Recent conceptual and technical improvements have resulted in clinically meaningful levels of gene transfer into repopulating hematopoietic stem cells. At the same time, evidence is accumulating that gene therapy may induce several kinds of unexpected side effects, based on preclinical and clinical data. To assess the therapeutic potential of genetic interventions in hematopoietic cells, it will be important to derive a classification of side effects, to obtain insights into their underlying mechanisms, and to use rigorous statistical approaches in comparing data. We here review side effects related to target cell manipulation; vector production; transgene insertion and expression; selection procedures for transgenic cells; and immune surveillance. We also address some inherent differences between hematopoiesis in the most commonly used animal model, the laboratory mouse, and in humans. It is our intention to emphasize the need for a critical and hypothesis-driven analysis of “transgene toxicology,” in order to improve safety, efficiency, and prognosis for the yet small but expanding group of patients that could benefit from gene therapy.
Effects and side effects in hematopoietic gene therapy
“This is a strange drop in my blood” (Goethe).
“It is the dose that makes the poison” (Paracelsus).
“What can go wrong, will go wrong” (Murphy).
Hematopoietic stem cells (HSCs) are important targets for somatic gene therapy, considering their availability for in vitro manipulation and their enormous biologic capacity.1,2 In selected entities, gene therapy involving manipulation of HSCs has now clearly shown clinical efficiency, opening up new perspectives for the entire field.3 4 However, it is a principle in pharmacology that no true effect is possible without inducing side effects. Prognosticating the type and incidence of side effects is an important step toward predicting the overall therapeutic benefit for a new modality.
The genetic modification of HSCs generates special concerns:
1. These cells are long-lived and might represent a reservoir for the accumulation of proto-oncogenic lesions.5
2. Current technology requires that HSCs have to be enriched and cultured in vitro to become accessible to genetic manipulation.
3. This also implies that the engineered graft represents only a small fraction (probably about 1%-10%) of the hematopoietic cell pool of a healthy individual. Infused cells may therefore be altered not only in terms of quality, but will also be heavily diluted by unmodified counterparts residing in the body. This may result in the establishment of a “strange drop in the blood,” which could correct diseases only if it were strongly enriched in vivo.
4. Therefore, achieving targeted amplification or preferential survival of engineered cells is one important key to success in hematopoietic gene therapy.2-4 However, clonal expansion, while limited by cellular senescence and exhaustion,6 has also been suggested as a risk factor contributing to cellular transformation, at least when occurring under nonphysiologic conditions of growth.7
5. HSCs, or at least the cell preparations enriched for HSCs, may not only reconstitute the entire myeloerythroid and lymphoid spectrum, but they may also differentiate into or fuse with other cell types, including endothelial; skeletal and heart muscle cells; hepatocytes; neurons; and epithelial of gut and lungs. However, the frequency of such events is controversial.8-12 The developmental potential of HSCs generates a huge repertoire of conceivable biologic conditions and anatomic sites where side effects may manifest. However, the likelihood of manifestations outside the hematopoietic system appears to be relatively low unless special triggers exist that drive fate-switching.11 12
6. Because of the high proliferative potential of HSCs, stable, heritable gene transfer is required for successful genetic modification. In the current “state-of-the-art” only viral vectors on the basis of retroviruses (including lentiviruses) mediate a predictable efficiency of stable transgene insertion with a predefined copy number.13 Chromosomal insertion guarantees transgene maintenance during clonal amplification. Episomally persisting viral vector systems such as those based on Epstein-Barr virus are still suboptimal14 because efficient gene transfer into HSCs is either not yet available or maintenance and expression of transgene copies are insufficiently investigated. Physicochemical methods result in a low probability for stable transgene insertion (< 10−4).15 Their efficiency may be increased when combined with endonucleases from retrotransposons or site-specific integrases.16 Adeno-associated viruses (AAVs) also have a low and variable rate of stable insertion.13 Recent advances in adenoviral vector technology may increase their potential for stable gene delivery.17 However, the utility of all of these alternative methods for transduction of HSCs with a defined and persisting transgene copy number is still unknown, as is the genetic risk associated with transgene insertion through these modalities.
7. The use of retroviral (including lentiviral) vectors implies that engineered cells of the same graft will vary with respect to transgene insertion sites (which are unpredictable and can affect both transgene and cellular gene expression), copy number per cell (which can be controlled more easily, but not entirely), and sequence (which can be modified in the error-prone process of reverse transcription). This produces a mixed chimerism of genetic modification in different stem cell clones, each with a theoretically distinct potential for eliciting side effects.
To facilitate the evaluation and discussion of side effects, we introduce a classification system at this point (Table1).
Categories of side effects in the genetic manipulation of hematopoietic stem cells
| Level . | Category . | Example . |
|---|---|---|
| 1 | Culture | Loss of homing potential |
| 2 | Vector | Fusiogenic properties of viral envelope proteins |
| 3 | Genotoxicity | Insertional mutagenesis |
| 4 | Phenotoxicity | Interference of transgene product with cellular signaling networks |
| 5 | Selection toxicity | Side effects of regimens used to selectively engraft or expand manipulated cells |
| 6 | Immune response | Elimination of modified transgenic cells by antibodies or cytotoxic T cells directed against transgene-encoded antigens |
| 7 | Interactions of 1-6 | Cooperation of 3, 4, and 5 |
| Level . | Category . | Example . |
|---|---|---|
| 1 | Culture | Loss of homing potential |
| 2 | Vector | Fusiogenic properties of viral envelope proteins |
| 3 | Genotoxicity | Insertional mutagenesis |
| 4 | Phenotoxicity | Interference of transgene product with cellular signaling networks |
| 5 | Selection toxicity | Side effects of regimens used to selectively engraft or expand manipulated cells |
| 6 | Immune response | Elimination of modified transgenic cells by antibodies or cytotoxic T cells directed against transgene-encoded antigens |
| 7 | Interactions of 1-6 | Cooperation of 3, 4, and 5 |
As the whole process of genetic manipulation of transplantable HSCs is complex (Figure1), problems may be encountered at different levels: (1) enrichment and culture of target cells (toxicity of cell manipulation); (2) vector production (vector toxicity); (3) insertion of foreign sequences or other alterations of the cellular genome (genotoxicity); (4) expression of transgenes (for which we would like to introduce the term “phenotoxicity”); (5) conditioning or selective drugs for enrichment of gene-manipulated cells (selection toxicity); (6) immune responses evoked by vector components or the transgene product (immunogenicity); and (7) aggravating interactions of some of these events.
Schematic overview of the procedures involved in the ex vivo manipulation of hematopoietic stem cells for gene therapy and preclinical approaches.
Schematic overview of the procedures involved in the ex vivo manipulation of hematopoietic stem cells for gene therapy and preclinical approaches.
Depending on the type, severity, and kinetics of side effects, patients may be asymptomatic or present with unclear symptoms, such as fever of unknown origin, signs of hemolysis, cytopenia of any lineage, immunodeficiency, autoimmune disorders, myelodysplasia, or, at worst, lymphoma, leukemia, or other types of malignancy. Some of these disorders, most of which are of only theoretical signficance at present, will occur only after prolonged periods of time18 19 and may be missed in preclinical studies with limited follow-up after genetic manipulation of HSCs. However, increasing the potency of the methods and the numbers of treatments may confront us with a growing number of reports.
Indeed, this review was prompted by our observation of a leukemia in a mouse study with prolonged follow-up after retroviral gene transfer into hematopoietic cells.20 Unfortunately, the first case of a malignant disorder following clinical retroviral vector-mediated gene transfer into human hematopoietic cells was observed shortly thereafter, manifesting 3 years after the infusion of retrovirally modified cells21,22 so that a once theoretical risk has become a real one. The uncertainty observed in the scientific and regulatory community following these reports23 24reflects a considerable need for systematic toxicology of genetic cell modifications.
Paracelsus, a founder of toxicology, has provided 3 golden rules for the assessment of side effects. The first is that poison is a question of dose.25 Dose issues are encountered at several levels in hematopoietic gene therapy (Figure2): the number of gene transfer particles to which the cells are exposed, the transgene copy number per cell, transcription rates, efficiency of RNA processing, protein features such as activity or stability of enzymes, the size of the target cell pool (generating a clonal repertoire due to the variations in transgene processing and integration), the life span of transplanted cells, and the number of patients treated.
Overview of dose issues in stem cell gene therapy.
The following items are indicated in the lettered circles: A, particle dosage; B, number of inserted genes; C, transcription; D, RNA processing; E, protein features; F, target pool size; G, cellular life span and plasticity; and H, number of patients.
Overview of dose issues in stem cell gene therapy.
The following items are indicated in the lettered circles: A, particle dosage; B, number of inserted genes; C, transcription; D, RNA processing; E, protein features; F, target pool size; G, cellular life span and plasticity; and H, number of patients.
Paracelsus' second rule is that a compound has a specific site (within the body) where it exerts the greatest effect.25 Applied to gene therapy, this indicates that cell type and its developmental plasticity really matter. The third rule is to use animal models for preclinical dose finding.25 Therefore, the limitations of animal models also have to be considered. Cell specificity and animal testing have been central items in gene therapy from the beginning. However, most studies focused on efficiency and were not designed to measure unexpected effects.
The present review summarizes recent insights into molecular mechanisms underlying side effects of genetic interventions in HSCs, following the classification of issues listed above (Table 1), and discusses consequences for the most commonly used animal model, the laboratory mouse.
Toxicity of cell manipulation
Under steady-state conditions (normal hematopoietic turnover and an intact bone marrow niche), the majority of HSCs cycles slowly, yet continuously.26-28 For genetic modification, HSCs are either harvested from peripheral blood or bone marrow.29The yield and biologic features of cells from these sources differ depending on the use of mechanical harvest versus cytokines (typically granulocyte colony-stimulating factor [G-CSF]) and/or chemotherapy, which may have direct implications for the efficiency of retroviral transduction and engraftment.29-31 Exposure to cytotoxic agents may compromise the engraftment potential of HSCs.32 Umbilical cord blood is a promising resource of stem cells, but the limited numbers of HSCs contained in cord blood may restrict a wider use in adults.33 34
Target cells of genetic manipulation usually have to be enriched to facilitate physical interaction with vector particles (Figure 1). Enrichment of HSCs for clinical use is most frequently achieved by immunoaffinity selection for the CD34 antigen. Developed for “mainstream” clinical applications, these processes for cell harvesting and enrichment have an excellent safety profile, and the engraftment potential of CD34-enriched cells is very good.35 However, according to our current understanding, long-term repopulating HSCs probably represent less than 1% of the CD34+ cell pool. Thus, the target pool size currently used for gene transfer is probably about 100-fold greater than actually required.
In theory, manipulating 10 000 HSCs (or maybe even much smaller numbers) should be sufficient to achieve a polyclonal transgenic hematopoiesis.27,36,37 This would reduce significantly the numbers of vector particles required for cell manipulation, the risk of random mutagenic events that are related to the number of transgene insertions (below), and probably also the costs of the procedure. However, methods required for further enrichment of HSCs, such as isolation of the CD34+CD38− population or their more primitive precursors,29,38-40 have not yet been established for routine clinical use. High-grade purification of HSCs based on flow cytometry sorting has been shown to be feasible, but concerns remain regarding the risk of contamination, fitness of the sorted cells, selective interference with short-term engraftment, and risks associated with cell expansion.39
Although short-term reconstitution may be promoted following cell expansion in vitro,41 current culture conditions may induce a selective loss of long-term HSCs.29 Several underlying mechanisms have been identified: commitment to differentiation (loss of pluripotency) or even apoptosis, a cell-cycle–associated loss of engraftment/homing properties, and differential susceptibility to natural killer cell–mediated rejection.29,42-44 Although engraftment with cultured cells alone has been rapid and sustained in clinical gene therapy studies,45,46 extended manipulations, such as prolonged culture or enrichment of cells expressing the transgene prior to infusion, may promote deficits in long-term reconstitution.29,41,47 Similar considerations apply for lymphocyte cultures.48 Long-term follow-up, which in humans encompasses many years, will be required to draw firm conclusions that HSC exhaustion is not triggered by the procedures used during HSC manipulation in vitro.29 Therefore, all efforts invested to maintain stem cell properties during in vitro culture are important. Improvements of HSC culture can be achieved by (1) the use of serum-free culture conditions,49 (2) the definition of appropriate cytokine combinations,50 (3) the manipulation of transcription factor levels such as HOXB4,51 (4) the introduction of other (such as extracellular matrix) molecules52-54 or appropriate stroma components,55,56 and (5) protocols allowing a return to cell-cycle quiescence prior to infusion.57,58 Moreover, new vector systems are being developed to reduce the need for stem cell proliferation prior to gene transfer.13 59-61
It may also be interesting to expand engineered cells in vitro following gene transfer. However, in at least one case, this attempt has been associated with an increased risk of malignant transformation of transduced murine hematopoietic cells.62 Although it is possible that the expansion culture promoted a specific side effect of the vector or packaging cells used in this study, further work is required to address the extent to which culture conditions support a preferential growth of mutants with proto-oncogenic lesions.
In summary, new procedures for HSC harvest, enrichment, gene transfer, and expansion culture need to be studied intensively before clinical application. Besides “conventional” mouse models,32immunodeficient mice38,63 or fetal sheep47supporting engraftment of primitive human hematopoietic cells and supporting studies in nonhuman primates64 serve as valuable models for this purpose.
Risks related to vector production
Conventional retroviral vectors based on mouse leukemia virus (MLV) and the more recently developed lentiviral vectors (such as those based on HIV-1) differ in many respects, particularly in their nuclear import strategies.13,60,61 While MLV vectors require cell division for chromosomal insertion, lentiviral vectors may also transduce nonproliferating cells. However, lentiviral transduction efficiency also declines according to cell-cycle stages in the order M > G1 > G0. Another feature of HIV-based lentiviral vectors is that complex transgene cassettes containing cryptic splice sites are more reliably transferred,65-68 which may be related to regulatory functions of the viral REV protein expressed during the packaging process. Because of the significant differences in the biologic properties of the viral proteins involved in the generation of replication-defective vectors, MLV and HIV vectors have distinct requirements for the design and culture of their respective producer cells.
Stable producer clones are more easily established with long terminal repeat (LTR)–driven vectors
Progress in the design of retroviral vectors developed on the basis of MLV has improved their performance with respect to vector production, gene transfer efficiency, and transgene expression.19,69-71 This vector system is the only one currently used in clinical trials for stable gene transfer into HSCs.2 One important advantage of MLV packaging components is their lack of cytotoxic effects, resulting in the ability to derive defined cloned producer cell lines with only 1 or 2 transgenes (“provector”) inserted to encode the vector. This ensures the highest degree of transgene stability that can be achieved with retroviral packaging cell technology. However, the establishment of stable cloned producers with a limited and defined transgene copy number is greatly alleviated when retroviral gene transfer is used to establish the provector. Thus, transcriptional control needs to originate from the long terminal repeat (LTR), as in wild-type retroviruses. This configuration implies an increased risk of activating neighboring cellular sequences in target cells (below) and for recombination with viral coding sequences in the packaging cells, potentially facilitating the accidental generation of replication-competent retrovirus (RCR) recombinants.19
Replication-competent retrovirus
Contamination of vector stocks with RCR can be detected by polymerase chain reaction (PCR), enzyme-linked immunosorbent assay (ELISA), or cell biologic assays. While the sensitivity of these methods can be very high, residual contamination of a clinical vector preparation as a matter of principle cannot be fully excluded. Important improvements in the design of vectors and packaging cells have greatly reduced the risk of generating RCR.69
The risk of developing a disease following accidental exposure to an MLV-related RCR depends heavily on the genetic background of the recipient and the integrity of the immune system. Replication-competent MLV with an amphotropic envelope protein was not found to represent a significant pathogen for immunocompetent or transiently immunosuppressed nonhuman primates.19 However, when CD34+-enriched cells were exposed to high titers of RCR-contaminated vector preparations in vitro and infused under conditions of strong immunosuppression, rhesus monkeys developed lymphomas within one year.72 This required the absence of an immune response against retroviral particles or infected cells, and was likely driven by insertional mutagenesis (below) due to massive virus replication within susceptible lymphoid cells.19,72 When inoculated into newborn mice, amphotropic MLV may also induce a spongiform encephalopathy, whose kinetics and anatomic distribution depend on the type of the envelope protein.73
Potential RCR originating from lentiviral packaging cells has not been described to date. Accordingly, the potential pathogenicity of such recombinants is unknown, yet expected to be unlike that of wild-type HIV as a result of the anticipated differences in Env proteins, regulatory elements for gene expression, and the absence of many HIV accessory genes. An established limitation of currently used stable lentivirus packaging lines is genetic instability, because they may undergo multiple superinfection events when cultured (due to the lack of subgroup interference with the VSV-G pseudotype used).74-76
Although no side effects have been reported in more than a decade of clinical experience even with early generations of retroviral producer cells,77 stringent safety testing and further technological improvements are still desirable for both retro- and lentivirus production systems. In the unlikely event of accidental exposure to RCR and their escape from immune control, it may be possible to suppress viral replication in patients using clinically approved inhibitors, unless resistance develops.78
Mobilization
In the absence of an RCR originating from the packaging cells, spread of a retroviral vector could be possible when naturally occurring viruses exist that can package the vector RNA and are transmitted in the human population. This concern appears to be irrelevant for MLV vectors,79 but needs to be considered for vectors developed on the basis of HIV or other lentiviruses.80 To prevent this problem, lentiviral vectors are typically designed with a so-called self-inactivating (SIN) LTR. This is achieved by placing the enhancer-promoter into an internal position between defective LTRs, eliminating transcription of the packaging signal required for incorporation of the vector RNA in virus particles.60 61
Transient transfection for vector production
So far, both lentiviral and previously investigated retroviral SIN vectors cannot be produced at sufficient titers from cloned packaging cell lines.74 Efficient production of SIN vectors has been achieved only following transient transfection of plasmid vector constructs into packaging cells.61,74,81 Although significant amounts of vector particles can be produced using this procedure,81 concerns remain unresolved regarding the type and incidence of plasmid recombinations, accidental transfer of plasmid DNA with vector particles,70 and the identity of the product obtained in independent production batches.
The infidelity of reverse transcription
A limitation common to all types of retroviral vectors is the possibility for transgene recombination or mutation occurring during the obligate step of reverse transcription. The retroviral enzyme reverse transcriptase converts RNA to double-stranded DNA with an infidelity of about 10−4, suggesting that mutations are introduced once per 10 kb of a retroviral RNA template.82This may reflect an evolutionary pressure to produce about one mutation per replication cycle, given a genome size of natural retroviruses in the range of 8 to 11 kb. The misincorporation rate is similar for vectors based on MLV and HIV.83 If we consider as a worst case scenario a proto-oncogene such as N-RAS with a size of 570 bp, one mutation could occur per 18 retrovirally transduced copies. For N-RAS, at least 3 activating mutations are known from a total of 1710 (3 × 570) possibilities for single-point mutations (http://www.expasy.ch/cgi-bin/niceprot.pl?p01111). Thus, about one oncogenic N-RAS mutant would be formed per 104reverse transcriptions. In a clinical setting, about 108 to 109 infectious particles are required per CD34+ cell preparation. Therefore, it may be important to define the oncogenic mutation frequency for a given cDNA, especially when dealing with transgenes encoding “signaling” molecules.
Much more frequent errors in transgene replication may result from sequence deletions or recombinations before or during reverse transcription.82,83 Regulatory genome sequences that can be required to achieve cell-type–specific gene expression84 and some clinically relevant cDNAs such asMDR1 or HSV-TK may contain cryptic splice sites that give rise to pregenomic splicing of the vector RNA in packaging cells.85,86 Interestingly, the frequency of these cryptic splicing events also depends on the packaging cell line.87
Also, intrastrand or interstrand recombinations are not uncommon during reverse transcription (retroviruses typically package 2 copies of a pregenomic RNA). These can be triggered by direct sequence repeats within the transgene,83,88 and again occur with similar frequency in vectors based on MLV or HIV.83 The vast majority of such events will simply reduce the efficiency of the gene transfer. However, it may be worthwhile to address potential hazards induced by aberrations of a given transgene prior to clinical testing. Attempts to reduce sequence repetitions, to eliminate unwanted splice sites, and to choose appropriate packaging cell clones greatly improve the fidelity of transferring intact transgene sequences.84,87,89 90
Another concern related to vector production is the accidental incorporation of cellular RNA in the retroviral particle. Acutely transforming retroviruses encoding cellular oncogenes have evolved through such events, again requiring recombination during reverse transcription.82 91 However, to create such an unwanted oncogene vector, further mutations triggered by multiple rounds of replication in virus/vector spread are typically required. Therefore, this risk appears extremely low with a replication-defective vector.
Risks related to transgene insertion
Complications resulting from transgene insertion (insertional mutagenesis) are a concern for all stable gene transfer methods. Retroviral insertion has some unique properties. The first resides in the fact that insertion is a default event in the retroviral life cycle,82,91 implying that the frequency of transgene insertion per cell can be predetermined by adjusting the multiplicity of infection.92 The second is that insertion tends to take place in euchromatin, possibly because of its improved accessibility.93 Consequently, the risk for insertion in transcriptionally active regions of chromosomal DNA is increased, as recently also demonstrated for HIV and derived vectors.94This implies a possibility for a cell-type–specific distribution, also assisted by host factors that participate in the preintegration complex.94 Retroviral integrases are not sequence-specific with respect to transgene insertion, yet prefer specific structural features (bended DNA).95 Thus, some yet unknown genetic loci may be at increased risk for retroviral insertion.94The third important feature of retroviral insertion is that it typically does not create subsequent recombinations within or outside the affected locus, although exceptions to this rule have been reported. Postintegration deletions may occur within repeats present in a single retroviral transgene, but these events appear to be rare.96 Mutations within and surrounding a retroviral genome during expansion or malignant transformation of a transduced cell have also been described.97,98 Finally, recombinations may occur between sequence-related, yet independently inserted, retroviral alleles.99 However, the incidence of such events in nontransformed cells is assumed to be low (although probably not as low as the error rate of the cellular replication machinery, which is in the range of 10−9 per base and replication). Compared with retroviral gene transfer, physicochemically transfected DNA, especially when forming transgene concatamers,15,100 as well as AAV gene transfer101 may be associated with increased risk of genetic instability, also involving flanking cellular sequences.
Incidence of recessive and dominant oncogenic insertions
With improving sequence information available from the murine and human genome projects, retroviral insertion events become increasingly mappable with regard to their exact to-the-base chromosomal location, relation to neighboring sequences, and potential interference with coding and regulatory regions.20-22 94
Previous assessments of the risk of untoward side effects from retrovirus insertion have been estimated to be rather low (between 10−6 and 10−8 per insertion event).97,102,103 In an experiment involving retrovirus infection of mouse embryonic carcinoma cells with a high copy number, the risk for inactivating a single gene locus (usually a recessive mutation) comprising 0.001% of a murine genome was determined to be in the range of 1 to 4 × 10−8 per insertion.97 The risk for producing a phenotype that could also be induced by dominant activation of oncogenes (growth-factor independence in TF-1 human leukemia cells) was in the range of 2 × 10−7 per insertion.102 However, these experiments focused on specific transforming events or mutation of specific target genes and involved cloning procedures to identify mutants, possibly reducing the sensitivity of the detection systems.
Based on the hypothesis of semirandom choice of target sequence, proto-oncogenic activation by a transgene insertion event would be expected to be more frequent. Considering that the entire human genome consists of approximately 3 × 109 base pairs (bp), a transforming insertion event frequency of 10−7 would mean that only a few hundred base pairs in the entire genome would allow oncogene activation. In the light of the fact that wild-type retroviruses have been demonstrated to interfere with genetic regulation from distances as far as 90 kbp upstream,104such numbers appear unrealistic.
Restricting the area of retrovirus insertion interference to a diameter of about 10 kbp around a given gene, the chance of a single insertion interfering with a defined allele is roughly 10−5. Between 100 and 200 proto-oncogenes or oncogenes have been “fished” from the murine genome by retrovirus insertional mutagenesis studies.105 106 With a margin of safety, the number of potential proto-oncogenes in the human genome is therefore probably not higher than 1000. The risk of an insertional event within 10 kbp of a potential proto-oncogene can therefore be estimated to range between 10−2 and 10−3.
At least 3 layers of safety, however, prevent such insertion events from being directly cancerogenic: first, retrovirus vector insertion is almost uniformly monoallelic,107 reducing the relevance of most recessive mutational events. This restricts the influence of insertional disturbance to the much more rare setting of dominant effects that are biologically active even if just one locus has been changed. Second, some signal alterations may trigger differentiation or apoptosis, impede engraftment, or otherwise reduce the survival probability of the affected cell clone. Third, and foremost, a single insertional mutation is, to our current knowledge, not sufficient to develop a malignant phenotype by itself.108 In the vector-associated incidents of murine and human leukemia that have recently been described,21 22 the insertional oncogene activation has at best contributed to a premalignant expansion of cells later developing the malignant clone because of additional genetic events. This underlines the need to screen for potential cooperation of insertional mutagenesis with side effects of the transgene or other circumstances contributing to clonal expansion of gene-modified cells (below).
An issue of unknown significance is whether multiple insertions in single cells will lead to a disproportionate increase in the risk for insertional mutagenesis, although the few available data suggest a linear relationship between insertion frequency and mutagenesis.97,102 It cannot be excluded that a high copy number of largely identical retrovirus transgenes distributed all over the genome may trigger chromosomal instability. In general, side effects observed under conditions of a high multiplicity of infection62,109 may not be relevant for a more carefully controlled transduction procedure.110 Considering these uncertainties, it appears reasonable to opt for the transfer of not more than 1 or 2 transgenes per cell. This represents the efficiency of currently available methods,36,111 but may in the future be more of an issue in vector systems with a higher efficiency of integration or high multiplicity of infection.112
In conclusion, the likelihood of oncogenic lesions induced by insertional mutagenesis alone would be expected to be relatively small when compared with some other established medical treatments, such as irradiation or chemotherapy with DNA-damaging agents.103Transformation of non-stem cells initiated by insertional mutagenesis does not seem to occur frequently: before 2002, no severe side effects related to insertional mutagenesis had been reported in more than a decade of clinical experience with retrovirus gene transfer into more committed hematopoietic cells and mature lymphocytes,1,2probably involving the manipulation of more than 1012cells. The number of cases in which these observations were made long-term is substantial, although the number of repopulating stem cells engrafted altogether probably did not exceed 10 to 100 per patient, putting the overall number of transgene insertion events in HSCs under long-term observation at approximately 104 to 105 worldwide.
Impact of vector design
The LTR configuration of conventional retroviral vectors comes with an increased risk to activate neighboring cellular sequences. The LTR establishes the enhancer-promoter regulating initiation of transcription and the polyadenylation signal giving rise to its termination on both ends of the transgene (Figure3). Although MLV enhancer-promoters are strongly active in most hematopoietic cells (albeit with pronounced differentiation dependence),71,113 the polyadenylation signal is relatively weak.114 Moreover, the major retroviral splice donor or related motifs in the transgene may interact with downstream splice acceptors of cellular genes. Combined with insufficient termination, these features generate a number of possibilities for activation of cellular sequences located downstream of the transgene insertion site (Figure 3).
Impact of vector design on the potential activation of a cellular gene located downstream of the transcriptional direction of the vector.
The gray boxes in panel A show a randomly inserted retroviral transgene with a conventional LTR architecture. Here, the enhancer (E) and promoter (P) are terminally repeated, and the polyadenylation [p(A)] signal is weak; a splice donor is present in the retroviral untranslated region and another one within the transgene cDNA. The desired vector transcript is shown as line D; potential aberrant transcripts are numbered and shown as dotted lines. Either of the vector's SDs may interact with a splice acceptor (SA) of a downstream-located cellular gene to generate alternative splice products 1 and 2. Aberrant transcripts 1 to 3 result from lack of termination; transcript 4, from activation of the 3′ promoter of the vector; and transcript 5, from a distant action of the vector's enhancer on a cellular promoter (which may also be located upstream and/or in inverse orientation to the insertion). Transcripts similar to 1 and 4 have been detected in the case of leukemia following retroviral gene marking in mice.20 The hypothetical vector shown in panel B was designed to prevent aberrant transcripts by deleting the enhancer-promoter from the LTR, inserting a strong splice acceptor upstream of the vectors coding sequences, deleting SD of the cDNA, utilizing a strong p(A) signal, and flanking the transgene cassette with insulator (INS) sequences that prevent long-distance enhancer interactions. Secondary prevention strategies shown in panels C and D make use of coexpressed selectable marker genes (“Impact of vector design”).
Impact of vector design on the potential activation of a cellular gene located downstream of the transcriptional direction of the vector.
The gray boxes in panel A show a randomly inserted retroviral transgene with a conventional LTR architecture. Here, the enhancer (E) and promoter (P) are terminally repeated, and the polyadenylation [p(A)] signal is weak; a splice donor is present in the retroviral untranslated region and another one within the transgene cDNA. The desired vector transcript is shown as line D; potential aberrant transcripts are numbered and shown as dotted lines. Either of the vector's SDs may interact with a splice acceptor (SA) of a downstream-located cellular gene to generate alternative splice products 1 and 2. Aberrant transcripts 1 to 3 result from lack of termination; transcript 4, from activation of the 3′ promoter of the vector; and transcript 5, from a distant action of the vector's enhancer on a cellular promoter (which may also be located upstream and/or in inverse orientation to the insertion). Transcripts similar to 1 and 4 have been detected in the case of leukemia following retroviral gene marking in mice.20 The hypothetical vector shown in panel B was designed to prevent aberrant transcripts by deleting the enhancer-promoter from the LTR, inserting a strong splice acceptor upstream of the vectors coding sequences, deleting SD of the cDNA, utilizing a strong p(A) signal, and flanking the transgene cassette with insulator (INS) sequences that prevent long-distance enhancer interactions. Secondary prevention strategies shown in panels C and D make use of coexpressed selectable marker genes (“Impact of vector design”).
Some of the mechanisms giving rise to activation of a cellular gene also apply to lentiviral or MLV vectors with a SIN architecture.115 However, the most frequent mechanism involved in retroviral insertional oncogene activation appears to be enhancer related, possibly working orientation independent and over large distances. Such a risk applies to almost any type of transgene configuration.
Considering the molecular mechanisms underlying activation of cellular genes, one could design vectors of improved safety. Such a vector should have a strong RNA termination/polyadenylation signal (serving as an “RNA insulator”)114,115; an internal position of the enhancer and promoter sequences that are excluded from functional interactions with neighboring sequences through the inclusion of dominant DNA insulators116; and a strong internal splice acceptor that largely prevents interaction of the retroviral splice donor with downstream sequences. If functioning as predicted, such a (hypothetical) construct depicted in Figure 3B would reduce the risk of insertional mutagenesis to the residual risk of disrupting genes. The latter may often be irrelevant unless haploinsufficiency becomes phenotypically relevant or loss of heterozygosity occurs through independent hits.
Another strategy to avoid insertional mutagenesis would be to achieve targeted insertion of transgenes into predefined “benign” cellular loci. Although conceptual progress has been achieved in the manipulation of retroviral integrase, experimental evidence for a stringent, sequence-specific targeting strategy is limited.117 A recent report indicates that physicochemical transfection procedures may be developed for targeted transgene insertion into defined genome loci in vivo (murine hepatocytes).16 It remains to be seen whether such technologies are free from genotoxic side effects and how they can be adapted to HSCs. Similar considerations apply to targeted transgene insertion technologies developed on the basis of AAV.118
Besides these primary prevention strategies, vectors could also be equipped with selectable marker genes to generate options for secondary prevention strategies. A drug-resistance marker could be used to reduce the clonal repertoire in vivo (Figure 3C) by ablating cells with low expression levels.119 However, if insertional oncogene activation enhances the fitness of cells during selection, this strategy may be counterproductive. Experiments addressing this issue have not been reported to our knowledge. Another option would be to include a negative selection marker in the transgene cassette. A conditional suicide gene (such as HSV-TK)90 may help to eliminate a malignant clone (Figure 3D), especially when combined with other antineoplastic treatments. However, this would also result in the loss of nontransformed transgenic cells. Before such an approach can be recommended, the potential immunogenicity of many suicide gene products and the limited preclinical experience with introducing suicide genes into HSCs has to be overcome.
Risks related to transgene expression
The ultimate goal of genetic therapy is to replace in situ a defective gene sequence, ideally by homologous recombination repair of the original locus. However, using available vectors and HSCs as targets, somatic gene transfer typically results in ectopic and nonregulated expression of the transgene, both with respect to the cell type affected and the level of expression achieved.
Depending on the type and assembly of cis elements used, expression levels generated by different vectors may differ by up to 3 orders of magnitude. Different variants of MLV enhancer-promoters and some cellular promoters have shown a great potential for multilineage and persistent transgene expression in hematopoietic cells in vivo, typically accounting for less than 1% of the total cellular protein content.71,113,119-123 Cellular control elements have been modified to provide lineage-specific expression with promising potency,65-67,124 and inducible expression has been achieved with designer promoters.68
The insertion site modulates all aspects of transgene expression, including duration, level, and differentiation dependence. With LTR-driven retroviral vectors, the majority of unselected clones shows fairly similar transgene expression levels. However, interclonal variability of transgene expression may be as high as 50-fold, and complete silencing can be observed in some HSCs and their progeny.113,120 122 Unless targeted insertion into the correct cellular allele or specific regulation is achieved, transgene expression will hardly ever be physiologic in every transduced cell.
According to Paracelsus' first rule (poison is a question of dose), it can be predicted that any transgene product has a defined therapeutic window compatible with the desired function and without the predominance of unwanted effects. Toxicity related to transgene expression may most frequently manifest in a competitive disadvantage, leading to the extinction of the affected cell (clone) and thus to a loss of efficiency. However, transgene interference with cellular decisions related to homing, proliferation, or differentiation may eventually result in the manifestation of new types of diseases. Currently, few observations are available that support these concerns. However, we have to be aware that up to now far less than 1% of the human cDNA pool and a necessarily minute fraction of all artificial sequences possible have been introduced into gene therapy research. Moreover, gene delivery systems have and will continue to become increasingly potent, also allowing the simultaneous transfer of 2 or more cDNAs with a single vector.
To support these considerations, 4 examples may be sufficient. Of the 4, 3 deal with the use of selectable marker genes, a key technology in hematopoietic gene therapy. These examples provide evidence for dose-dependent toxicity (HOXB4), an as yet uncertain contribution to a severe side effect (MDR1), and evidence for context-dependent side effects (dLNGFR). These and a final example (CD40L) highlight the importance of developing vectors for spatially or temporally controlled expression of transgenes.
Ectopic expression of HOXB4: dose-dependent side effects?
Retroviral vector–mediated expression of HOXB4, encoding a homeodomain transcription factor involved in the regulation of hematopoietic pool size, has been shown to promote polyclonal and regulated expansion of engineered HSCs.51,125 In contrast to many other homeobox genes, ectopic expression of HOXB4 in hematopoietic cells did not lead to overt alterations of differentiation or uncontrolled expansion of gene-modified cells in mice.126 The interest in HOXB4 gene transfer for cell therapy has been reinforced by the finding that murine embryonic stem (ES) cell–derived hematopoiesis can be partially converted to repopulation competence in adult hosts upon transient or stable activation of HOXB4expression.127
In human HSCs transplanted into immunodeficient mice, ectopic expression of HOXB4 promoted the expansion of primitive hematopoietic cells.128,129 However, high levels of HOXB4 expressed from “stronger” vectors impeded myeloid and lymphoid differentiation of human hematopoietic cells.129 In line with these data, impaired repopulation of lymphatic tissues was observed in a study using HOXB4-engineered hematopoietic cells derived from a somatic cloning procedure.130 These studies taken together argue that the effects of HOXB4 are highly dependent on the dose and the kinetics of its ectopic expression. Importantly, activation of HOXB4-interacting partners such as PBX1 (possibly by insertional mutagenesis) may be sufficient to promote transformation of HSCs with constitutive ectopic expression of HOXB4.131 Thus, a potential therapeutic use of HOXB4 may require an exact definition of a therapeutic window and may depend on the ability of regulated expression.
Murine leukemia following MDR1 gene transfer: phenotoxicity, genotoxicity, or both?
Adenosine triphosphate binding cassette (ABC) transporter pumps encoded by multidrug resistance 1 (MDR1) orABCG2 are naturally expressed in primitive hematopoietic cells, explaining their inherent competence for extruding some fluorescent dyes and other amphiphilic compounds.132,133Increasing expression levels of such pumps may promote a survival advantage in the presence of high doses of some chemotherapeutic agents,134,135 and independently antagonize some proapoptic signals, as shown forMDR1.132,136 137
Interestingly, ectopic expression of ABCG2 was associated with impaired differentiation of myeloid cells in mice.138It is yet unclear to what extent this effect is dose related. The results with MDR1 have been controversial. Numerous studies, including a transgenic mouse model, have shown the ability to overexpress MDR1 in hematopoietic cells without overt alterations of cell functions (other than the acquired drug-resistance phenotype).85,134,135,139,140 Applications in dogs,141 nonhuman primates,110 and clinical trials45,46 142 have been safe, with occasional evidence for increased pump activity, although gene transfer efficiency was likely very low.
However, myeloproliferative disorders have also been observed in different strains of mice using retroviral vector–mediated transfer ofMDR1 into hematopoietic cells,62,109 and disease induction was promoted by prolonged expansion of cells in vitro prior to transplantation.62 Interestingly, this disease was associated not only with ectopic expression of MDR1, but also with an unusually high transgene copy number (in many cases exceeding 10 copies per clone, which is quite unusual even in mouse studies). Therefore, the most straightforward explanation is that excess MDR1 expression in this study may have been pathogenic. Besides, sequences other than MDR1 could have been expressed from insertion of intact or rearranged vectors. This aspect needs to be clarified, as the genetic integrity of the inserted transgenes has not been investigated, and the disease was so far observed only with a specific vector backbone (based on a first generation vector derived from Harvey murine sarcoma virus containing, in addition to an engineered “splice corrected” cDNA, considerable amounts of viral gene remnants that are not required for proper vector function).62,109 Moreover, it has not been reported whether the otherwise well-designed control vectors used had a similar high copy number in the producer and target cells.62,109Thus, it remains formally unclear whether the disease was dependent on side effects of very high MDR1 expression (driven from multiple transgenic alleles in the mouse model);137 the expression of vector sequences other than MDR1 (potentially driven from rearranged vectors); or an increased risk for insertional mutagenesis or genomic instability under conditions of high copy numbers per genome. It is also quite possible that some or all of these factors acted together to produce the myeloproliferation.
Another open question is whether MDR1 overexpression may promote engraftment of gene-modified HSCs,45 althoughMDR1 expression alone would not be sufficient to overcome a culture-dependent loss of engraftment capacity.143 Taken collectively, these data indicate that defining a therapeutic window for ectopic expression of MDR1 or other efflux pumps in hematopoietic cells may be difficult. If future research will not facilitate the definition of safe conditions of transgenicMDR1 expression, alternative metabolic selection markers may be more promising (Table 2 and references therein).
Classes of selectable marker genes and corresponding drugs
| Principle . | Utility . | Example for gene . | Drug/agent category . | Agent . | Reference no. . |
|---|---|---|---|---|---|
| Surface tag | In vitro | dLNGFR | Monoclonal antibody | NA | 120,121,123, 144-149 |
| Drug resistance | In vitro and in vivo | MGMT | Cytotoxic agent | Temozolomide, BCNU | 137, 150-152 |
| DHFR | Cytotoxic agent | MTX | 137 | ||
| MDR1 | Cytotoxic agent | Paclitaxel, etoposide | 45,46,134,135,139-142 | ||
| Growth promoting | In vitro and in vivo | FK-mpl | Chemical dimerizer | AP1903 | 153,154 |
| SAG | Inducer of protein function | Estrogens | 155 | ||
| HOXB4 | Controlled expression required | NA | 51,125-131 |
| Principle . | Utility . | Example for gene . | Drug/agent category . | Agent . | Reference no. . |
|---|---|---|---|---|---|
| Surface tag | In vitro | dLNGFR | Monoclonal antibody | NA | 120,121,123, 144-149 |
| Drug resistance | In vitro and in vivo | MGMT | Cytotoxic agent | Temozolomide, BCNU | 137, 150-152 |
| DHFR | Cytotoxic agent | MTX | 137 | ||
| MDR1 | Cytotoxic agent | Paclitaxel, etoposide | 45,46,134,135,139-142 | ||
| Growth promoting | In vitro and in vivo | FK-mpl | Chemical dimerizer | AP1903 | 153,154 |
| SAG | Inducer of protein function | Estrogens | 155 | ||
| HOXB4 | Controlled expression required | NA | 51,125-131 |
This table does not provide a comprehensive overview of the genes developed as selectable markers. Its purpose is rather to show the different principles with a few selected examples. NA indicates currently not available as good manufacturing practice product.
Context-dependent toxicity of a cell-surface marker:dLNGFR
The cytoplasmically deleted low-affinity nerve growth factor receptor (dLNGFR, also abbreviated ΔLNGFR, LNGFR, tNGFR, or NGFR) was derived from p75 neurotrophin receptor (p75NTR) to develop a clinically applicable cell-surface marker for hematopoietic cells.144 Although dLNGFR has been used by several laboratories to tag gene-modified cells,145 few data have been published regarding the ability for long-term marking (> 1 year) of HSCs and their progeny.146 A nonhuman primate study reported a failure to mark long-term repopulating HSCs with dLNGFR, however without investigating potential mechanisms.147 On the other hand, use of dLNGFRin clinical trials with gene-modified T cells has been shown feasible and safe.148,149 However, a recent mouse experiment20 in conjunction with an earlier study in fibroblasts156 offered the hypothesis thatdLNGFR expression in myeloid cells may promote their transformation in an unusual, highly context-dependent manner. It is this proposed context dependence that renders the discussion of this issue interesting.
p75NTR is a member of the tumor necrosis factor (TNF) receptor superfamily that can bind all known neurotrophins (NTs) including nerve growth factor (NGF).157p75NTR is usually not expressed in hematopoietic cells, with the exception of some B-cell subsets.158 The cytoplasmic domain of p75NTR contains a proapoptotic juxtamembrane region and a death domain.157 These sequences were deleted in dLNGFR before their precise function was known in an attempt to create an inert surface marker.144 The deletion may weaken the anchoring in the cell membrane, and therefore the shedding of dLNGFR,145 which is still able to bind NTs in vivo,159 may affect the local extracellular cytokine milieu. Moreover, deletion of the intracellular domain renders dLNGFR structurally similar to naturally occurring antiapoptotic decoy receptors of the TNF-receptor family, which can act as dominant-negative inhibitors of proapoptotic intracellular pathways.160
In cells expressing TrkA, TrkB, orTrkC, which encode tyrosine kinase receptors for different NTs, association of p75NTR creates a heterodimeric receptor complex with increased ligand affinity that is not dependent on the presence of the cytoplasmic residues.157 It is noteworthy that coexpression of either one of the Trk receptors with ap75NTR mutant that lacked most of the intracellular domain, a construct basically identical with dLNGFR, resulted in transformation of fibroblasts when NTs were added to the culture.156 This growth-promoting role dLNGFR is clearly dependent on the coexpression of a Trk gene and the presence of NTs (Figure4). The same configuration occurred in the murine monocytic leukemia originating in association with retroviral insertional up-regulation ofEvi1 in hematopoietic cells, which provides circumstantial evidence but no formal proof of a contributing role ofdLNGFR.20
Proposed context-dependence of side effects elicited by dLNGFR.
Situation A represents a physiologic situation that can be observed in neuronal and some other cell types.157 Situation B was shown to promote the transformation of fibroblasts in vitro.156 Situation C represents the ideal context for cell marking.144
Proposed context-dependence of side effects elicited by dLNGFR.
Situation A represents a physiologic situation that can be observed in neuronal and some other cell types.157 Situation B was shown to promote the transformation of fibroblasts in vitro.156 Situation C represents the ideal context for cell marking.144
Evi1 encodes a Zinc-finger transcription factor that has been implicated in the pathogenesis of human myelodysplastic syndromes and acute myeloid leukemia (AML). Ectopic expression of Evi1impairs granulocytic differentiation, but leads to only mild alterations of hematopoiesis in transgenic mouse models.161 We proposed a specific interaction of dLNGFR and Evi1 in the induction of the leukemic clone, possibly reflecting a bias for a lineage (ie, monocytic) in whichTrkA expression and NGF signaling were also present and functionally relevant.20 158
If this hypothesis can be confirmed, it would represent an example for cooperation of random insertional mutagenesis (genotoxicity) and transgene-related side effects (phenotoxicity) in the induction of leukemia. Alternatively or in addition, Evi1 may have induced expression of TrkA,162 and the interaction with dLNGFR may have promoted the transformation of a monocytic precursor. Also, a protein related to Evi1 has been shown to play a role in Trk-signaling of C elegans,163 opening further possibilities for transforming loops.
The potential risk associated with the use of dLNGFRin HSCs is underlined by observations that signals generated through oncogenic versions of Trk receptors may contribute to the pathogenesis of human AML.164-166 Therefore, dLNGFR does not appear to be a perfect choice for the manipulation of cells with a broad plasticity such as HSCs. However, as side effects of dLNGFR are proposed to be context dependent, its use in restricted cell lineages lacking cooperating signal transducers can be justified (considering Paracelsus' second rule). Interestingly, variants of dLNGFR have been developed that are deficient in ligand binding167 in order to reduce the probability of side effects. Similar concerns of context-dependent side effects and potential for cooperation with randomly activated oncogenes apply to many other therapeutic or marker genes.
Problems resulting from unregulated expression:CD40L
Finally, the mode of transgene expression is an important determinant of potential toxicity. This has been exemplified in an attempt to develop gene therapy for inherited deficiency of the CD40 ligand (X-linked hyper-IgM syndrome). Ectopic constitutive, but not naturally regulated, expression of CD40L, although at low level, produced abnormal proliferative responses in developing murine T lymphocytes, apparently through dysregulated intercellular interactions during thymic maturation and selection.168 For many applications of hematopoietic gene therapy it is worth repeating the conclusion of this study: “Current methods of gene therapy may prove inappropriate for disorders involving highly regulated genes in essential positions in proliferative cascades.”168
These 4 examples should be sufficient to underline the importance of a systematic risk assessment of the transgenes under consideration. Special attention should be paid to molecules that are involved in cellular signaling networks, such as those required for correction of some inherited disorders3,168,169 or those generated as surface tags120,123,144 or artificially inducible proteins that promote cellular proliferation or differentiation decisions.153-155 We would propose that such transgenes should be tested under conditions of high, intermediate, and low constitutive expression,129 preferentially achieved with vector design and not with variation of transgene dosage. Preclinical assay systems available for such work range from cell-culture–based model systems to animal studies and functional genomics or proteomics (Figure5).
Schematic overview of potential interactions between genotoxicity (alteration of cellular genes by vector insertion) and phenotoxicity (side effects of transgene expression), and experimental approaches allowing their detection.
Interaction may either occur in cis (on the same DNA molecule) or in trans (through mobile factors).
Schematic overview of potential interactions between genotoxicity (alteration of cellular genes by vector insertion) and phenotoxicity (side effects of transgene expression), and experimental approaches allowing their detection.
Interaction may either occur in cis (on the same DNA molecule) or in trans (through mobile factors).
Risks related to conditioning, use of selective drugs, and cell amplification
Following genetic modification of HSCs in vitro, their engraftment and contribution to hematopoiesis in vivo are dependent upon the methods used for conditioning or selective amplification. Conditioning eliminates host cells prior to infusion of gene-modified cells. Irradiation or cytotoxic agents induce a moderate to severe (myeloablative) lymphohematotoxicity. However, these regimens can be complicated by severe long-term toxicity. Nonmyeloablative regimens with sublethal toxicity have become increasingly well investigated170 and begin to show great promise for HSC-mediated gene therapy.4,171 In animal models, high doses of donor cells32 and application of G-CSF to the recipient before nonmyeloablative conditioning172 have been shown to promote engraftment. However, it is unclear whether chimerism will be maintained in a stable manner in the long term when nonmyeloablative protocols are performed in an autologous clinical setting. Here, donor-dependent immune functions have no facilitating role to promote engraftment of the transplant; tolerance may be incomplete and engineered cells usually do not have a spontaneous selective advantage. An alternative, potentially more specific and thus less toxic approach to conditioning is the use of monoclonal antibodies directed against stem cell antigens or more common leukocyte antigens.173 Although it is likely that side effects associated with conditioning regimens will be reduced significantly in the near future, this issue will continue to be an important aspect of the risk-benefit evaluation for stem cell–based gene therapy.
Importantly, several diseases could be successfully treated with a moderate rate of chimerism (5%-30%). A selective survival advantage of engineered HSCs can be promoted upon transfer and expression of appropriate selectable marker genes. Table 2 summarizes 3 different categories of such genes that have a well-documented efficiency in animal models. Potential side effects resulting from the expression of selectable marker genes have been reviewed above (“Risks related to transgene expression”).
For most of these selectable marker genes, drugs are required to trigger their function. Therefore, side effects associated with these drugs represent another important aspect of the preclinical and clinical evaluation. Some of these agents have a well-documented toxicity profile in humans; others represent experimental agents with limited clinical experience. In this context, it is interesting to note that the most powerful selection system currently available for gene-modified hematopoietic cells requires the use of DNA-damaging agents.137,150-152 Although potentially less toxic alternatives for selective amplification of gene-modified cells have been proposed,153-155 expansion of hematopoietic cells promoted by these gene functions may be incomplete, lineage restricted, and unstable, suggesting preferential action at the level of progenitor cells as opposed to HSCs. This implies a need for repetitive use of the corresponding drugs over prolonged periods of time, or induction of a distorted hematopoiesis with unclear long-term consequences.
Clonal amplification of transgenic cells is another important variable.7 The risk for accumulating mutations that are not related to gene transfer increases with the life span and the number of generations of the engineered cell. In most conditions of human bone marrow transplantation, the size of the graft's stem cell dose implies a modest pressure for expansion and a high likelihood for polyclonal reconstitution. This is underlined by results from nonhuman primate studies of gene-marking.36,111 With the advent of genetic selection strategies, a risk related to forced expansion of individual clones may become more relevant. On the other hand, single clones of transduced HSCs may provide a perfectly normal hematopoiesis with persistence of transgene expression in all hematopoietic lineages, at least in mice.120 123 This supports the idea that clones with favorable insertion sites and “neutral” transgenes are not necessarily at increased risk for transformation, even when undergoing massive expansion. The minimal number of HSCs that stably support primate hematopoiesis remains to be defined.
Risks related to immune surveillance
A further category of side effects is related to innate or acquired immunity against vector components or immune surveillance of engineered cells. A recent review proposed that certain gene transfer procedures may set “danger” signals that result in an increased likelihood of an immune reaction.174 However, severe inflammatory reactions elicited by viral proteins in the vector preparation, as observed with early generations of adenoviral vectors administered in vivo,175 are unlikely following a single administration of ex vivo–manipulated hematopoietic cells. In principle, a transient exposure to antigens may be caused by remnants of vector particles or culture media components on infused cells even if the transgene does not encode viral antigens.176 This risk appears small with conventional retroviral transduction protocols in which cells are cultured for at least a day following the final exposure to vector particles. However, with the use of adenovirus177 or lentivirus vectors178 the time in culture after the final round of vector exposure may be shortened, which could increase the probability of contamination with viral antigens.
Repetitive infusion of engineered cells may be complicated by sensitization to antigens originating from components of culture media, vector particles, or transgene expression, potentially resulting in clearance of transgenic cells179 or even severe acute adverse reactions. This potential problem could be solved by appropriate preparation of cells and recipient prior to infusion or simply by single use of engineered cells (given that the vector system does not generate antigens to which pre-existing immunity exists). Also, sensitization could be diagnosed prior to repeated infusion of cells.
Moreover, it will be interesting to determine whether some clinical settings, such as those resulting from repeated infections, may trigger cellular or humoral innate immune functions to clear incoming gene-modified cells or create an unfavorable cytokine milieu. If so, a transient blockade of these mechanisms, as proposed in the context of preclinical adenoviral gene therapy,180 may improve the “take” of gene-modified cells.
Immune responses mounted against transgene antigens may develop with some latency. This concern is of particular relevance when introducing artificial or xenogenic sequences (as in the use of some selectable marker genes) and when correcting inherited genetic disorders in so-called CRIM (cross-reactive immunologic material)-negative patients. Although bone marrow transplantation may promote tolerance to multiple or individual antigens,181-183 this does not necessarily occur following nonmyeloablative conditioning regimens. Immune-mediated rejection of transgenic cells expressing the xenogenic marker enhanced green fluorescent protein occurred in a study with nonhuman primates.184 Disturbingly, one affected animal developed hemolytic anemia after rejecting the transgenic cells.184Further investigations are needed to determine whether autoimmunity can be induced as a side effect of sensitization against transgenic cells. For more advanced applications of gene therapy as well as for allogeneic transplantation, tolerance induction is a key issue of future research.
Combination and interactions of risk factors
After listing this collection of potential problems, it is important to mention that combinatorial side effects of transgene insertion (genotoxicity), transgene expression (phenotoxicity), and cell expansion (selection toxicity) may be required to produce malignant transformation. Monocytic leukemia observed afterdLNGFR marking in mice and serial bone marrow transplantation may serve as a paradigm.20 It is possible that at least some clones observed in the CD40L-induced lymphoproliferation168 or in theMDR1-associated myeloproliferative disease62 109 had a similar history involving insertional mutagenesis in addition to transgene side effects and/or forced cell expansion. Insertional activation (in cis) of an oncogene or an otherwise “innocent” transcription factor may change the cellular program (in trans), which in turn may cooperate with the transgene product to induce an undesired phenotype (Figure 5). Such program alterations may also influence expression levels of the transgene, jointly acting to promote the initial survival of a premalignant clone.
Similar considerations apply for the serious adverse event recently observed in a clinical gene therapy trial.21,22 Ten children with X-linked severe combined immunodeficiency (X-SCID) were successfully treated by retroviral transfer of the interleukin 2 receptor common γ-chain into CD34+ cells and reinfusion of cells without conditioning. This transgene was required to correct the underlying genetic deficiency and provides (in X-SCID patients with this deficiency) a powerful selective advantage during T-cell maturation.3 The initial outcome of gene therapy was better than that typically achieved with allogeneic bone marrow transplantation.3 At 3 years after cell infusion, one patient presented with clinical signs of an acute lymphoblastic leukemia (ALL) caused by a monoclonal proliferation of γδ T cells. The clone had one insertion of an intact vector copy that occurred in one LMO2 allele (readily identified by LAM-PCR)21,36 inducing ectopic expression of this proto-oncogene. LMO2 is known to be involved in the pathogenesis of ALL, but probably not sufficient to cause malignancy (for which 4 to 6 independent genetic “hits” seem to be required).21,22,108,185 Although no evidence for aberrant signaling through the transgenic common γ-chain has been reported, its physiologic function obviously was sufficient to promote a strong expansion of the clone. Although no detailed analysis is available at this point, it can be postulated that additional genetic hits could have subsequently promoted malignant progression: the malignant clone (expressing a mature T-cell phenotype) was more mature than the initially transduced cell population (which must have lacked T cells), and a severe varizella zoster virus infection was reported to have coincided with first clinical symptoms of hematologic abnormalities. Moreover, a cytogenetic abnormality was observed that did not involve the insertion site. Another level of concern was a genetic background of childhood cancer.21 22
Given that an unfavorable concert of oncogenic factors is required for tumor manifestation (as in the model presented in Figure6), such a serious adverse event is not expected to be found in any clinical scenario for retroviral gene therapy, nor can we predict its frequency in gene therapy for X-SCID patients. Besides the specific clinical setting and features of the transgene (cDNA and regulatory regions), target cell features represent an important variable. These determine the overall susceptibility to gene transfer, which loci are open for transgene insertion, and how many of these may contribute to malignant transformation.103 Further points to consider include the number of cell generations following gene transfer,103 the expansion conditions that may sometimes suppress balancing proapoptotic signals, the exposure to mutagenic hazards that are independent of the genetic manipulation, and the endogenous capacity for DNA repair and proliferation control. Finally, systemic responses to transformed cell clones add another level of complexity (Table3).
Not every insertional oncogene activation is expected to result in clinical manifestation of malignancy (hypothetical curve).
Transformation-promoting events are symbolized by flashes; the one marked with i represents insertional mutagenesis; the one labeled t indicates transgene side effects; u1 and u2 indicate subsequent unrelated hits. Clone size, time course, and the probability of extinction represent arbitrary values.
Not every insertional oncogene activation is expected to result in clinical manifestation of malignancy (hypothetical curve).
Transformation-promoting events are symbolized by flashes; the one marked with i represents insertional mutagenesis; the one labeled t indicates transgene side effects; u1 and u2 indicate subsequent unrelated hits. Clone size, time course, and the probability of extinction represent arbitrary values.
How cell type may determine the risk of insertional mutagenesis
| Level . | Before insertion . | After insertion . |
|---|---|---|
| Genome | Number of pre-existing mutations | Potential for secondary mutations |
| Number and size of activated, potentially disruptible “dangerous” loci susceptible to transgene insertion | Proliferation capacity/propensity | |
| Cell-transgene interactions, direct | Host factors participating in preintegration complex | Recognition of transgene regulatory elements and expression level of transgene |
| Cell-cycle status | Type and number of mechanisms supporting transgene side effect | |
| Cellular response, indirect | Response balance to “danger” signals | |
| Potential for escape from senescence | ||
| Number of generations following genetic modification | ||
| Level of differentiation |
| Level . | Before insertion . | After insertion . |
|---|---|---|
| Genome | Number of pre-existing mutations | Potential for secondary mutations |
| Number and size of activated, potentially disruptible “dangerous” loci susceptible to transgene insertion | Proliferation capacity/propensity | |
| Cell-transgene interactions, direct | Host factors participating in preintegration complex | Recognition of transgene regulatory elements and expression level of transgene |
| Cell-cycle status | Type and number of mechanisms supporting transgene side effect | |
| Cellular response, indirect | Response balance to “danger” signals | |
| Potential for escape from senescence | ||
| Number of generations following genetic modification | ||
| Level of differentiation |
Bearing this in mind, we need to consider appropriate preclinical models, including animal experiments, in order to derive clear statistics indicating the importance of individual risk factors and the probability of their combinations. For scientific, economical, and ethical reasons, studies will often rely on work with cell lines and laboratory mice. In this respect, it is important to discuss features of the mouse model that distinguish it from human hematopoiesis.
The mouse model compared with human hematopoiesis
The outstanding role of the laboratory mouse for modeling human development and disease has received further support by the recent findings of the mouse genome project.186 Nevertheless, the differences between the hematopoietic systems of mice and humans must be carefully evaluated to diagnose with certainty reactive and neoplastic blood cell disorders and to improve the predictive value of the animal model.
Even in humans, the classification of preleukemic states such as the myelodysplastic syndromes is still controversial.187Very recently, preleukemia and its progression to leukemia have been classified in mice,188,189 in analogy to the French-American-British (FAB) scheme developed for human leukemia.190 Murine leukemias may be experimentally induced with specific genetic alterations.191 Examples for preleukemic alterations are ectopic expression ofBCR-ABL,192N-RAS,193BCL2,194 or Evi1,161and the ICSBP-knock-out mouse.195 The latency period between leukemia induction by application of x-rays and/or inoculation of MLVs and leukemia manifestation could be regarded as a preleukemic condition.
The susceptibility of mice to develop leukemia varies according to strain and its contamination with MLVs. Unless a genetic predisposition is involved (such as endogenous RCRs), spontaneous leukemia occurs only sporadically in older animals. It can be induced with high incidence by irradiation and inoculation of newborn or immunodeficient animals with MLVs. RCRs transform cells by insertional mutagenesis, which has been useful in the identification of tumor-associated genes.82,91,104-106,196,197 In some cases acute leukemia can be induced following a rapid polyclonal expansion of progenitor cells when a mouse is infected with a retrovirus complex that cotransfers a replication-defective retrovirus encoding an oncogene.82 91
The mouse has a significantly higher daily hematopoietic cell turnover (especially of red blood cells and platelets) compared with humans. Accordingly, the complete bone cavities are used for hematopoiesis, and there are very few or no fat cells interspersed.198,199Reactive or malignant increases of hematopoietic tissue rapidly lead to extramedullary hematopoiesis, typically starting in the spleen.200-202 Thus, splenomegaly with expansion of red pulp caused by leukemic infiltrations (> 20% blasts) and regression of the white pulp (periarteriolar lymphatic sheath and lymph follicles) are characteristic and early findings in murine leukemia.188,199 In advanced disease, normal architecture of spleen is totally abolished and widely replaced by masses of blasts. Liver involvement in AML is characterized by leukemic infiltrates in periportal areas. Therefore, unlike human leukemia, bone marrow is only variably, and spleen is constantly, involved in murine leukemia.200
The mouse model may not be fully predictive for human leukemia development when considering differences in HSC turnover. Leukemia development often involves genetic alterations of true HSCs.5 However, it is unclear whether this also applies to oncogenesis related to retroviral manipulations. The pool size of murine HSCs is tightly regulated, although with considerable genetic and age-dependent variability.203 Abkowitz et al postulate a similar size of the HSC pool in mice and cats (approximately 12 000 per animal), and possibly also in humans.27 The study also suggests a conservation of the replicative activity per lifetime between murine and human HSCs.27 Thus, mice would present with a higher density and shortened cycling times of HSCs within the bone marrow.
However, if the burden of insertional mutagenesis also involves less primitive progenitor and precursor cells, typical mouse experiments performed with a relatively small total number of hematopoietic cells would underestimate the risk (about 106transplanted per mouse, compared with at least 108 cells in a clinical trial). Moreover, the life span of this animal is short (2 years), and for practical reasons, observation periods rarely exceed 6 to 12 months, further reducing the chance to detect slowly developing dysplasias. Bone marrow transplantation and inappropriate cell culture strongly reduce the pool size of HSCs.51 Serial bone marrow transplantations generate monoclonal or oligoclonal hematopoiesis in mice, suggesting an enormous pressure for massive amplification of individual HSCs.51,120,123,204,205 Such a forced expansion may promote the manifestation of dysplastic or overt leukemic clones.7,206 207 Thus, although the mouse model suffers from a poor sensitivity to detect rare mutagenic events related to integrating, nonreplicating vectors, in case such events do occur, experimental conditions can be adjusted to promote their manifestation.
It has been demonstrated that rodent cells can be more easily transformed than human cells, and several functional differences of signaling pathways involved in human and rodent models of transformation have been identified.108 One has been linked to a stronger activity of the telomerase function in mature rodent cells. Thus, only 2 to 3 hits are typically required for transformation of a rodent cell: 1 that dysregulates apoptosis and 1 or 2 further hits that alter cell-cycle control and provide proliferative stimuli. In humans, yet another event promoting telomerase activity is required for malignant transformation of mature epithelial and fibroblastic cells, but it is still unclear whether this would be needed to transform a human HSC.5 108 If so, human cells would be more resistant to transformation by random vector insertions and transgene side effects.
Summarizing these considerations, it appears justifiable to develop mouse models in which the sensitivity for detecting side effects related to genetic manipulations is increased by the choice of the experimental conditions, such as in vitro expansion62; hemolysis202 or bleeding206; serial bone marrow transplantation20,207; or introduction of a proto-oncogenic genetic lesion.161,191-195 Also, “humanization” of the mouse genome may be helpful to obtain models of increased predictive value.108,186 Currently available “large”-animal models would be even more relevant for human gene therapy, but obviously do not allow broader genotoxicity screening.64 Moreover, tumor manifestations usually take many years in nonhuman primates. It may be better to design oversensitive mouse models as a “worst case” scenario, thereby generating clear statistics that reflect the impact of defined risk factors.
Conclusions
Successfully exploiting the enormous potential of gene therapy targeting hematopoietic cells requires an open eye for side effects. Proof of principle in animal models may be spectacular, but is not all that is relevant for developing safe clinical applications. As with the use of drugs or irradiation, dose finding is a required next step. One realistic option to improve safety without loss of efficiency is to translate better procedures for stem cell purification into clinical use. The resulting reduction in the target cell pool likely represents a straightforward way to reduce the risk of insertional mutagenesis. The associated long-term goal is to reconstitute hematopoiesis with just a few clones of genetically characterized transgenic stem cells. Beyond the issue of target cells, dose finding takes into account the interaction of multiple features in vectors, transgenes, and clinical scenarios, which can be reflected in the design of preclinical models. Improved vector design may result in greater target site specificity of insertion and a transgene composition that has a reduced risk of activating cellular genes. Another important goal is to derive a risk classification of transgenes and clinical scenarios, considering all conditions potentially contributing to side effects. This may help to avoid premature generalization of side effects that occurred under specific circumstances. A database of vector insertion sites and statistical analyses on the clonality of reconstitution will be key to understanding the dynamics of these processes. Results obtained in long-term follow-up, especially, will allow us to interpret the impact of the combined action of the transgene and the vector insertion site on cell biology, a factor that until now, because of lack of accessibility, has not been studied in detail.
Individual institutions may provide significant contributions, but a combined international effort will probably be needed to accumulate the amount of data required. Revisions of the existing regulatory guidelines may be helpful only if they promote reasonable standards of comparison and agreements on basic experimental approaches in preclinical research. We propose to consider the collaborative development of preclinical proto-oncogenic worst case models as one basis for dose finding. Thus, developing tools for the best feasible genetic treatment will profit from a dialectic approach that anticipates adverse events by active, hypothesis-driven investigation. While doing so, we need to continue with many of the current clinical trials on the basis of the risk evaluation at hand. It should not be forgotten that for a number of patients, after carefully weighing risks and benefits, the worst case scenario of gene therapy may be to not receive it.
We thank the reviewers and the editors for important suggestions.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/blood-2002-07-2314.
Supported by grants from the Deutsche Forschungsgemeinschaft (KFO 110; WI1955/1-1); the European Union (QLK3-2001-01265, QLRT-2001-00427); the VolkswagenStiftung; and the National Institutes of Health (D.A.W.; grant no. HL 53586).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christopher Baum, Experimental Cell Therapy, Department of Hematology and Oncology, Hannover Medical School, Carl-Neuberg-Straße 1, D-30625 Hannover, Germany; e-mail:baum.christopher@mh-hannover.de.


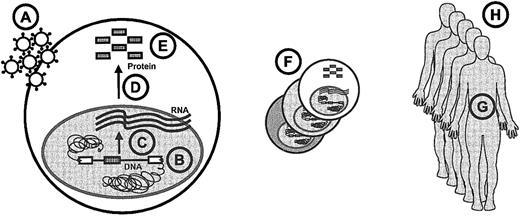
![Fig. 3. Impact of vector design on the potential activation of a cellular gene located downstream of the transcriptional direction of the vector. / The gray boxes in panel A show a randomly inserted retroviral transgene with a conventional LTR architecture. Here, the enhancer (E) and promoter (P) are terminally repeated, and the polyadenylation [p(A)] signal is weak; a splice donor is present in the retroviral untranslated region and another one within the transgene cDNA. The desired vector transcript is shown as line D; potential aberrant transcripts are numbered and shown as dotted lines. Either of the vector's SDs may interact with a splice acceptor (SA) of a downstream-located cellular gene to generate alternative splice products 1 and 2. Aberrant transcripts 1 to 3 result from lack of termination; transcript 4, from activation of the 3′ promoter of the vector; and transcript 5, from a distant action of the vector's enhancer on a cellular promoter (which may also be located upstream and/or in inverse orientation to the insertion). Transcripts similar to 1 and 4 have been detected in the case of leukemia following retroviral gene marking in mice.20 The hypothetical vector shown in panel B was designed to prevent aberrant transcripts by deleting the enhancer-promoter from the LTR, inserting a strong splice acceptor upstream of the vectors coding sequences, deleting SD of the cDNA, utilizing a strong p(A) signal, and flanking the transgene cassette with insulator (INS) sequences that prevent long-distance enhancer interactions. Secondary prevention strategies shown in panels C and D make use of coexpressed selectable marker genes (“Impact of vector design”).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/6/10.1182_blood-2002-07-2314/4/m_h80634104003.jpeg?Expires=1769117289&Signature=KL9hMfSuw~RO68meqXe0Yc~bMBAiFGu0EdumDX~33vaBnMjkFK9R-2lqiNUOEYOqBCTJ7tJd8crSf861dk-HdqyO8msIURmJpSOS1xukrwEY3fs3lDjiqU-vF2sxknVMFRJJ3qKASj1NXck8G-7engYzErm7Qh05MxJ5vqy26suf8We3Ba94Bqu5MOWEulw0EIcrEZSjVujWcA4pBvb3pK9cAiAYQjE9~0QoQLK0icHEINxOVzgpLD-zFXYx7wO~2ggrmiL1INsySY3bOctoEiGzFdhAzVZO9xGEhsZW98f6tdv8n6qQFgx~4j0dUrYLRX5185WefhnlYYkOFvvCeA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
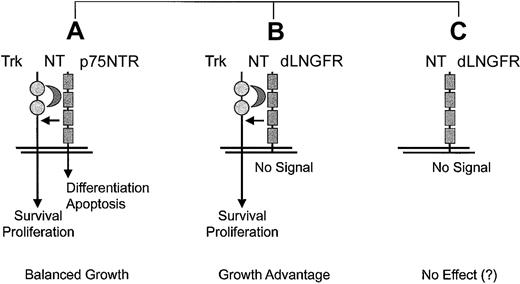
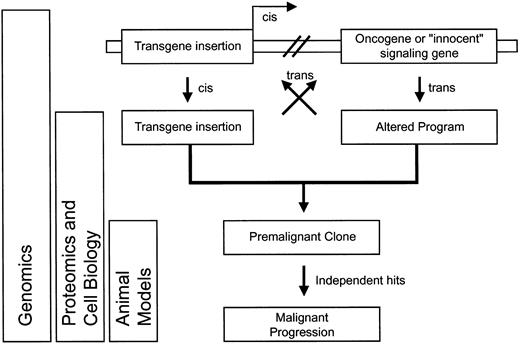

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal