TCR-BV13S2 family within the CD4 subset expanded on stimulation with leukemic B cells with a monoclonal CDR3 profile (Figures 2B, 4A). BV13S2-BJ2S5 accounted for approximately 4% of all BJs before activation with the leukemic cell, but after stimulation 100% of all JBs were BV-RRSGSLV-BJ2S5 rearranged (Table4).
TCR-BV13S2 gene transcripts in patient CLL403
| Population . | BV . | NDN . | BJ . | Gene segments . | Frequency . |
|---|---|---|---|---|---|
| CD4/native BV13S2 | C A S S | V Y R A | T E A F F G | BJ1S1 | 2 of 23 |
| C A S | R F D S S S | T E A F F G | |||
| C A S S | K Q G S W | Q P Q H F G | BJ1S5 | 4 of 23 | |
| C A S S | Q G G | E Q F F G | BJ2S1 | 5 of 23 | |
| C A S S | S Q T S G R L Y | E Q F F G | |||
| C A S | R L P G G D | E Q F F G | |||
| C A S | R P L G A G T A V R G | E Q F F G | |||
| C A S S | F S R V | T G E L F F G | BJ2S2 | 3 of 23 | |
| C A S S | V E A | G E L F F G | |||
| C A S S | Y P Q L K | D T Q Y F G | BJ2S3 | 6 of 23 | |
| C A S S | S I L A G G A | D T Q Y F G | |||
| C A S | D D | T Q Y F G | |||
| C A S S | S D | D T Q Y F G | |||
| C A S S | T R L I D S Q E | T Q Y F G | |||
| C A S S | P R G R K T | Y F G | BJ2S4 | 1 of 23 | |
| C A S S | Y S G A G A Y E | Q Y F G | BJ2S5 | 1 of 23 | |
| C A S S | D G G R N | Y E Q Y F G | BJ2S7 | 1 of 23 | |
| CD4/activated BV13S2 | C A S | R R S G S L V | T Q Y F G | BJ2S5 | 5 of 5 |
| Population . | BV . | NDN . | BJ . | Gene segments . | Frequency . |
|---|---|---|---|---|---|
| CD4/native BV13S2 | C A S S | V Y R A | T E A F F G | BJ1S1 | 2 of 23 |
| C A S | R F D S S S | T E A F F G | |||
| C A S S | K Q G S W | Q P Q H F G | BJ1S5 | 4 of 23 | |
| C A S S | Q G G | E Q F F G | BJ2S1 | 5 of 23 | |
| C A S S | S Q T S G R L Y | E Q F F G | |||
| C A S | R L P G G D | E Q F F G | |||
| C A S | R P L G A G T A V R G | E Q F F G | |||
| C A S S | F S R V | T G E L F F G | BJ2S2 | 3 of 23 | |
| C A S S | V E A | G E L F F G | |||
| C A S S | Y P Q L K | D T Q Y F G | BJ2S3 | 6 of 23 | |
| C A S S | S I L A G G A | D T Q Y F G | |||
| C A S | D D | T Q Y F G | |||
| C A S S | S D | D T Q Y F G | |||
| C A S S | T R L I D S Q E | T Q Y F G | |||
| C A S S | P R G R K T | Y F G | BJ2S4 | 1 of 23 | |
| C A S S | Y S G A G A Y E | Q Y F G | BJ2S5 | 1 of 23 | |
| C A S S | D G G R N | Y E Q Y F G | BJ2S7 | 1 of 23 | |
| CD4/activated BV13S2 | C A S | R R S G S L V | T Q Y F G | BJ2S5 | 5 of 5 |
cDNA from the populations indicated was PCR-amplified with BV-BC primers and was cloned into vector (see “Materials and methods”). The TCR joining segments (BJ regions) were eluted by comparing to the BJ gene segments in the GenBank. The NDN region nucleotides were defined as any nucleotide that could not have been accounted for by any of the germline BV or BJ sequences.
BV is TCR β variable region segment gene.
In patient CLL 402, the BV10 family within the CD8 subset was not overexpressed in native T cells (Figure 1A), but a dramatic expansion was noted after stimulation with autologous leukemic B cells (Figure2A). The CDR3 pattern was monoclonal before and after leukemic cell activation (Figure 3B). Sequencing revealed identical nucleotide sequences of all BJ2S1 before and after activation (Table5).
TCR-BV CDR3 length profile of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL402.
See Figure 2A.
TCR-BV CDR3 length profile of native CD4 and CD8 T cells and after stimulation with the autologous leukemic B cells in patient CLL402.
See Figure 2A.
TCR-BV10 gene transcripts of patient CLL 402
| Population . | BV . | NDN . | BJ . | Gene segments . | Frequency . |
|---|---|---|---|---|---|
| CD8/native BV10 | C A S | G V G G | N E Q F F G | BJ1S1 | 14 of 15 |
| C A S | L E G G S | Y S N Q P Q H F G | BJ1S5 | 1 of 15 | |
| CD/8 activated BV10 | C A S | G V G G | N E Q F F G | BJ2S1 | 15 of 15 |
| Population . | BV . | NDN . | BJ . | Gene segments . | Frequency . |
|---|---|---|---|---|---|
| CD8/native BV10 | C A S | G V G G | N E Q F F G | BJ1S1 | 14 of 15 |
| C A S | L E G G S | Y S N Q P Q H F G | BJ1S5 | 1 of 15 | |
| CD/8 activated BV10 | C A S | G V G G | N E Q F F G | BJ2S1 | 15 of 15 |
cDNA from the populations indicated was PCR-amplified with BV-BC primers and cloned into vector (see “Materials and methods”). The TCR joining segments (BJ regions) were eluted by comparison with the BJ gene segments in the GenBank. The NDN-region nucleotides were defined as all nucleotides that could not have been accounted for by any of the germline BV or BJ sequences.
BV indicates TCR-β variable region segment gene.
Discussion
Specific interaction of T cells with a putative antigen occurs through the recognition of MHC–peptide complexes through the T-cell receptor. The CDR3 region of the TCR entails direct contact with the MHC peptide complex. An antigen-driven immune response leads to the appearance of recurrent transcripts with identical junction regions—that is, a monoclonal expansion within a given TCR-BV family.
Analyzing T cells from tumor-infiltrating lymphocytes applying TCR-CDR3 analyses using nucleotide sequencing or length polymorphism revealed the presence of overexpressed TCR-BV families and monoclonal/oligoclonal T-cell populations.11,12,26,28Monoclonal and oligoclonal expansions have been reported among T cells in patients with B-cell malignancies.13,14,29 In a CLL patient an oligoclonally expanded BV19+CD8+T-cell subset was shown to react specifically with the autologous leukemic B cells.13 In myeloma patients clonal expansions of T cells recognizing the tumor-derived idiotypic immunoglobulin have also been found. Patients with such idiotype-reactive T cells seemed to have better prognoses.30
In the present study, 3 patients previously shown to have leukemia-specific T cells3 were analyzed for TCR-BV usage. The results clearly demonstrate that TCR-BV overexpression in vivo is not necessarily an indication of monoclonal or oligoclonal T-cell expansion but that it requires TCR-CDR3 length analyses to confirm monoclonality or oligoclonality. Moreover, within a TCR-BV family with a polyclonal CDR3 length distribution pattern, a small monoclonal or oligoclonal T-cell population might be hidden, revealing itself only after activation with autologous leukemic B cells. Monoclonal expansions among in vitro–stimulated T cells by leukemic B cells were found within the CD4 and the CD8 T-cell subsets, and different TCR-BV families were expanded. These results indicate that CLL patients may house leukemia cell-specific T cells within the CD4 and CD8 subsets that probably recognize different leukemia-associated antigens or different epitopes of the same antigen.
The increase of certain TCR-BV families after in vitro activation by tumor cells may be attributed to culture conditions or to superantigen activation.31 In T cells cultured in medium alone or in the presence of PHA, however, there was no change in the TCR-BV pattern (data not shown). Thus, the monoclonal or oligoclonal TCR-BV pattern seen after in vitro stimulation with the autologous tumor B cells strongly suggests selection by tumor cell-associated antigens. Polyclonal expansions of some but not other TCR-BV families after leukemia cell stimulation might be explained by the parallel production of weak superantigens in such conditions. Note, for instance, the parallel expansions in CD4 T cells of BV1, BV12, and BV17 in patients 402 and 403.
To detect minor tumor-specific T-cell populations and to confirm monoclonality, sequencing is needed. In patient CLL 403, a monoclonal BV13S2-BJ2S5 T-cell population was only detected after activation with the leukemic cells. The in vivo overexpressed BV13S2 population had other CDR3 sequences than that induced in vitro by the leukemic cells, indicating that the TCR-BV expansion detected in vivo may or may not reflect T cells recognizing autologous tumor cells. In patient CLL 402, a minor clonal expansion of the TCR-BV10 population in vivo was further expanded after in vitro activation using the same CDR3 sequence. After the activation of T cells in vitro with autologous carcinoma cells, an expanded TCR-BV21 population with a unique BV21-D-J2S7 junctional region was found not to have been detected in in vivo T cells.32 In hairy cell leukemia (HCL), 2 different BV2 T-cell clones and one BV8S3 T-cell clone were generated in vitro using CD40L-activated HCL cells to stimulate autologous T cells. These clones could not be detected in vivo among native spleen and blood T cells.33 In melanoma patients vaccinated with Melan-A peptide overexpression of BV16 in vitro and in vivo, CD8 T cells with identical CDR3 sequences were found.34
The exact mechanisms behind TCR-BV expansions in vivo of patients with malignant diseases are unclear. It may seem obvious, however, from the present study that all T-cell expansions in vivo are not caused by the recognition of tumor cells. Some, particularly in CD8 T cells, may be age related,35 others may be caused by viral infections,36 and so on. In support of the assumption that at least some TCR-BV expansions may be due to leukemia cell recognition are results using Ad-CD54–transduced leukemic B cells. After infusion of such autologous Ad-CD54–transduced leukemia cells, a marked, select increase in TCR-BV5 T cells was noted in patients.37 38
The presence of a monoclonal or an oligoclonal TCR-CDR3 profile in the blood of patients with malignant diseases has been suggested to reflect a tumor-specific T-cell response.10 However, our data indicate that monoclonality or oligoclonality of T cells is not necessarily a reflection of a tumor response. Furthermore, because of serial dilutions of cDNA, PCR amplification, and further dilutions during CDR3 length analysis, weak tumor-specific TCR-BV families might disappear whereas strong non–tumor-specific TCR-BV families might appear as single peak(s). This novel observation indicates that only BVs that have been expanded after tumor cell activation showing a monoclonal or an oligoclonal CDR3 length pattern may, with some degree of certainty, reflect a tumor-specific T-cell response. Our data suggest that the use of TCR-BV analyses to monitor a tumor response should be used with caution. Specific tumor activation in vitro might be required to achieve an acceptable degree of certainty.
In summary, this study indicates that CLL patients might have leukemia-specific T cells within CD4 and CD8 populations. Response expansions in vitro were high in CD8 but low in CD4, indicating different stages of memory status in the 2 subsets. Thus, TCR-BV analyses of native peripheral blood T cells should be used with caution as a measure for the presence of tumor-specific T cells. Functional capabilities of clonally expanded T cells in the B-cell leukemia context and their specificity remain to be established. Immunization attempts in vivo using B-cell leukemia cells would now seem a logical further step. A second one may concentrate on exploring in particular the possible CTL activities associated with the in vivo expanded, in vitro responsive, and further propagated TCR-BV–restricted CD8 T-cell “clones.”
We thank Ms Gerd Ståhlberg for skillful secretarial assistance.
Prepublished online as Blood First Edition Paper, October 3, 2002; DOI 10.1182/blood-2002-03-0746.
Supported by grants from the Cancer Society in Stockholm, the Swedish Cancer Society, the King Gustaf Vth Jubilee Fund, the Cancer and Allergy Foundation, the Torsten and Ragnar Söderberg Foundation, the Gunnar Nilsson Foundation, the Karolinska Institute Foundation, and Sigbritt and Arne Lundbergs Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Håkan Mellstedt, Cancer Center Karolinska, Department of Oncology (Radiumhemmet), Karolinska Hospital, SE-17176 Stockholm, Sweden; e-mail: hakan.mellstedt@ks.se.

![Fig. 2. Change in TCR-BV gene expression (Δ value) of CD4 and CD8 T cells after stimulation with autologous leukemic cells (percentage of a given TCR-BV family after stimulation subtracted from that of unstimulated cells [baseline]). / (A) CLL 402; (B) CLL403; (C) CLL411. A relative increase of more than 25% or a decrease (only for panel 2B) is indicated above the column by the percentage figure. *Subsets in which CDR3 length analyses were performed (Figures 3-5).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/3/10.1182_blood-2002-03-0746/4/m_h80333780002.jpeg?Expires=1769112326&Signature=qMex6sJXsXLga5toC-h7z66kvuJWwT1wjkdmEkdndrWcPKSQAdlS1rNoyHX1wYFoeGfiTwEpIs0jJFR8pSqvoXfrDYfrHA2ZH5q3F6KZpM-DeG784mfxtlZ~-4c2XW32ya1uUYLlGZk5MRhS9tP-btLlUkF7hkLw9zZKnDsq17uC~SmNIdz~ue0iaC5YX4yLE2ffKS5qfTOZJ3sIa9WlfkaYnN3546EsylO41~eQlVkxKWLExWA0nVQzANsyYwaOTFSTt~3Gte3bK0RXY-OrAAThHZpZRu5yI7sfB5dlRJvHgUnCIsjyrzMmR0Gx3G8coDToVO~qlNbE6cPxSIxLRg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal