Gene transfer experiments in nonhuman primates have been shown to be predictive of success in human clinical gene therapy trials. In most nonhuman primate studies, hematopoietic stem cells (HSCs) collected from the peripheral blood or bone marrow after administration of granulocyte colony-stimulating factor (G-CSF) + stem cell factor (SCF) have been used as targets, but this cytokine combination is not generally available for clinical use, and the optimum target cell population has not been systematically studied. In our current study we tested the retroviral transduction efficiency of rhesus macaque peripheral blood CD34+ cells collected after administration of different cytokine mobilization regimens, directly comparing G-CSF+SCF versus G-CSF alone or G-CSF+Flt3-L in competitive repopulation assays. Vector supernatant was added daily for 96 hours in the presence of stimulatory cytokines. The transduction efficiency of HSCs as assessed by in vitro colony-forming assays was equivalent in all 5 animals tested, but the in vivo levels of mononuclear cell and granulocyte marking was higher at all time points derived from target CD34+ cells collected after G-CSF+SCF mobilization compared with target cells collected after G-CSF (n = 3) or G-CSF+Flt3-L (n = 2) mobilization. In 3 of the animals long-term marking levels of 5% to 25% were achieved, but originating only from the G-CSF+SCF–mobilized target cells. Transduction efficiency of HSCs collected by different mobilization regimens can vary significantly and is superior with G-CSF+SCF administration. The difference in transduction efficiency of HSCs collected from different sources should be considered whenever planning clinical gene therapy trials and should preferably be tested directly in comparative studies.
Introduction
Nonhuman primates are invaluable models for evaluation of gene therapy protocols prior to their implementation in human studies. The ability to transduce primitive human hematopoietic stem cells (HSCs) has been poorly predicted by murine and in vitro models, but has been more closely correlated with results in rhesus macaque and other nonhuman primate autologous transplantation assays. This is likely due to similarities in hematopoietic stem cell kinetics, cytokine responsiveness, and retroviral receptor levels between humans and nonhuman primates.1 Performance of transduction on a surface coated with the carboxy terminal fragment of fibronectin (FN) and inclusion of a combination of cytokines including stem cell factor (SCF), megakaryocyte growth and development factor (MGDF), andfms-like tyrosine kinase 3 ligand (Flt3-L) has been shown to greatly improve retroviral gene transduction in nonhuman primates, with long-term marking levels of 15% to 20% possible.2,3 Most importantly, advances in nonhuman primate gene transfer strategies have been translated into the first successful gene therapy clinical trial in patients with X-linked severe combined immunodeficiency syndrome (SCID).4 5
One aspect of gene transfer into hematopoietic stem cells that has received less attention since the advent of more efficient retroviral transduction protocols is the source of target HSCs used in therapeutic or marking clinical trials. So far target HSCs for clinical gene transfer or therapy protocols have been collected via bone marrow (BM) harvest,4-10 granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral blood (PB) leukapheresis,8,11-13 or umbilical cord blood collection.14,15 In 1994, Bodine et al showed that mobilized peripheral blood cells collected after administration of G-CSF+SCF were transduced better with retroviral vectors compared with the BM cells of mice treated with 5-fluorouracil (5-FU).16 We later extended these studies to the rhesus macaque model and found that the level of retroviral transduction was comparable in long-term engrafting cells collected from PB after 5 days of G-CSF+SCF administration or collected from BM 14 days after G-CSF+SCF administration.17 In the same set of experiments we also showed that CD34+ cells collected from BM primed with G-CSF+SCF administration were superior targets for retroviral transduction compared with steady state BM CD34+ cells, using transduction conditions without Flt3-L, MGDF, or fibronectin fragment support, now known to be suboptimal.
The combination of G-CSF+SCF has proven to be a more effective mobilizing regimen compared with G-CSF alone in murine,18-20 canine,21 and nonhuman primate22-25 models, and in several randomized clinical trials the combined use of G-CSF+SCF led to increased median yield of CD34+ cells per leukapheresis.26-30 It has been suggested that these cells might be better targets for gene therapy compared with G-CSF–mobilized cells,31 but to date this regimen has not been used in any clinical gene therapy trial.
Due to serious anaphylactic reactions occurring in patients treated with SCF in combination with other agents for aplastic anemia, this cytokine is no longer available, even for approved clinical protocols in the United States, although it is marketed in several other countries. In contrast, G-CSF has an extensive safety profile, both as an agent for accelerating recovery from neutropenia after myelotoxic treatments (chemotherapy or radiation) and as the primary agent for peripheral blood stem cell mobilization for transplantation purposes. It also has been reported recently that Flt3-L administered in combination with G-CSF for 5 days is a potent mobilizer of HSCs into the peripheral blood of mice32 and nonhuman primates,33 and more recently Rosenzweig et al used G-CSF+Flt3-L–mobilized PB CD34+ cells for retroviral gene transfer in nonhuman primate experiments, although in the latter study maximum number of colony-forming cells in the blood were obtained after 7 days of administration of the cytokines.34 In this study we directly compared the levels of long-term gene marking achievable in the nonhuman primate model using G-CSF+SCF–mobilized PB CD34+ HSC targets versus CD34+ cells obtained by mobilization with clinically more practical and available mobilization regimens, including G-CSF alone or the combination of G-CSF+Flt3-L, each regimen administered for 5 doses.
Materials and methods
Collection of PB HSCs from rhesus macaques
Young rhesus macaques (Macaca mulatta) used in these studies were housed and handled in accordance with the guidelines set by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council (DHHS publication No. NIH 85-23), and the protocol was approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute. The animals received recombinant human (rhu) G-CSF (10 μg/kg; Amgen, Thousand Oaks, CA) alone or in combination with either recombinant polyethylene glycol conjugated human SCF (200 μg/kg; Amgen) or recombinant human Flt3-L (200 μg/kg; Immunex, Seattle, WA) as daily subcutaneous injections for 3 days, and twice daily on the fourth day. Mobilized PB cells were collected by leukapheresis on day 5 as described.25 Mononuclear cells were isolated using density gradient centrifugation over lymphocyte separation media (ICN Biomedicals/Cappel, Aurora, OH). CD34+ enrichment was performed using the 12.8 Immunoglobulin-M (IgM) anti-CD34 biotinylated antibody and MACS streptavidin microbeads (Miltenyi Biotec, Auburn, CA). The degree of progenitor enrichment was calculated from CFU assays performed before and after column purification.
Vectors and transduction procedures
G1Na and LNL6 are Moloney murine leukemia virus–derived retroviral vectors that carry an identical bacterial neomycin phosphotransferase (neo) gene.35 A 16-bp polylinker insertion 5′ of the neo gene allows quantitative assessment of marking from the 2 vectors within one polymerase chain reaction (PCR).36 The biologic titers of these vectors were assessed by making serial dilutions of the vectors and then monitoring the transfer of G418 resistance to HeLa cells. The biologic titer of both vectors were equivalent and between 2-5 × 105 biologically active vector particles/mL. For transduction, retroviral supernatant was harvested from subconfluent producer cells cultured for 12-18 hours in Dulbecco modified Eagle medium (DMEM) (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA), 4 mMl-glutamine, penicillin (50 mg/mL), and streptomycin (50 mg/mL) at 37°C in 5% CO2. Fresh vector supernatant was passed through a 0.22-mm filter (Millipore, Bedford, MA) to remove cellular debris before transduction.
CD34-enriched cells were cultured at a starting concentration of 1-2 × 105 cells/mL in filtered vector supernatant, supplemented with 100 ng/mL rhuSCF, 100 ng/mL rhuFlt-3 L, and 100 ng/mL rhuMGDF (Amgen), in flasks previously coated with the CH-296 fragment of fibronectin (Retronectin, TaKara, Shiga, Japan) per manufacturer's instructions. Every 24 hours, nonadherent cells were collected, spun down, and resuspended in fresh vector supernatant and cytokines, and added back to the same fibronectin-coated flask. At the end of 96 hours cultured cells were removed from the plates with trypsin, collected, frozen viably in 50% autologous serum mixed with 40% DMEM and 10% dimethyl sulfoxide (DMSO), placed on dry ice, and then transferred to a liquid nitrogen container.
Each animal was remobilized after a period of 6 weeks, with the alternative cytokine regimen, and CD34+ cells were obtained and purified using the same procedures. This remobilization interval was chosen as likely beyond the time period impacted by a prior mobilization cycle.37 CD34+ cells were transduced under the same conditions as described above, but with the alternative vector, and then cryopreserved. One week later, the animals received 500 cGy × 2 total body irradiation, and both aliquots of transduced cells were thawed and reinfused via a central venous catheter. Twenty-four hours later, the animals were started on G-CSF at 5 μg/kg intravenously daily until the total white blood cell count reached 6000/μL. Standard supportive care included blood product transfusions, fluid and electrolyte management, and antibiotic administration as needed. Hematopoietic recovery was monitored by daily complete blood counts.
Sample collection
PB samples were collected at the time of recovery, monthly through 6 months after transplantation, and then every 3 months. From each blood sample, mononuclear cells were isolated by density gradient centrifugation over lymphocyte separation medium, and granulocytes were obtained as previously described.38
CFU assays
PB CD34-enriched cells from before and after each transduction and mononuclear cells obtained from bone marrow samples after transplantation were plated and analyzed by CFU assays using MethoCult M4230 methylcellulose (MC) media (StemCell Technologies, Vancouver, BC, Canada) supplemented with 5 U/mL rhu erythropoietin (Amgen), 10 ng/mL rhuGM-CSF (Sandoz, East Hanover, NJ), 10 ng/mL rhuIL-3, (Sandoz), and 100 ng/mL rhuSCF (Amgen) at 37°C in 5% CO2. Between days 10 and 14, colonies of more than 50 cells were counted, and between 16 and 32 individual CFUs were plucked from the plates at each time point for PCR analysis. Colonies were plucked into 50 μL distilled water, digested with 20 μg/mL proteinase K (Qiagen, Valencia, CA) at 55°C for 1 hour, followed by 99°C for 10 minutes, and later assessed for the presence of vector sequences by PCR as described below.
PCR analysis
Genomic DNA was extracted using the QIAamp DNA blood Midi kit (Qiagen). The primers and conditions used for neo PCR andβ-actin PCR have been previously described.8All neo and β-actin PCR reactions were run under conditions optimized to yield linear results in the range of the intensity of the in vivo samples. For the outer reaction, 100-200 ng DNA was used, and 18-20 cycles were performed for the inner reaction, based on the level of in vivo marking. For every PCR analysis, negative controls included DNA from normal rhesus PB samples extracted with the same methodology and a reagent control. Serial dilutions of G1Na DNA (containing 2 copies of integrated vector per cell) into normal rhesus PB DNA were used as positive controls for generating the control regression curve. Band intensity was quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). neo band intensity was normalized for amplifiable DNA content based on theβ-actin signal, and the overall contribution of each vector to in vivo marking was calculated by plotting the signal intensity of each band on a standard curve derived from known copy number controls amplified concurrently.
Southern blot analysis
Ten micrograms of genomic DNA was digested with KpnI (Boehringer-Mannheim, Indianapolis, IN), which cuts once within each viral long-terminal repeat, and thus can distinguish integrated retroviral G1Na and LNL6 DNA. The DNA fragments were transferred to a nylon membrane and hybridized with a radiolabeledneo gene–specific probe generated by PCR using the following primers: forward primer 5′-TCC ATC ATG GCT GAT GCA ATG CGG C-3′ and reverse primer 5′-GAT AGA AGG CGA TGC GCT GCG AAT CG-3′.39
Statistical analysis
Analysis of significance using the 2-tailed Student ttest and regression analysis were carried out using SigmaPlot (SPSS Science, Chicago, IL) and Excel software (Microsoft, Seattle, WA).
Results
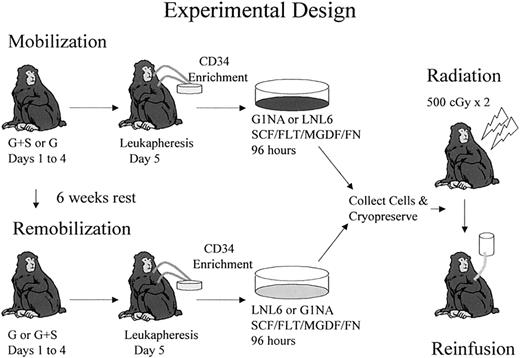
The experimental design for comparison of G-CSF+SCF versus either G-CSF alone or G-CSF+Flt3-L–mobilized CD34+ cells as targets for retroviral transduction is shown in Figure1 and explained in “Materials and methods.” Three animals were used to compare G-CSF+SCF with G-CSF alone, and 2 animals were used to compare G-CSF+SCF with G-CSF+Flt3-L. Table 1 summarizes the order in which each mobilization regimen was administered and the marking vector used to transduce each population of CD34+ cells. The order of administration of cytokines was reversed in each consecutive animal to control for any impact of prior mobilization on remobilization.37 The G1Na and LNL6 vectors have comparable titers on HeLa cells, and in multiple prior studies have been shown to have equivalent transduction efficiency of rhesus hematopoietic stem and progenitor cells.3,39 40
Experimental design.
Each animal initially received 5 doses of either G-CSF+SCF or G-CSF before leukapheresis. CD34+ cells were transduced with either G1Na or LNL6 vectors for 96 hours. Each animal was mobilized with the alternative regimen 6 weeks later, but this time transduction was done with the other vector. Both transduced aliquots were frozen at the end of transduction and then thawed and reinfused to the monkey after 500 cGy × 2 total body irradiation. The same experimental design was used to compare G-CSF+SCF– versus G-CSF+Flt3-L–mobilized cells.
Experimental design.
Each animal initially received 5 doses of either G-CSF+SCF or G-CSF before leukapheresis. CD34+ cells were transduced with either G1Na or LNL6 vectors for 96 hours. Each animal was mobilized with the alternative regimen 6 weeks later, but this time transduction was done with the other vector. Both transduced aliquots were frozen at the end of transduction and then thawed and reinfused to the monkey after 500 cGy × 2 total body irradiation. The same experimental design was used to compare G-CSF+SCF– versus G-CSF+Flt3-L–mobilized cells.
Summary of CD34+ enrichment, CFU transduction efficiency, cell expansion, and engraftment kinetics of animals
| Cytokine regimen . | Vector . | CD34+ enrichment* . | Transduction efficiency† . | Initial no.of cells . | No. of cells at the end of culture . | Exp‡ . | No. of infused cells/kg . | Eng1-153 (d) . | F/U1-155 (mo) . |
|---|---|---|---|---|---|---|---|---|---|
| Animal RQ2314 | 7 | 18 | |||||||
| G-CSF | G1Na | 100 | 89% (25 of 28) | 1.4 × 107 | 2.6 × 107 | 1.8 | 6.2 × 106 | ||
| G-CSF+SCF | LNL6 | 151 | 96% (27 of 28) | 2.1 × 107 | 2.6 × 108 | 12.4 | 6.2 × 107 | ||
| Animal RQ2284 | 9 | 15 | |||||||
| G-CSF+SCF | G1Na | 73 | 100% (16 of 16) | 3.0 × 107 | 3.5 × 107 | 1.2 | 7.9 × 106 | ||
| G-CSF | LNL6 | 93 | 90% (27 of 30) | 2.2 × 107 | 2.2 × 107 | 1.0 | 4.8 × 106 | ||
| Animal RQ2237 | 7 | 12 | |||||||
| G-CSF | LNL6 | 113 | 100% (32 of 32) | 1.9 × 107 | 2.4 × 107 | 1.3 | 4.9 × 106 | ||
| G-CSF+SCF | G1Na | 95 | 100% (32 of 32) | 4.5 × 107 | 1.1 × 108 | 2.5 | 2.2 × 107 | ||
| Animal RQ2277 | 7 | 12 | |||||||
| G-CSF+SCF | G1Na | 155 | 100% (32 of 32) | 4.0 × 107 | 2.1 × 108 | 5.2 | 5.0 × 107 | ||
| G-CSF+Flt3-L | LNL6 | 95 | 100% (32 of 32) | 1.6 × 107 | 3.1 × 107 | 1.9 | 7.3 × 106 | ||
| Animal RQ2265 | 14 | 12 | |||||||
| G-CSF+Flt3-L | G1Na | 127 | 100% (32 of 32) | 2.9 × 107 | 4.6 × 107 | 1.6 | 1.0 × 107 | ||
| G-CSF+SCF | LNL6 | 72 | 100% (32 of 32) | 3.9 × 107 | 9.2 × 107 | 2.3 | 2.1 × 107 |
| Cytokine regimen . | Vector . | CD34+ enrichment* . | Transduction efficiency† . | Initial no.of cells . | No. of cells at the end of culture . | Exp‡ . | No. of infused cells/kg . | Eng1-153 (d) . | F/U1-155 (mo) . |
|---|---|---|---|---|---|---|---|---|---|
| Animal RQ2314 | 7 | 18 | |||||||
| G-CSF | G1Na | 100 | 89% (25 of 28) | 1.4 × 107 | 2.6 × 107 | 1.8 | 6.2 × 106 | ||
| G-CSF+SCF | LNL6 | 151 | 96% (27 of 28) | 2.1 × 107 | 2.6 × 108 | 12.4 | 6.2 × 107 | ||
| Animal RQ2284 | 9 | 15 | |||||||
| G-CSF+SCF | G1Na | 73 | 100% (16 of 16) | 3.0 × 107 | 3.5 × 107 | 1.2 | 7.9 × 106 | ||
| G-CSF | LNL6 | 93 | 90% (27 of 30) | 2.2 × 107 | 2.2 × 107 | 1.0 | 4.8 × 106 | ||
| Animal RQ2237 | 7 | 12 | |||||||
| G-CSF | LNL6 | 113 | 100% (32 of 32) | 1.9 × 107 | 2.4 × 107 | 1.3 | 4.9 × 106 | ||
| G-CSF+SCF | G1Na | 95 | 100% (32 of 32) | 4.5 × 107 | 1.1 × 108 | 2.5 | 2.2 × 107 | ||
| Animal RQ2277 | 7 | 12 | |||||||
| G-CSF+SCF | G1Na | 155 | 100% (32 of 32) | 4.0 × 107 | 2.1 × 108 | 5.2 | 5.0 × 107 | ||
| G-CSF+Flt3-L | LNL6 | 95 | 100% (32 of 32) | 1.6 × 107 | 3.1 × 107 | 1.9 | 7.3 × 106 | ||
| Animal RQ2265 | 14 | 12 | |||||||
| G-CSF+Flt3-L | G1Na | 127 | 100% (32 of 32) | 2.9 × 107 | 4.6 × 107 | 1.6 | 1.0 × 107 | ||
| G-CSF+SCF | LNL6 | 72 | 100% (32 of 32) | 3.9 × 107 | 9.2 × 107 | 2.3 | 2.1 × 107 |
Degree of progenitor cell enrichment by CD34 selection was calculated by enumerating total CFUs before and after column purification.
Transduction efficiency calculated by the percentage ofβ-actin-positive colonies that were also neopositive at the end of transduction, as determined by PCR of individual colonies.
Expansion: ratio of the number of cells at the end of 4 days of transduction (number of infused cells) to the number of cells plated on the first day.
Engraftment: days to reach absolute neutrophil count greater than 500/μL.
Follow-up: duration (months) of follow-up after transplantation.
There were no significant differences in the efficiency of transduction of committed progenitors, as defined using the in vitro CFU assay, found in G-CSF+SCF–mobilized CD34+ cells compared with G-CSF alone or G-CSF+Flt3-L (Table 1), and no overall discrepancies between results achieved using the G1Na versus the LNL6 vector. Comparable with our recent experiments,40 the transduction of committed progenitors was extremely efficient in all CD34+ populations, but our prior studies have indicated that CFU transduction may not correlate with transduction of more primitive in vivo repopulating cells.38-40
The number of CD34+ cells collected by leukapheresis was higher in each experimental group after mobilization with G-CSF+SCF (3.20 ± 1.21 × 107) versus mobilization with G-CSF alone (1.83 ± 0.40 × 107; P = .157) or G-CSF+SCF (3.95 ± 0.07 × 107) versus mobilization with G-CSF+Flt3-L (2.25 ± 0.91 × 107;P = .248), although the differences did not reach statistical significance given the small numbers of animals in each group. Cells collected following G-CSF+SCF administration expanded more robustly during the 4-day transduction period compared with cells mobilized with G-CSF alone (5.4 versus 1.4, respectively;P = .350) or with G-CSF+Flt3-L (3.7 versus 1.7, respectively; P = .366). Consequently, all 5 animals received a higher number of transduced G-CSF+SCF–mobilized cells compared with transduced G-CSF–mobilized cells (3.06 ± 2.80 × 107 versus 0.53 ± 0.08 × 107, respectively; P = .249) or G-CSF+SCF–mobilized cells compared with G-CSF+Flt3-L–mobilized cells (3.55 ± 2.05 × 107 versus 0.86 ± 0.19 × 107, respectively; P = .339) per kilogram of body weight. Due to a small number of animals in each experimental group, a limitation inherent to these types of primate experiments, statistical significance was not achieved in these comparisons despite the clear observed trends.
All animals recovered their peripheral blood counts without significant morbidity, and there was a trend toward faster engraftment in the animals that received higher total cell doses, as we have previously observed.38
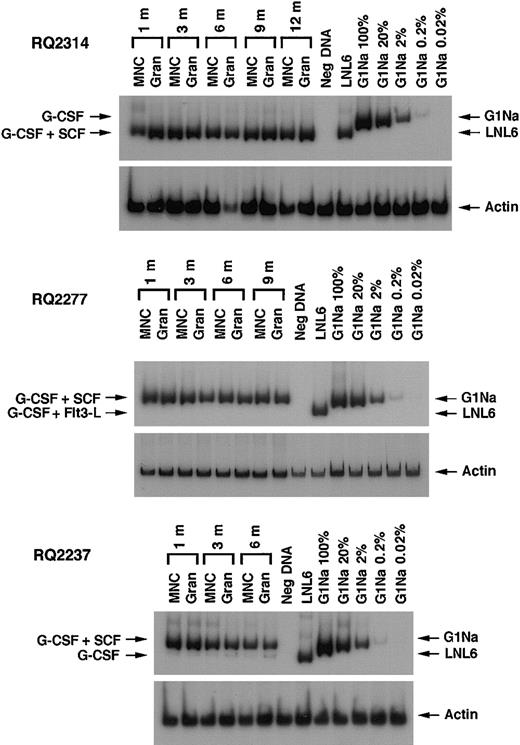
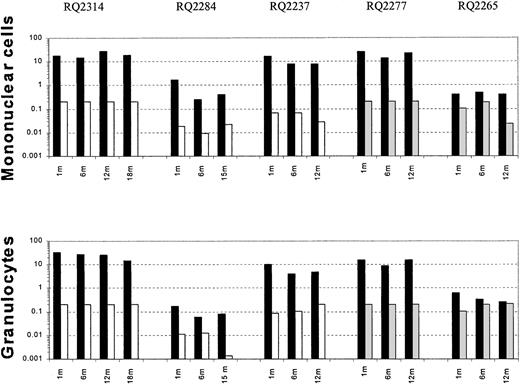
After transplantation, semiquantitative PCR analysis of peripheral blood samples allowed assessment of the relative contribution of marked cells derived from CD34+ target cells collected using different cytokine regimens. Figure 2shows representative PCR gels from animals RQ2314, RQ2277, and RQ2237. In animals RQ2314 and RQ2277, there was detectable neomarking derived only from the G-CSF+SCF–mobilized target cells and not from the G-CSF or the G-CSF+Flt3-L fractions. In animal RQ2237, the presence of marked cells derived from both G-CSF+SCF targets and G-CSF alone targets can be appreciated in both mononuclear cells and in granulocytes collected from peripheral blood at different time points after transplantation, but the level of marking from G-CSF+SCF–mobilized cells is on average 206 and 48 times higher, respectively, in mononuclear cells and granulocytes, compared with marking from G-CSF–mobilized cells. A summary of marking levels in PB mononuclear cells and granulocytes over time, assuming one vector integration per cell, is depicted on a logarithmic scale in Figure3. Animals RQ2284 and RQ2265 had overall lower marking levels, but the G-CSF+SCF target cells still resulted in better marking levels compared with G-CSF alone or G-CSF+Flt3-L.
PCR analysis of in vivo marking.
Representative PCR of neo and β-actin sequences in PB mononuclear cells (MNCs) and granulocytes (Gran) at different time points after engraftment in 3 animals. Concurrentβ-actin PCR was performed for each sample to quantitate the amount of amplifiable template DNA. Serial dilutions of G1Na DNA (2 copies of integrated vector per cell) into normal rhesus PB DNA were used as positive controls. In animals RQ2314 and RQ2277 the only amplified band corresponds to the vector that was used to transduce G-CSF+SCF–mobilized cells. In animal RQ2237 in addition to the band corresponding to G-CSF+SCF aliquot, a very faint band can be seen that corresponds to transduced G-CSF–mobilized cells.
PCR analysis of in vivo marking.
Representative PCR of neo and β-actin sequences in PB mononuclear cells (MNCs) and granulocytes (Gran) at different time points after engraftment in 3 animals. Concurrentβ-actin PCR was performed for each sample to quantitate the amount of amplifiable template DNA. Serial dilutions of G1Na DNA (2 copies of integrated vector per cell) into normal rhesus PB DNA were used as positive controls. In animals RQ2314 and RQ2277 the only amplified band corresponds to the vector that was used to transduce G-CSF+SCF–mobilized cells. In animal RQ2237 in addition to the band corresponding to G-CSF+SCF aliquot, a very faint band can be seen that corresponds to transduced G-CSF–mobilized cells.
Summary of in vivo gene marking.
Level of peripheral blood mononuclear cell and granulocyteneo gene marking as assessed by semiquantitative PCR at certain time points after transplantation, shown on a logarithmic scale. In each animal black bars (▪) represent the marking level originating from G-CSF+SCF–mobilized target cells; white bars (■), G-CSF–mobilized target cells; and gray bars (░), G-CSF+Flt3-L–mobilized cells.
Summary of in vivo gene marking.
Level of peripheral blood mononuclear cell and granulocyteneo gene marking as assessed by semiquantitative PCR at certain time points after transplantation, shown on a logarithmic scale. In each animal black bars (▪) represent the marking level originating from G-CSF+SCF–mobilized target cells; white bars (■), G-CSF–mobilized target cells; and gray bars (░), G-CSF+Flt3-L–mobilized cells.
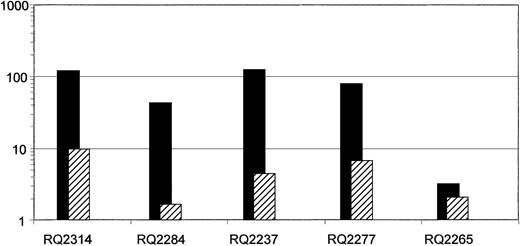
In Figure 4 we show the comparison between the ratio (G-CSF+SCF/G-CSF alone or G-CSF+SCF/G-CSF+Flt3-L fraction) of the number of infused cells (ie, number of cells collected at the end of 4 days of transduction) per kilogram weight of the animals versus the ratio of the marking achieved from each target cell group in each animal. Taking into account the number of infused cells, the ratio of marking remains significantly higher originating from the G-CSF+SCF–mobilized CD34+ cells compared with the ratio of infused cells, especially in the 3 animals that had high level gene marking.
Comparison of the ratio of in vivo gene marking versus the ratio of infused cells on a logarithmic scale.
In each animal the ratio of the average neo in vivo marking of first-year PB samples originating from G-CSF+SCF–mobilized cells to G-CSF alone or G-CSF+Flt3-L–mobilized cells were calculated (▪). For each animal this ratio was compared with the ratio of infused cells (ie, number of cells collected at the end of transduction) from G-CSF+SCF–mobilized cells to G-CSF alone or G-CSF+Flt3-L–mobilized cells (▨). Note that although the number of cells infused from the G-CSF+SCF aliquot was always higher in each animal compared with the other aliquot, the difference in gene marking levels between the aliquots was in every case significantly greater, except in animal RQ2265, which overall had low level marking. Monkeys RQ2314, RQ2284, and RQ2237 were mobilized with G-CSF+SCF versus G-CSF alone, and monkeys RQ2277 and RQ2265 were mobilized with G-CSF+SCF versus G-CSF + Flt3-L.
Comparison of the ratio of in vivo gene marking versus the ratio of infused cells on a logarithmic scale.
In each animal the ratio of the average neo in vivo marking of first-year PB samples originating from G-CSF+SCF–mobilized cells to G-CSF alone or G-CSF+Flt3-L–mobilized cells were calculated (▪). For each animal this ratio was compared with the ratio of infused cells (ie, number of cells collected at the end of transduction) from G-CSF+SCF–mobilized cells to G-CSF alone or G-CSF+Flt3-L–mobilized cells (▨). Note that although the number of cells infused from the G-CSF+SCF aliquot was always higher in each animal compared with the other aliquot, the difference in gene marking levels between the aliquots was in every case significantly greater, except in animal RQ2265, which overall had low level marking. Monkeys RQ2314, RQ2284, and RQ2237 were mobilized with G-CSF+SCF versus G-CSF alone, and monkeys RQ2277 and RQ2265 were mobilized with G-CSF+SCF versus G-CSF + Flt3-L.
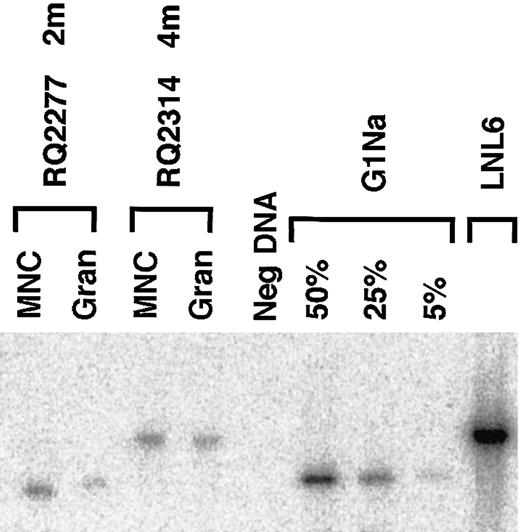
The level and ratio of marking originating from the 2 transduced fractions were confirmed by genomic Southern blotting of peripheral blood samples from selected time points in the 2 animals with the highest level of marking (RQ2314 and RQ2277). In the blot shown in Figure 5 the only detectableneo marking in each of these 2 animals corresponds to the vector that was used to transduce the G-CSF+SCF–mobilized cells, and there is no detectable level of marking from the other fraction.
Southern blot analysis of genomic marking.
In animals RQ2277 and RQ2314 the Southern blot analysis of PB mononuclear cells and granulocytes after engraftment shows only the vector that was used to transduce the G-CSF+SCF fraction (G1Na in animal RQ2277, and LNL6 in animal RQ2314). LNL6 DNA and serial dilutions of G1Na DNA into normal rhesus DNA were used as controls. Due to residual env sequences in LNL6 that are not present in G1Na, the expected fragment size is 3.0 kb for LNL6 and 2.3 kb for G1Na.
Southern blot analysis of genomic marking.
In animals RQ2277 and RQ2314 the Southern blot analysis of PB mononuclear cells and granulocytes after engraftment shows only the vector that was used to transduce the G-CSF+SCF fraction (G1Na in animal RQ2277, and LNL6 in animal RQ2314). LNL6 DNA and serial dilutions of G1Na DNA into normal rhesus DNA were used as controls. Due to residual env sequences in LNL6 that are not present in G1Na, the expected fragment size is 3.0 kb for LNL6 and 2.3 kb for G1Na.
We also confirmed the functional high level of marking in these 2 animals long after transplantation by PCR analysis of CFU assays of BM mononuclear cells plated with and without G418 at 1.0 mg/mL active concentration, which is effective in inhibiting 98%-100% untransduced control rhesus cells plated in parallel.3 We counted the number of colonies from MC plates that were each plated with 105/mL of BM mononuclear cells. In animal RQ2314, 20 months after transplantation there were on average 78 colonies in each G418− plate and 6 colonies in each G418+plate; among the colonies plucked from G418+ plates, 27 of 29 were positive for LNL6 vector by PCR (G-CSF+SCF fraction) and only 2 of 29 colonies were positive for G1Na (G-CSF fraction). In animal RQ2277, 16 months after transplantation there were on average 51 colonies in each G418− plate and one colony in each G418+ plate; among colonies plucked from G418+plates, 16 of 18 were positive for G1Na (G-CSF+SCF fraction) and only 2 of 18 colonies were positive for LNL6 (G-CSF+Flt3-L fraction).
Discussion
In this study we directly compared the level of in vivo marking of long-term repopulating cells obtained using 3 different mobilization regimens for collection of target CD34+ cells. As expected, based on previous nonhuman primate23-25 and human clinical studies,26-30 the number of CD34+ cells collected after G-CSF+SCF mobilization was higher than the number of cells collected using G-CSF alone. There is less prior information on the relative efficacy of G-CSF+Flt3-L mobilization, but we also found G-CSF+SCF to result in a higher number of cells collected compared with G-CSF+Flt3-L in the 2 animals tested when each combination was administered for 5 doses. The cells collected following G-CSF+SCF mobilization expanded more in vitro. Despite a similar level of transduction efficiency of progenitors as assessed by PCR for vector in CFUs present at the end of the 4-day culture period, the in vivo results using the different target cell mobilization regimens were quite different. In all 5 animals the marking of circulating mononuclear cells and granulocytes was significantly higher from the G-CSF+SCF–mobilized cells compared with either G-CSF alone or G-CSF+Flt3-L, and in 3 animals we achieved stable long-term marking of 5%-25%.
There are at least 3 possible explanations for the much higher marking levels achieved using G-CSF+SCF–mobilized target cells. The first 2 are simply numeric: a larger number of HSCs mobilized with this regimen, or improved survival or expansion of these cells in vitro following transduction, if the actual transduction frequency remains the same, would result in higher in vivo marking given the competitive repopulation design. The third explanation would be actual differences in transduction efficiency of primitive HSCs. Although a higher number of starting cells or a higher number of infused cells from G-CSF+SCF aliquot collected at the end of 4 days of transduction might have contributed in part to the differences observed in marking levels, we believe that this is unlikely to be the only reason for the improved in vivo marking. As we demonstrated in Figure 4, even if we normalize the marking level achieved for the number of infused CD34+cells per kilogram weight of the animals, the level of marking remains significantly higher originating from the G-CSF+SCF–mobilized CD34+ cells. The relative frequency of actual repopulating stem cells present within the total CD34+ population may have been higher within G-CSF+SCF–mobilized cells and account for the differences seen,41 but in vivo NOD/SCID engraftment assays do not suggest this to be the case.42 Therefore, we suggest that there is some intrinsic difference in the target HSCs mobilized with G-CSF+SCF that permits more efficient transduction.
One of the major obstacles in retroviral gene transduction of HSCs is the quiescent nature of these cells, because successful transduction of HSCs by retroviral vectors requires mitotic divisions to allow entry of preintegration viral complexes into the nucleus.43-45Primitive CD34+ hematopoietic stem cells present in mobilized peripheral blood, whether with G-CSF or G-CSF+SCF, are primarily in the G0 phase of the cell cycle.39,46 Using different combinations of stimulatory cytokines in culture media allows primitive hematopoietic cells to enter cell cycle and hence become susceptible to retroviral transduction, but this occurs at the cost of differentiation and loss of engraftment potential.47,48 Gene transfer investigators have spent years testing many different combinations of cytokines, and recent improvements in gene transfer efficiency into primitive repopulating cells assayed in murine xenograft models or nonhuman primates have occurred using combinations including stem cell factor and Flt3-L, along with traditional cytokines such as interleukin-3 (IL-3) and IL-6 or, most recently, thrombopoietin/MGDF.2,38 49 Presumably these combinations induce the most primitive cells to cycle without initiating terminal differentiation.
Duarte and Frank50 transduced the MO7e primitive hematopoietic cell line, known to express the SCF receptor, with the G-CSF receptor gene. Treating these cells with a combination of G-CSF and SCF resulted in a significant reduction in the percentage of cells in G0/G1 compared with cells treated with G-CSF alone.50 They also observed a marked shortening of the duration of the G0/G1 phase of the cell cycle following G-CSF+SCF cotreatment compared with cells treated with G-CSF alone, mediated in part by decreased expression of p27kip-1. It is possible that primitive cells obtained from different sources vary in an intrinsic program governing cell division in vitro or are differentially susceptible to extrinsic cytokine stimulation. This has been demonstrated for primitive cells in cord blood or fetal liver compared with adult marrow or mobilized blood.51 We hypothesize that stem cells present in the G-CSF+SCF–mobilized blood might enter the cell cycle more frequently or rapidly than stem cells present after G-CSF or G-CSF+Flt3-L administration, and thus be susceptible to transduction by retroviral vectors.
The induction of cycling in vitro can be a double-edged sword. A number of investigators reported loss of engraftment potential with cytokine stimulation in vitro and linked this loss specifically to cells that were actively cycling.46,47,52,53 The potential reversibility of this engraftment defect was first shown by Habibian and coworkers, who reported dramatic fluctuations in the engraftment phenotype of murine HSCs linked to their progress through and then out of the cell cycle.54 We have recently extended this observation to the nonhuman primate model and gene transfer applications by demonstrating that cells transduced for 4 days in the presence of stimulatory cytokines regained engraftment ability if they were allowed to exit S/G2/M by maintenance in SCF alone for 48 hours on fibronectin-coated plates.39 One potential factor explaining our current results could be that G-CSF+SCF–mobilized stem cells enter cell cycle more rapidly in vitro, permitting transduction, but then come back out of cycle and thus by 96 hours have regained some engraftment potential. HSCs mobilized with G-CSF alone or G-CSF+Flt3-L instead perhaps enter cell cycle more slowly and are therefore both more difficult to transduce and could even be hindered in engraftment because at the time of reinfusion they may have finally entered cell cycle. Our ability to test these hypotheses is limited by the poor phenotypic characterization of rhesus primitive engrafting cells, thus cell cycle analysis on purified rhesus HSCs has not been feasible, but some of these issues could be explored in the NOD/SCID model using human cells.42
One other factor that might contribute to our results is the level of amphotropic retrovirus receptor (amphoR) on target cells from different sources. Orlic et al55 have shown that a low level of amphoR mRNA in mouse HSCs correlated with a low level of amphotropic retrovirus transduction of these cells. They also reported that primitive c-KitHi HSCs have a higher level ofamphoR mRNA when marrow was collected after treatment with G-CSF+SCF compared with steady state, and that this higher level correlated with improved transduction efficiency. They found very low levels of amphoR mRNA in adult bone marrow Lin−CD34+ CD38− cells, compared with cord blood or fetal liver,56 but receptor levels have not been studied in the context of cytokine therapy. Although this phenomenon has not been tested directly in the human and nonhuman primate experiments regarding actual repopulating cell transduction, it seems likely that it may be at least partially responsible for some of the differences observed in the in vivo results using different sources of HSCs.
These studies all point to the fact that HSCs collected from alternative sources might have different properties both at the time of collection and at the end of transduction and, consequently, variable abilities to be transduced or to engraft at the end of culture. Highly proliferative characteristics of umbilical cord blood and increased amphotropic receptor levels, along with an in vivo advantage for corrected lymphoid cells, might explain the relatively encouraging results obtained in a trial for adenosine deaminase deficiency using autologous cord blood CD34+ cells.14,15The recent reports of successful gene therapy for X-linked SCID used marrow target cells, but from very young children,4,5 10and these cells may also have improved transduction capabilities based on the properties discussed above.
We believe our findings might provide an explanation for the low level of in vivo gene transduction results obtained so far in human clinical gene therapy trials using primitive hematopoietic stem cell targets in G-CSF–mobilized peripheral blood. Over the last several years advances in vector design, culture conditions, and cytokines used in vitro in gene transduction protocols have significantly improved the level of gene transfer using G-CSF+SCF–primed blood or bone marrow, or bone marrow from young children, but based on our results, it seems that more research is needed in elucidating the biology of CD34+cells collected through different sources and the effect it might have on the final results. The importance of reconsidering the use of SCF in mobilization regimens, at least in gene therapy applications, seems warranted, because the mast cell release toxicity generally can be avoided by premedication with antihistamines before SCF administration, and patients undergoing a brief treatment mobilization could receive all SCF doses in a monitored setting.
We thank Dr David Woo at Amgen for supplying rhuG-CSF and rhuSCF, Dr Hiroshi Miyazaki at Kirin for supplying rhuMGDF, and Dr Stewart Lyman at Immunex for supplying Flt3-L.
Prepublished online as Blood First Edition Paper, November 7, 2002; DOI 10.1182/blood-2002-08-2663.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Cynthia E. Dunbar, Hematology Branch, NHLBI, NIH, Building 10, Rm 7C103, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: dunbarc@nhlbi.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal