T-acute lymphoblastic leukemias (T-ALLs) derive from human T-lymphoid precursors arrested at various early stages of development. Correlation of phenotype and T-cell receptor (TCR) status with RAG-1 and pTα transcription in 114 T-ALLs demonstrated that they largely reflect physiologic T-lymphoid development. Half the TCRαβ lineage T-ALLs expressed a pre-TCR, as evidenced by RAG-1, pTα, and cTCRβ expression, absence of TCRδ deletion, and a sCD3−, CD1a+, CD4/8 double-positive (DP) phenotype, in keeping with a population undergoing β selection. Most TCRγδ T-ALLs were pTα, terminal deoxynucleotidyl transferase (TdT), and RAG-1lo/neg, double-negative/single-positive (DN/SP), and demonstrated only TCRβ DJ rearrangement, whereas 40% were pTα, TdT, and RAG-1 positive, DP, and demonstrated TCRβ V(D)J rearrangement, with cTCRβ expression in proportion. As such they may correspond to TCRαβ lineage precursors selected by TCRγδ expression, to early γδ cells recently derived from a pTα+ common αβ/γδ precursor, or to a lineage-deregulated αβ/γδ intermediate. Approximately 30% of T-ALLs were sCD3/cTCRβ− and corresponded to nonrestricted thymic precursors because they expressed non–T-restricted markers such as CD34, CD13, CD33, and CD56 and were predominantly DN, CD1a, pTα, and RAG-1 low/negative, despite immature TCRδ and TCRγ rearrangements. TCR gene configuration identified progressive T-lymphoid restriction. T-ALLs, therefore, provide homogeneous expansions of minor human lymphoid precursor populations that can aid in the understanding of healthy human T-cell development.
Introduction
T lymphocytes are derived from pluripotent hemopoietic progenitors that migrate from the fetal liver or bone marrow to the thymus, where most T-cell development takes place. This process is associated with progressive restriction of developmental potential, with the earliest recognizable thymic precursor retaining T, natural killer (NK), and dendritic cell (DC)—and possibly B lymphoid and myeloid—potential but having lost the capacity for erythroid or megakaryocytic differentiation.1 The early stages of human T-lymphoid development are relatively poorly understood because of difficulty in obtaining sufficient quantities of homogeneous material and because comparison with murine development has been hampered by different immunophenotypic selection criteria. The early stages of murine thymic development occur in the minor (less than 1% of thymocytes) sCD3−, CD4/CD8-double-negative (DN) population and have been defined by surface (s) phenotype, T-cell receptor (TCR) gene configuration and in vitro developmental potential. DN thymocytes are classified on the basis of CD44 and CD25 expression: DN1 (CD44+/hi, CD25−), DN2 (CD44+/hi, CD25+), DN3 (CD44−/lo, CD25+), and DN4 (CD44−/lo CD25−).2,3 TCR rearrangement starts at the DN2 stage with the TCRδ locus, closely followed by TCRγ and TCRβ, and rearrangements of all 3 loci are largely completed in DN3 thymocytes.4 Successful TCRβ rearrangement in the presence of pTα allows expression of a pre-TCR in association with sCD3, progression to the CD4/CD8 double-positive (DP) stage, and massive thymocyte expansion, a process known as β selection. This is followed by TCRα rearrangement, TCRδ deletion, and replacement of the pre-TCR by TCRαβ. Low-level surface expression of the pre-TCR leads to difficulties in its immunophenotypic detection.5 In humans, the earliest thymic precursor demonstrates T/NK/DC potential and is defined by a CD34+/CD33+/CD7++/CD45RA+/sCD3−/CD2−/CD5−/CD1a−phenotype. It can be distinguished from bone marrow progenitors by the expression of CD45RA and CD7. Progressive restriction to a CD5+ CD1a− T/NK precursor, followed by T-restricted potential at the CD5+, CD1a+ stage of CD34+, sCD3− DN thymocyte development has been described.6 This is followed by intermediate single positivity (ISP) for CD4, immediately before the DP transition. TCRδ rearrangements start at the CD5+, CD1a− stage, and TCRγ and TCRβ rearrangements start at the CD1a+stage, just before the start of cTCRβ expression and β selection at the CD4 ISP/DP transition.7 TCRδ rearrangements start with VD or DD and progress to DJ and V(D)J, before TCRδ deletion in αβ lineage precursors. The limits of the TCRδ locus can be defined by the δRec and ψJα segments, because all functional V and J segments outside these limits have been found to be rearranged in TCRαβ lineage cells, including certain Vα/δ segments that can also rearrange to Jδ8 TCRβ rearrangements start with DJ, followed by V(D)J, and are subjected to allelic exclusion.9 The TCRγ locus does not contain D segments, but downstream Vγ segments such as Vγ9 and upstream JP1/2 segments are preferentially used in immature, fetal-type rearrangements.10 The 2 most 3′ Vγ segments, Vγ10 and Vγ11, are pseudogenes, because of the absence of splicing of their leader introns.11 Progressive opening has also been described for the TCRα locus.12 Compared with the αβ lineage, understanding of the early stages of TCRγδ lymphoid development is less complete, largely because of the absence of recognized lineage-specific surface markers other than the TCR. Mature TCRγδ cells are predominantly CD4/CD8 DN or CD8 SP and, unlike αβ T cells, variably express CD2.13 Maturation of γδ lymphocytes can occur in the thymus, where they represent approximately 1% of lymphocytes, but it is not restricted to this site. The factors that determine whether a thymic precursor differentiates toward the TCRαβ or TCRγδ lineage have been extensively studied and reviewed,14,15 but the relative roles of a stochastic versus an instructive process are not yet entirely clear, and both are likely to be operational. Several arguments suggest that rearrangement of a functional TCRγδ orientates toward the γδ lineage, whereas expression of the pre-TCR directs cells toward the αβ lineage. Expression of a functional pre-TCR is fundamental to αβ lineage development, because the loss of pTα, TCRβ, CD3, or RAG-1/2 leads to a block at the DN3 stage.16,17 The extent of the block is variable; it is more severe for CD3 and RAG1 than for TCRβ. This is because TCRγδ allows some, albeit inefficient, development from the DN3 block to the DP stage.18,19 Whether TCRγδ contributes to αβ lineage development in the absence of pre-TCR abnormalities is unclear. Human T-acute lymphoblastic leukemias (T-ALLs) represent malignant proliferations arrested at variable early stages of T-lymphoid development. It has, however, often been suggested that the process of leukemic transformation leads to immunophenotypic and genotypic deregulation, limiting the use of these precursor populations for the study of human lymphopoiesis.20 Comparison of pTα and RAG-1 transcripts with extensive immunophenotyping and TCR genotyping allowed us to show that T-ALLs largely represent physiological T-lymphoid maturation as we currently understand it. As such, T-ALLs can provide valuable tools for the analysis of human early T-cell development.
Patients, materials, and methods
Patients and cell lines
Diagnostic samples of peripheral blood or bone marrow from 114 cases of T-ALL were analyzed, with informed consent. Biphenotypic acute leukemia, as defined by the European Group for the Immunological Characterization of Leukemias (EGIL),21 were excluded. They included 46 children younger than 15 years (mean, 8.1 years) and 68 adults from 15 to 78 years (mean, 20.4 years); the male-female ratio was 2.4:1. All samples had more than 80% blasts. Patients came from 16 clinical centers, and most were treated on national pediatric (FRALLE, coordinator A. Baruchel) and adult (LALA, coordinator D. Fière) protocols. Cells underwent Ficoll gradient centrifugation before immunophenotyping and DNA and RNA extraction, which was performed directly or after freezing in dimethyl sulfoxide (DMSO).22Cytospins were also used for immunocytochemistry. Five diagnostic AML, 9 healthy bone marrow, and 6 healthy peripheral blood mononuclear cell (PBMC) fractions were used as controls. The HPB-ALL cell line, used to normalize RAG-1 and pTα transcript levels, was grown in RPMI 1640 and 10% fetal calf serum (FCS).
Immunophenotype
Immunophenotyping was performed in the diagnostic center on fresh material and was completed from cryopreserved material. Diagnostic panels included at least the T (CD2, CD5, CD7, cCD3, mCD3, CD4, CD8, CD1a, TCR αβ, and TCR γδ), the B (CD19, CD20, CD22, cIgM, terminal deoxynucleotidyl transferase [TdT], CD10, cCD79a), and myeloid (CD13, CD33, CD117, myeloperoxidase [MPO]) cell markers. TCRαβ expression was detected with BMA031 phycoerythrin (PE; Immunotech, Marseilles, France).23 TCRγδ expression was assessed with the Immu510 fluorescein isothiocyanate (FITC) pan-γδ antibody (Immunotech).24 Intracytoplasmic CD3 (cCD3), immunoglobulin M (IgM), and nuclear TdT were analyzed by flow cytometry after permeabilization with a commercial kit (Harla Sera-Lab, Loughborough, England). Detection of cTCRβ was undertaken by indirect labeling of permeabilized cells with the βF1 antibody (Bioadvance, Emerainville, France) and fluorescent goat anti-mouse (Immunotech). βF1 recognizes a TCRβ epitope that is not expressed at the cell surface.25 Flow cytometric analysis was compared with immunocytochemical detection using the Vectastain ABC avidin-biotin kit (Vector Laboratories, Burlingame, CA). Samples with more than 20% labeled cells after correction for the proportion of blasts and of healthy T lymphocytes were considered positive.
TCR rearrangements
DNA and RNA were extracted from fresh or cryopreserved cells, as previously described.22 TCRγ rearrangements were assessed by fluorescence multiplex polymerase chain reaction (PCR) amplification.26 The TCRδ and TCRβ multiplex PCRs were developed within the Biomed-2 BMH4-CT98-3936 Concerted Action.27 Briefly, 100 ng DNA was amplified for 35 cycles in the presence of 0.2 μM each primer, 2 mM MgCl, and 1 U Taq Gold (Perkin Elmer). TCRβ gene configuration was assessed with a 3-tube multiplex PCR, 2 of which contained 27 Vβ family-specific upstream primers with either 9 (PCR A) or 4 (PCR B) downstream Jβ primers. The third (PCR C) contained all 13 Jβ primers and Dβ1 and Dβ2 upstream primers (Figure 1). TCRβ and TCRδ PCR products were analyzed by heteroduplex analysis.27 T-ALLs were classified as TCRβ germline if all PCRs were negative, immature if only DJ rearrangements were observed, and mature if a clonal V(D)J rearrangement was identified on at least one allele. TCRδ rearrangements were assessed by Southern blot analysis, multiplex PCR analysis, or both. DNA was digested withEcoRI, HindIII, or BglII and were hybridized sequentially with 32P-labeled Jδ1, δRec, and ψJα probes.28 The single-tube TCRδ multiplex PCR was performed using Vδ1-6 and Dδ2-specific upstream primers and Dδ3 and Jδ1-4-specific downstream primers. The sensitivity of detection of clonality using these techniques is approximately 1% to 10%. Identification of Vδ and Jδ segment usage was based on the size of the Southern blot-rearranged bands with Jδ1 and/or the size of the multiplex PCR heteroduplex products, confirmed by specific monoplex PCR and/or multiplex multifluorescence PCR. TCRδ multifluorescence typing was performed with unlabeled Jδ Dδ primers and Vδ Vδ2, and Dδ2 labeled primers, using Biomed-2 conditions. Assessment of TCRδ deletion was based on loss of Jδ1, δRec, and ψJα signals. Unidentified rearrangements were those clearly detected by Southern blot analysis but with no apparent clonal PCR product. Complete TCRβ and TCRδ rearrangements are referred to as V(D)J, because no attempt was made to identify the presence of delta segments within clonal PCR products unless stated. TCRδ status was assessed by Southern blot and PCR in 85 cases and by PCR alone in 28 cases. Twenty of these were classified as rearranged because at least one clear clonal band was seen. Eight (4 pre-αβ, 3 TCRαβ SP, and 1 TCRγδ) were negative by multiplex heteroduplex PCR analysis and as such could either have a germline TCRδ configuration, a biallelic deletion, or TCRδ rearrangements that could not be detected by the primers used. Evaluation of the frequency of these configurations is therefore likely to be slightly underestimated in Table 1.
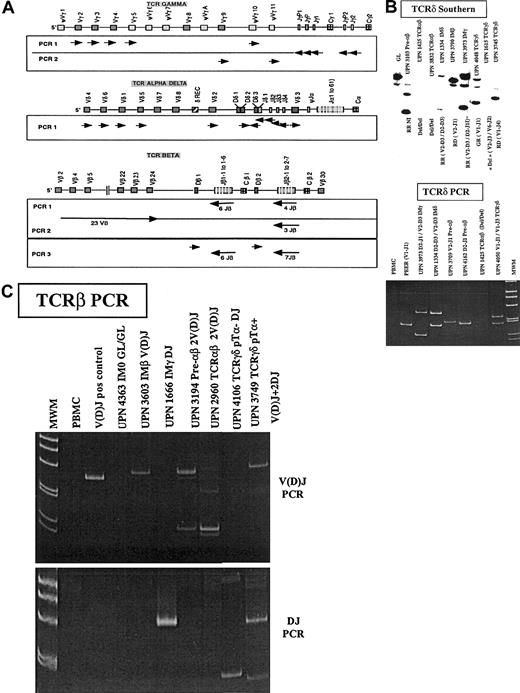
TCRαδ, TCRγ, and TCRβ loci and TCRδ and TCRβ profiles in T-ALL.
(A) Schematic representation (not to scale) of the TCRαδ, TCRγ, and TCRβ loci, after the IMGT (International ImMunoGeneTics) database http://imgt.cines.fr (initiator and coordinator, Marie-Paule le Franc, Montpellier, France).66 Orientation of the PCR primers are shown as arrows. (B) Representative Southern blot patterns obtained with a Jδ1 probe (top) and TCRδ heteroduplex PCR (bottom). GL indicates gemline; R, rearranged; Del, deleted. (C) Representative TCRβ PCR profiles. The immunophenotypic group is indicated for each patient, and the type of rearrangement is shown. TCRβ PCR detects V(D)J and DJ rearrangements.
TCRαδ, TCRγ, and TCRβ loci and TCRδ and TCRβ profiles in T-ALL.
(A) Schematic representation (not to scale) of the TCRαδ, TCRγ, and TCRβ loci, after the IMGT (International ImMunoGeneTics) database http://imgt.cines.fr (initiator and coordinator, Marie-Paule le Franc, Montpellier, France).66 Orientation of the PCR primers are shown as arrows. (B) Representative Southern blot patterns obtained with a Jδ1 probe (top) and TCRδ heteroduplex PCR (bottom). GL indicates gemline; R, rearranged; Del, deleted. (C) Representative TCRβ PCR profiles. The immunophenotypic group is indicated for each patient, and the type of rearrangement is shown. TCRβ PCR detects V(D)J and DJ rearrangements.
Immunophenotypic, transcriptional, and genotypic profiles in T-ALL
| . | sCD3− cTCRβ− . | sCD3− cTCRβ+ . | sCD3+ TCR− . | sCD3+TCR αβ+ . | sCD3+ TCR γδ+ . | Total . | |
|---|---|---|---|---|---|---|---|
| IM . | Pre-αβ . | . | TCRαβ DP . | TCRαβ SP/DN . | TCRγδ . | ||
| No. T-ALLs (%) | 33 (29) | 27 (24) | 9 (8) | 11 (10) | 11 (10) | 23 (20) | 114 |
| Phenotype | |||||||
| CD34 (%) | 22/33 (66) | 2/27 (7) | 3/9 (33) | 4/10 (40) | 4/11 (36) | 6/22 (27) | 41/112 (37) |
| CD117 (%) | 2/33 (6) | 0/23 (0) | 0/5 (0) | 0/9 (0) | 0/9 (0) | 0/16 (0) | 2/93 (2) |
| CD133-150 (%) | 9/33 (27) | 0/27 (0) | 0/9 (0) | 0/9 (0) | 1/9 (11) | 2/22 (9) | 12/109 (11) |
| CD333-150 (%) | 12/33 (36) | 1/26 (4) | 0/9 (0) | 0/9 (0) | 1/9 (11) | 2/21 (10) | 16/107 (15) |
| CD56 (%) | 9/26 (35) | 0/15 (0) | 0/6 (0) | 0/8 (0) | 0/8 (0) | 1/13 (8) | 10/76 (13) |
| CD5 (%) | 25/33 (75) | 26/27 (96) | 9/9 (100) | 11/11 (100) | 11/11 (100) | 22/23 (96) | 104/113 (92) |
| CD2 (%) | 16/33 (45) | 24/27 (89) | 7/9 (78) | 11/11 (100) | 9/11 (82) | 10/23 (43) | 77/114 (68) |
| CD4/8 DN (%) | 25/33 (75) | 1/27 (4) | 2/9 (22) | 0/11 (0) | 3/11 (27) | 7/22 (32) | 38/113 (33) |
| CD4 SP (%) | 5/33 (15) | 2/27 (7) | 1/9 (11) | 0/11 (0) | 6/11 (55) | 5/22 (23) | 19/113 (17) |
| CD8 SP (%) | 2/33 (6) | 5/27 (19) | 0/9 (0) | 0/11 (0) | 2/11 (18) | 3/22 (14) | 12/113 (11) |
| CD4/8 DP (%) | 1/33 (3) | 19/27 (70) | 6/9 (67) | 11/11 (100) | 0/11 (0) | 7/22 (32) | 44/113 (39) |
| CD1a (%) | 6/32 (19) | 23/27 (85) | 7/9 (78) | 5/11 (45) | 5/11 (45) | 6/23 (26) | 51/113 (45) |
| CD10 (%) | 8/32 (25) | 12/26 (46) | 2/9 (22) | 4/11 (36) | 2/11 (18) | 11/23 (48) | 39/112 (35) |
| TdT (%) | 21/29 (72) | 20/21 (95) | 6/6 (100) | 10/11 (91) | 7/10 (70) | 10/14 (71) | 74/91 (81) |
| cTCRβ (%) | 0/33 (0) | 27/27 (100) | 5/9 (56) | 7/7 (100) | 8/8 (100) | 2/15 (13) | 49/99 (50) |
| PTα/RAG-13-151 | |||||||
| pTα (%) | 6/31 (19) | 25/25 (100) | 7/9 (78) | 7/7 (100) | 5/9 (56) | 8/21 (38) | 58/102 (57) |
| RAG-1 (%) | 6/29 (21) | 22/22 (100) | 7/9 (78) | 5/5 (100) | 6/8 (75) | 6/18 (33) | 52/91 (57) |
| TCRδ PCR and/or Southern3-152 | |||||||
| GL/GL cases (%) | 4/33 (12) | 0/233-153 (0) | 0/9 (0) | 0/10 (0) | 0/83-153 (0) | 0/223-153 (0) | 4/105 (5) |
| R/(R-G-Del) cases (%) | 27/33 (82) | 22/27 (81) | 8/9 (89) | 4/10 (40) | 2/11 (18) | 22/23 (96) | 83/113 (75) |
| Del/Del cases (%) | 2/33 (6) | 1/23 (4) | 1/9 (11) | 6/10 (60) | 6/8 (75) | 0/22 (0) | 16/105 (15) |
| VD or DD alleles (%) | 18/41 (44) | 1/19 (5) | 0/12 (0) | 0/10 (0) | 0/11 (0) | 2/35 (6) | 21/114 (19) |
| DJ alleles (%) | 16/41 (39) | 2/19 (11) | 1/12 (8) | 1/10 (10) | 0/11 (0) | 2/35 (6) | 21/114 (19) |
| V(D)J alleles (%) | 7/41 (17) | 16/19 (84) | 11/12 (92) | 3/10 (30) | 4/11 (36) | 31/35 (89) | 66/108 (61) |
| TCRγ PCR | |||||||
| Neg cases (%) | 12/33 (36) | 0/27 (0) | 0/9 (0) | 0/11 (0) | 0/11 (0) | 0/23 (0) | 12/114 (11) |
| Immature cases (3′V-5′J)3-155 (%) | 16/33 (49) | 15/27 (56) | 5/9 (56) | 6/11 (55) | 7/11 (64) | 15/23 (65) | 64/114 (56) |
| End-stage cases (5′V-3′J)3-155 (%) | 5/33 (15) | 12/27 (44) | 4/9 (44) | 5/11 (45) | 4/11 (36) | 8/23 (35) | 38/114 (33) |
| TCRβ PCR | |||||||
| Neg cases (%) | 18/33 (55) | 0/26 (0) | 0/8 (0) | 0/9 (0) | 0/10 (0) | 1/19 (5) | 19/104 (18) |
| Only DJ cases (%) | 7/33 (21) | 1/263-154 (4) | 2/8 (25) | 3/9 (33) | 0/10 (0) | 7/19 (37) | 20/104 (19) |
| At least one V(D)J case (%) | 8/33 (24) | 25/26 (96) | 6/8 (75) | 6/9 (67) | 10/10 (100) | 11/19 (58) | 65/104 (63) |
| . | sCD3− cTCRβ− . | sCD3− cTCRβ+ . | sCD3+ TCR− . | sCD3+TCR αβ+ . | sCD3+ TCR γδ+ . | Total . | |
|---|---|---|---|---|---|---|---|
| IM . | Pre-αβ . | . | TCRαβ DP . | TCRαβ SP/DN . | TCRγδ . | ||
| No. T-ALLs (%) | 33 (29) | 27 (24) | 9 (8) | 11 (10) | 11 (10) | 23 (20) | 114 |
| Phenotype | |||||||
| CD34 (%) | 22/33 (66) | 2/27 (7) | 3/9 (33) | 4/10 (40) | 4/11 (36) | 6/22 (27) | 41/112 (37) |
| CD117 (%) | 2/33 (6) | 0/23 (0) | 0/5 (0) | 0/9 (0) | 0/9 (0) | 0/16 (0) | 2/93 (2) |
| CD133-150 (%) | 9/33 (27) | 0/27 (0) | 0/9 (0) | 0/9 (0) | 1/9 (11) | 2/22 (9) | 12/109 (11) |
| CD333-150 (%) | 12/33 (36) | 1/26 (4) | 0/9 (0) | 0/9 (0) | 1/9 (11) | 2/21 (10) | 16/107 (15) |
| CD56 (%) | 9/26 (35) | 0/15 (0) | 0/6 (0) | 0/8 (0) | 0/8 (0) | 1/13 (8) | 10/76 (13) |
| CD5 (%) | 25/33 (75) | 26/27 (96) | 9/9 (100) | 11/11 (100) | 11/11 (100) | 22/23 (96) | 104/113 (92) |
| CD2 (%) | 16/33 (45) | 24/27 (89) | 7/9 (78) | 11/11 (100) | 9/11 (82) | 10/23 (43) | 77/114 (68) |
| CD4/8 DN (%) | 25/33 (75) | 1/27 (4) | 2/9 (22) | 0/11 (0) | 3/11 (27) | 7/22 (32) | 38/113 (33) |
| CD4 SP (%) | 5/33 (15) | 2/27 (7) | 1/9 (11) | 0/11 (0) | 6/11 (55) | 5/22 (23) | 19/113 (17) |
| CD8 SP (%) | 2/33 (6) | 5/27 (19) | 0/9 (0) | 0/11 (0) | 2/11 (18) | 3/22 (14) | 12/113 (11) |
| CD4/8 DP (%) | 1/33 (3) | 19/27 (70) | 6/9 (67) | 11/11 (100) | 0/11 (0) | 7/22 (32) | 44/113 (39) |
| CD1a (%) | 6/32 (19) | 23/27 (85) | 7/9 (78) | 5/11 (45) | 5/11 (45) | 6/23 (26) | 51/113 (45) |
| CD10 (%) | 8/32 (25) | 12/26 (46) | 2/9 (22) | 4/11 (36) | 2/11 (18) | 11/23 (48) | 39/112 (35) |
| TdT (%) | 21/29 (72) | 20/21 (95) | 6/6 (100) | 10/11 (91) | 7/10 (70) | 10/14 (71) | 74/91 (81) |
| cTCRβ (%) | 0/33 (0) | 27/27 (100) | 5/9 (56) | 7/7 (100) | 8/8 (100) | 2/15 (13) | 49/99 (50) |
| PTα/RAG-13-151 | |||||||
| pTα (%) | 6/31 (19) | 25/25 (100) | 7/9 (78) | 7/7 (100) | 5/9 (56) | 8/21 (38) | 58/102 (57) |
| RAG-1 (%) | 6/29 (21) | 22/22 (100) | 7/9 (78) | 5/5 (100) | 6/8 (75) | 6/18 (33) | 52/91 (57) |
| TCRδ PCR and/or Southern3-152 | |||||||
| GL/GL cases (%) | 4/33 (12) | 0/233-153 (0) | 0/9 (0) | 0/10 (0) | 0/83-153 (0) | 0/223-153 (0) | 4/105 (5) |
| R/(R-G-Del) cases (%) | 27/33 (82) | 22/27 (81) | 8/9 (89) | 4/10 (40) | 2/11 (18) | 22/23 (96) | 83/113 (75) |
| Del/Del cases (%) | 2/33 (6) | 1/23 (4) | 1/9 (11) | 6/10 (60) | 6/8 (75) | 0/22 (0) | 16/105 (15) |
| VD or DD alleles (%) | 18/41 (44) | 1/19 (5) | 0/12 (0) | 0/10 (0) | 0/11 (0) | 2/35 (6) | 21/114 (19) |
| DJ alleles (%) | 16/41 (39) | 2/19 (11) | 1/12 (8) | 1/10 (10) | 0/11 (0) | 2/35 (6) | 21/114 (19) |
| V(D)J alleles (%) | 7/41 (17) | 16/19 (84) | 11/12 (92) | 3/10 (30) | 4/11 (36) | 31/35 (89) | 66/108 (61) |
| TCRγ PCR | |||||||
| Neg cases (%) | 12/33 (36) | 0/27 (0) | 0/9 (0) | 0/11 (0) | 0/11 (0) | 0/23 (0) | 12/114 (11) |
| Immature cases (3′V-5′J)3-155 (%) | 16/33 (49) | 15/27 (56) | 5/9 (56) | 6/11 (55) | 7/11 (64) | 15/23 (65) | 64/114 (56) |
| End-stage cases (5′V-3′J)3-155 (%) | 5/33 (15) | 12/27 (44) | 4/9 (44) | 5/11 (45) | 4/11 (36) | 8/23 (35) | 38/114 (33) |
| TCRβ PCR | |||||||
| Neg cases (%) | 18/33 (55) | 0/26 (0) | 0/8 (0) | 0/9 (0) | 0/10 (0) | 1/19 (5) | 19/104 (18) |
| Only DJ cases (%) | 7/33 (21) | 1/263-154 (4) | 2/8 (25) | 3/9 (33) | 0/10 (0) | 7/19 (37) | 20/104 (19) |
| At least one V(D)J case (%) | 8/33 (24) | 25/26 (96) | 6/8 (75) | 6/9 (67) | 10/10 (100) | 11/19 (58) | 65/104 (63) |
Only 5 cases (4 IM, 1 TCRαβ) expressed CD13 and CD33.
Five cases were classified as noninformative for pTα and 16 for RAG-1, including 2 IM, 6 pre-αβ, 6 TCRαβ, and 4 TCRγδ. pTα and RAG-1 expression were closely correlated, with each expressed in 57% of cases overall; 47 expressed both transcripts, and 37 expressed neither. pTα expression in the absence of RAG-1 was only seen in 2 IM, and isolated RAG-1 expression was seen in 2 IMβ and 1 DN TCRαβ. pTα and RAG-1 negativity was restricted to IM, TCRγδ, and TCRαβ SP/DN T-ALLs.
TCRδ configurations refer to T-ALL cases unless specified as (rearranged) alleles. GL indicates germline; R, rearranged; Del, deleted. TCRδ rearrangement on at least one allele was seen by Southern blot analysis in 75% of cases. In total, it was possible to identify 108 TCRδ rearrangements in 85 T-ALLs. These were classified as non–T-restricted VD ior DD, immature DJ, or complete V(D)J rearrangements.
Four pre-αβ, 3 TCRαβ, and 1 TCRγδ were TCRδ multiplex PCR negative and were not analyzed by Southern blot, thus preventing distinction between a germline, bilateral deletion, and an unidentified rearranged profile.
Because the TCRγ locus does not contain D segments, we classified T-ALLs as demonstrating an immature TCRγ recombination if a rearrangement involving Vγ9, Vγ10, Vγ11, JP, and/or JP1/2 (3′Vγ-5′Jγ) was observed on at least one allele, and as demonstrating end-stage if only Vγf1-Jγ1/2 rearrangements (5′Vγ-3′Jγ) were observed. TCRγ rearrangement on at least one allele was seen in 89% of cases. Most rearranged TCRγ alleles involved the Vγf1-Jγ1/2 (60%), Vγ10, and Vγ11 pseudogenes (22%), and Vγ9 (18%). Twenty-six JP1/2 and 2 JP (Vγf1) rearrangements were identified.
These cases probably correspond to a failure of the Vβ family-specific primers to recognize the functional rearrangement, corresponding to a 9% false-negative rate.
Quantification of pTα and RAG-1 transcripts
RNA and cDNA quality was assessed by quantification of the Abl housekeeping gene on an ABI PRISM 7700 (Applied Biosystems), and samples with Ct values greater than 32 (threshold, 0.1) were considered uninterpretable. cDNA synthesis and real-time quantitative (RQ)–PCR were performed using conditions standardized within the Europe Against Cancer program (www.ifrjr.nord.univ-mrs.fr/mrd-leukemia). Amplification efficiency was assessed by the slope obtained from logarithmic dilutions of a positive cell line, HPB-ALL. The specificity of amplification was also assessed by polyacrylamide gel electrophoresis (PAGE) analysis. Results were normalized for RNA quality and amplifiability relative to Abl. pTα was amplified using exon 1 and 2 primers, and, as such, only full-length, pTαa transcripts were detected.29 Primers and probes used were as follows: Abl sense, TggAgATAACACTCTAAgCATAACTAAAggT; antisense, gATgTAgTTgCTTgggACCCA; probe, Fam-CCATTTTTggTTTgggCTTCACACCATT-Tamra; RAG1 sense, AGCCTGCTGAGCAAGGTACC; antisense, GAACTGAGTCCCAAGGTGGG; probe, Fam-AGCCAGCATGGCAGCCTCTTTCC-Tamra; pTα sense, TTGGGTGTCCAGCCCTACC; antisense, GCCATAGGTGAAGGCATCCA; probe, Fam-CAGCCGGCAATGGCAGTGCA-Tamra.
Results
TCR classification of T-ALL
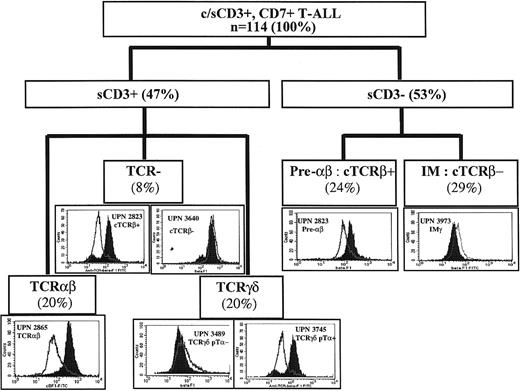
All 114 T-ALLs expressed CD7 and cytoplasmic (cCD3) or surface CD3 (sCD3). All were myeloperoxidase (MPO) negative by cytochemistry, immunophenotype, or both. All sCD3− cases and 39 of 54 sCD3+ cases were assessed for cytoplasmic TCRβ (cTCRβ) expression by flow cytometry, using the βF1 antibody (Figure 2). Fifty-three were also analyzed by immunocytochemistry, with no significant discrepancies. Absence of staining of nonpermeabilized T lymphocytes confirmed that only intracytoplasmic TCRβ was recognized.25 cTCRβ was seen in 50% of cases overall, including all TCRαβ T-ALLs tested, 45% of sCD3− cases, 13% of TCRγδ cases, and 56% of sCD3+ TCR− cases (Table 1). T-ALLs were subdivided on the basis of sCD3, TCRαβ, TCRγδ, and cTCRβ expression. sCD3+ cases included TCRαβ (20% overall), TCRγδ (20%) or TCR− (8%). Because the classification of sCD3+, TCR− cases requires an understanding of the CD3-associated receptor, these cases will not be described in detail here. sCD3− cases (53%) were divided into cTCRβ+ (pre-αβ) (24%) and cTCRβ−(immature [IM]) (29%). To identify progressive T-lymphoid lineage restriction, we classified the 33 IM T-ALLs on the basis of their TCR profiles into 4 IM0 with germline TCRδ, TCRγ, and TCRβ, 8 IMδ with only TCRδ rearrangement, 13 IMγ with TCRδ and TCRγ but absent or incomplete TCRβ rearrangement, and 8 IMβ with complete TCRβ rearrangement (Table 2).
Expression of cytoplasmic TCRβ in different subsets of T-ALL.
Isotype control on permeabilized cells is unshaded, and βF1 staining is in black. UPN indicates unique patient number. The proportion of cases in each category is shown in parentheses. c indicates cytoplasmic; s, surface.
Expression of cytoplasmic TCRβ in different subsets of T-ALL.
Isotype control on permeabilized cells is unshaded, and βF1 staining is in black. UPN indicates unique patient number. The proportion of cases in each category is shown in parentheses. c indicates cytoplasmic; s, surface.
Immunophenotypic, transcriptional, and genotypic profiles of immature and TCR-γδ lineage T-ALLs
| . | IM* . | Pre-αβ . | TCRγδ† . | ||||
|---|---|---|---|---|---|---|---|
| IM0 . | IMδ . | IMγ . | IMβ . | — . | pTα+ . | PTα− . | |
| TCRδ status | GL | R | R | R/Del | — | — | — |
| TCRγ status | GL | GL | R | R | — | — | — |
| TCRβ status | GL | GL/DJ | GL/DJ | V(D)J | — | — | — |
| T-ALLs | 4 | 8 | 13 | 8 | 27 | 8 | 13 |
| Phenotype | |||||||
| CD34, % | 100 | 75 | 46 | 75 | 7 | 50 | 17 |
| CD117, % | 0 | 13‡ | 0 | 13‡ | 0 | 0 | 0 |
| CD13/33, % | 75 | 63 | 62 | 25 | 4 | 38 | 8 |
| CD56, % | 25 | 50 | 40 | 33 | 0 | 17 | 0 |
| CD5, % | 75 | 63 | 77 | 100 | 96 | 100 | 100 |
| CD2, % | 75 | 50 | 38 | 63 | 89 | 38 | 53 |
| CD4/8 DN, % | 100 | 88 | 85 | 38 | 4 | 12 | 53 |
| CD4 SP, % | 0 | 0 | 15 | 38 | 7 | 25 | 25 |
| CD8 SP, % | 0 | 13 | 0 | 13 | 19 | 0 | 21 |
| CD4/8 DP, % | 0 | 0 | 0 | 13 | 70 | 63 | 8 |
| CD1a, % | 50 | 0 | 8 | 43 | 85 | 38 | 15 |
| CD10, % | 0 | 0 | 15 | 75 | 46 | 75 | 30 |
| TdT, % | 75 | 57 | 64 | 86 | 95 | 100 | 50 |
| CTCRβ, % | 0 | 0 | 0 | 0 | 100 | 25 | 0 |
| pTα/RAG-1 | |||||||
| RAG-1hi, % | 0 | 12 | 0 | 71 | 100 | 100 | 0 |
| PTα, % | 25 | 12 | 8 | 43 | 100 | — | — |
| TCRδ1-153 | |||||||
| Unidentified alleles | 0 | 1 | 0 | 2 | 11 | 2 | 0 |
| Identified alleles/T-ALL | 0 | 1.63 | 1.61 | 1 | 0.67 | 1.37 | 1.62 |
| Dδ2-Dδ3, % | — | 62 | 5 | 0 | 5 | 0 | 5 |
| Vδ2-Dδ3, % | — | 8 | 38 | 13 | 0 | 0 | 5 |
| Dδ2-Jδ1, % | — | 22 | 48 | 37 | 11 | 0 | 10 |
| Vδ1-Jδ1, % | — | 0 | 9 | 13 | 50 | 45 | 52 |
| Vδ(2-6)-Jδ1, % | — | 8 | 0 | 37 | 34 | 18 | 23 |
| Vδ(1-6)-Jδ(2-4), % | — | 0 | 0 | 0 | 0 | 37 | 5 |
| TCRγ | |||||||
| Identified alleles/T-ALL | 0 | 0 | 1.77 | 2 | 1.85 | 2 | 1.85 |
| Vf1-Jγ1/2, % | — | — | 26 | 75 | 66 | 75 | 54 |
| Vγ9-11/Jγ1/2, % | — | — | 52 | 19 | 20 | 25 | 25 |
| Vγ-JP/JP1/2, % | — | — | 22 | 6 | 14 | 0 | 21 |
| TCRβ | |||||||
| GL/GL, % | 100 | 75 | 69 | 0 | 0 | 0 | 10 |
| Identified alleles/T-ALL | 0 | 0.25 | 0.38 | 1.75 | 1.63 | 2.2 | 0.77 |
| DJ, % | — | 100 | 100 | 20 | 34 | 22 | 80 |
| V(D)J, % | — | 0 | 0 | 80 | 66 | 78 | 20 |
| . | IM* . | Pre-αβ . | TCRγδ† . | ||||
|---|---|---|---|---|---|---|---|
| IM0 . | IMδ . | IMγ . | IMβ . | — . | pTα+ . | PTα− . | |
| TCRδ status | GL | R | R | R/Del | — | — | — |
| TCRγ status | GL | GL | R | R | — | — | — |
| TCRβ status | GL | GL/DJ | GL/DJ | V(D)J | — | — | — |
| T-ALLs | 4 | 8 | 13 | 8 | 27 | 8 | 13 |
| Phenotype | |||||||
| CD34, % | 100 | 75 | 46 | 75 | 7 | 50 | 17 |
| CD117, % | 0 | 13‡ | 0 | 13‡ | 0 | 0 | 0 |
| CD13/33, % | 75 | 63 | 62 | 25 | 4 | 38 | 8 |
| CD56, % | 25 | 50 | 40 | 33 | 0 | 17 | 0 |
| CD5, % | 75 | 63 | 77 | 100 | 96 | 100 | 100 |
| CD2, % | 75 | 50 | 38 | 63 | 89 | 38 | 53 |
| CD4/8 DN, % | 100 | 88 | 85 | 38 | 4 | 12 | 53 |
| CD4 SP, % | 0 | 0 | 15 | 38 | 7 | 25 | 25 |
| CD8 SP, % | 0 | 13 | 0 | 13 | 19 | 0 | 21 |
| CD4/8 DP, % | 0 | 0 | 0 | 13 | 70 | 63 | 8 |
| CD1a, % | 50 | 0 | 8 | 43 | 85 | 38 | 15 |
| CD10, % | 0 | 0 | 15 | 75 | 46 | 75 | 30 |
| TdT, % | 75 | 57 | 64 | 86 | 95 | 100 | 50 |
| CTCRβ, % | 0 | 0 | 0 | 0 | 100 | 25 | 0 |
| pTα/RAG-1 | |||||||
| RAG-1hi, % | 0 | 12 | 0 | 71 | 100 | 100 | 0 |
| PTα, % | 25 | 12 | 8 | 43 | 100 | — | — |
| TCRδ1-153 | |||||||
| Unidentified alleles | 0 | 1 | 0 | 2 | 11 | 2 | 0 |
| Identified alleles/T-ALL | 0 | 1.63 | 1.61 | 1 | 0.67 | 1.37 | 1.62 |
| Dδ2-Dδ3, % | — | 62 | 5 | 0 | 5 | 0 | 5 |
| Vδ2-Dδ3, % | — | 8 | 38 | 13 | 0 | 0 | 5 |
| Dδ2-Jδ1, % | — | 22 | 48 | 37 | 11 | 0 | 10 |
| Vδ1-Jδ1, % | — | 0 | 9 | 13 | 50 | 45 | 52 |
| Vδ(2-6)-Jδ1, % | — | 8 | 0 | 37 | 34 | 18 | 23 |
| Vδ(1-6)-Jδ(2-4), % | — | 0 | 0 | 0 | 0 | 37 | 5 |
| TCRγ | |||||||
| Identified alleles/T-ALL | 0 | 0 | 1.77 | 2 | 1.85 | 2 | 1.85 |
| Vf1-Jγ1/2, % | — | — | 26 | 75 | 66 | 75 | 54 |
| Vγ9-11/Jγ1/2, % | — | — | 52 | 19 | 20 | 25 | 25 |
| Vγ-JP/JP1/2, % | — | — | 22 | 6 | 14 | 0 | 21 |
| TCRβ | |||||||
| GL/GL, % | 100 | 75 | 69 | 0 | 0 | 0 | 10 |
| Identified alleles/T-ALL | 0 | 0.25 | 0.38 | 1.75 | 1.63 | 2.2 | 0.77 |
| DJ, % | — | 100 | 100 | 20 | 34 | 22 | 80 |
| V(D)J, % | — | 0 | 0 | 80 | 66 | 78 | 20 |
Immature (IM) sCD3 cTCRβ-negative cases were divided into those demonstrating no TCR rearrangement (IM0), those with only TCRδ rearrangement (IM δ), those with TCRγ and TCRδ rearrangement (IMγ), and those with TCRβ V(D)J on at least one allele (IMβ). IMγ and IMδ demonstrated either no or only DJ TCRβ rearrangement.
TCRγδ were divided on the basis of pTα expression.
Both CD117+ cases were CD13/33 and, notably, CD34−.
TCR rearrangements are expressed as rearranged alleles. All IM cases were assessed for all 3 TCR loci. One TCRγδ negative by TCRδ PCR and not analyzed by Southern blot was presumed to have one unidentified rearrangement and one deleted allele. A significant proportion of, particularly pre-αβ, cases demonstrated Jδ1 rearrangements that were not detected by multiplex PCR and, as such, potentially represented Vα-Dδ3/Jδ1, δRec-Dδ3/Jδ1, transrearrangements, or translocations involving Dδ3 or Jδ1.
— indicates not applicable.
IM T-ALLs resemble multipotent thymic precursors
IM T-ALLs demonstrated a distinctive immunophenotype (Tables 2 and3). Expression of the CD13 and CD33 (myeloid) antigens or the CD56 (NK lineage) marker were frequent and relatively specific to this category, as was CD5 negativity. CD34 expression was more frequent in IM T-ALLs, though, surprisingly, it was maintained on 25% to 50% of TCR, but not pre-TCR, expressing T-ALLs. CD2 negativity was found in more than 50% but was rare in all other categories other than TCRγδ.30 31 Most were CD4/CD8 DN and CD1a−, although a proportion were CD4/CD8 SP. They included the only T-ALLs to have a germline configuration of TCRγ or TCRδ loci. Those rearrangements that did occur were frequently incomplete, involved pseudogenes such as Vγ10 or Vγ11, or corresponded to non-T-restricted TCRδ VD or DD rearrangements, often classified as illegitimate. Absence of cTCRβ expression therefore identifies an immature category of T-ALLs that could include expansions of nonlineage-restricted thymic precursors. IM0 T-ALLs expressed CD34/117, CD2, CD5, and TdT but not CD10. Two expressed CD1a and both CD13 and CD33—one in conjunction with pTα. The 8 IMδ differed immunophenotypically by frequent CD56 positivity, CD2 negativity, and absence of CD1a expression. The predominant TCRδ rearrangement was Dδ2-Dδ3. TCRβ rearrangements were rare and incomplete. All 12 TCRγ− T-ALLs were IM and included the 4 TCRδ germline cases, reflecting the fact that TCRδ rearrangement precedes TCRγ. Most immature T-ALLs belonged to the IMγ category. They demonstrated predominantly ongoing TCRγ rearrangements, with only one showing biallelic Vγf1-Jγ1/2. TCRδ rearrangements were mainly Dδ2-Jδ1 or Vδ2-Dδ3, and complete V(D)J was rare. TCRβ rearrangements were more frequent than in IMδ but, by definition, remained incomplete. In keeping with a later stage of maturation arrest, half were CD34/117 or CD13/33 negative, and fewer than half expressed CD1a or CD10. As with IMδ, CD2 negativity and CD56 positivity were frequent. Absence of TCRβ rearrangement was virtually restricted to IM0, IMδ, and IMγ cases. IM T-ALLs, which had undergone complete TCRβ rearrangement (IMβ) on at least one allele, were phenotypically and genotypically similar to pre-αβ T-ALLs. Half the TCRδ rearrangements were complete, and one case had undergone biallelic TCRδ deletion. TCRγ rearrangements were predominantly biallelic Vγf1-Jγ1/2 and 80% of TCRβ rearranged alleles were complete. Most expressed CD4/CD8, and CD1a/CD10 expression was frequent. These cases presumably correspond either to out-of-frame TCRβ rearrangements or to maturation arrest just after the completion of rearrangement before detectable protein expression. Most IM0, IMδ, and IMγ T-ALLs were pTα and RAG-1 negative (Figure3), with only 1 IMδ expressing both weakly. The level of pTα and RAG-1 expression increased markedly in IMβ.
Individual details of IM T-ALLs
| UPN . | Type . | TCR configuration . | Immunophenotype . | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stn Jδ1 . | PCR δ . | δ Allele 1 . | δ Allele 2 . | γ Allele 1 . | γ Allele 2 . | PCR β . | CD34 . | CD13 . | CD33 . | CD56 . | CD2 . | CD5 . | CD4 . | CD8 . | CD1a . | CD10 . | TdT . | ||
| 1147 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Neg | Neg | Pos | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 546 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Pos | Neg | Neg | Neg | Neg | Neg | Pos |
| 281 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Pos | Pos | Neg | Neg | Pos | Neg | Pos |
| 2586 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Neg | Pos | Neg | Neg | Pos | Neg | Pos |
| 1978 | IMδ | ND | 1P | D2-D3 | ND | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Neg | Pos | Neg | Neg | Neg | ND | Pos |
| 3954 | IMδ | GRR | 2P | V2-J1 | D2-J1 | Neg | Neg | Neg | Pos | Pos | Neg | Pos | Neg | Pos | Neg | Neg | Neg | ND | Pos |
| 1334 | IMδ | RR | 2P | V2-D3 | D2-D3 | Neg | Neg | Neg | Pos | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Neg | Neg |
| 3776 | IMδ | RR | 2P | D2-D3 | D2-D3 | Neg | Neg | 1 DJ | Neg* | Neg | Neg | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 2889 | IMδ | Gr | Neg | NI | GL | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 738 | IMδ | GG | 2P | D2-J1 | D2-J1 | Neg | Neg | 1 DJ | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 3594 | IMδ | ND | 2P | D2-D3 | D2-D3 | Neg | Neg | Neg | Pos | Neg | Pos | ND | Pos | Pos | Neg | Neg | Neg | Neg | ND |
| 4156 | IMδ | ND | 2P | D2-D3 | D2-D3 | Neg | Neg | Neg | Pos | Neg | Neg | ND | Pos | Pos | Neg | Pos | Neg | Neg | Pos |
| 3188 | IMγ | RR | 2P | D2-J1 | D2-J1 | Vf1-J1/2 | Vf1-JP1/2 | Neg | Neg | Neg | Neg | ND | Neg | Pos | Neg | Neg | Neg | Neg | Pos |
| 1439 | IMγ | RRr | 2P | V2-D3 | D2-J1 | Vf1-J1/2 | V11-J1/2 | Neg | Pos | Neg | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 1488 | IMγ | GR | 1P | D2-J1 | GL | V11-J1/2 | Neg | Neg | Pos | Neg | Pos | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Neg |
| 4161 | IMγ | ND | 1P | D2-J1 | ND | Vf1-J1/2 | V10-J1/2 | Neg | Neg | Neg | Neg | ND | Pos | Pos | Neg | Neg | Neg | Neg | ND |
| 634 | IMγ | RR | 2P | V2-D3 | D2-D3 | Vf1-JP | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Pos | Neg | Neg | Neg | Pos | Pos |
| 1666 | IMγ | RR | 2P | V1-J1 | D2-J1 | V10-J1/2 | V10-J1/2 | 1 DJ | Neg | Pos | Pos | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Pos |
| 160 | IMγ | ND | 1P | D2-J1 | ND | V10-J1/2 | Vf1-J1/2 | Neg | Pos | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 4158 | IMγ | ND | 2P | V2-D3 | D2-J1 | V9-J1/2 | V9-J1/2 | Neg | Neg | Neg | Neg | ND | Neg | Neg | Neg | Neg | Neg | Neg | ND |
| 3973 | IMγ | RR | 2P | D2-J1 | V2-D3 | V10-JP1/2 | V11-JP1/2 | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 226 | IMγ | RR | 1P | V2-D3 | V2-D3 | V10-JP1/2 | V9-J1/2 | 1 DJ | Pos | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Pos | Pos |
| 1434 | IMγ | DD | 1P | V2-V3 | Del | Vf1-J1/2 | Vf1-J1/2 | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Neg | Pos | Neg | Pos |
| 3891 | IMγ | ND | 1P | D2-J1 | ND | V11-J1/2 | Neg | 2 DJ | Neg | Neg | Pos | Neg | Neg | Pos | Neg | Neg | Neg | Neg | Neg |
| 2289 | IMγ | RR | 2P | V1-J1 | V2-D3 | V10-J1/2 | V9-J1/2 | 1 DJ | Pos | Neg | Pos | Pos | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 844 | IMβ | RD | Neg | NI | Del | Vf1-J1/2 | V11-J1/2 | 1 V(D)J | Pos | Neg | Neg | Pos | Neg | Pos | Neg | Neg | Neg | Pos | Pos |
| 3790 | IMβ | RD | 1P | V2-J1 | Del | Vf1-J1/2 | V10-J1/2 | 1 V(D)J | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Pos | Neg | Pos |
| 1029 | IMβ | RR | 2P | V2-D3 | D2-J1 | Vf1-J1/2 | V11-JP1/2 | V(D)J+2 DJ | Pos | Neg | Neg | Pos | Neg | Pos | Neg | Neg | ND | Pos | Neg |
| 3603 | IMβ | ND | 1P | D2-J1 | ND | Vf1-J1/2 | V10-J1/2 | 2 V(D)J | Pos | Pos | Neg | ND | Pos | Pos | Pos | Neg | Pos | Pos | ND |
| 1769 | IMβ | GR | 1P | V1-J1 | GL | Vf1-J1/2 | Vf1-J1/2 | 2 V(D)J | Neg* | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Pos | Pos | Pos |
| 1456 | IMβ | RR g | 2P | V6-J1 | D2-J1 | Vf1-J1/2 | Vf1-J1/2 | 2 V(D)J | Pos | Pos | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Pos |
| 1435 | IMβ | DD | Neg | Del | Del | Vf1-J1/2 | Vf1-J1/2 | 1 V(D)J | Pos | Neg | Neg | Neg | Pos | Pos | Neg | Pos | Neg | Pos | Pos |
| 1021 | IMβ | RR g | 1P | V5-J1 | NI | Vf1-J1/2 | Vf1-J1/2 | V(D)J+1 DJ | Pos | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Pos | Pos |
| UPN . | Type . | TCR configuration . | Immunophenotype . | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stn Jδ1 . | PCR δ . | δ Allele 1 . | δ Allele 2 . | γ Allele 1 . | γ Allele 2 . | PCR β . | CD34 . | CD13 . | CD33 . | CD56 . | CD2 . | CD5 . | CD4 . | CD8 . | CD1a . | CD10 . | TdT . | ||
| 1147 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Neg | Neg | Pos | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 546 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Pos | Neg | Neg | Neg | Neg | Neg | Pos |
| 281 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Pos | Pos | Neg | Neg | Pos | Neg | Pos |
| 2586 | IM0 | GG | Neg | GL | GL | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Neg | Pos | Neg | Neg | Pos | Neg | Pos |
| 1978 | IMδ | ND | 1P | D2-D3 | ND | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Neg | Pos | Neg | Neg | Neg | ND | Pos |
| 3954 | IMδ | GRR | 2P | V2-J1 | D2-J1 | Neg | Neg | Neg | Pos | Pos | Neg | Pos | Neg | Pos | Neg | Neg | Neg | ND | Pos |
| 1334 | IMδ | RR | 2P | V2-D3 | D2-D3 | Neg | Neg | Neg | Pos | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Neg | Neg |
| 3776 | IMδ | RR | 2P | D2-D3 | D2-D3 | Neg | Neg | 1 DJ | Neg* | Neg | Neg | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 2889 | IMδ | Gr | Neg | NI | GL | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 738 | IMδ | GG | 2P | D2-J1 | D2-J1 | Neg | Neg | 1 DJ | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 3594 | IMδ | ND | 2P | D2-D3 | D2-D3 | Neg | Neg | Neg | Pos | Neg | Pos | ND | Pos | Pos | Neg | Neg | Neg | Neg | ND |
| 4156 | IMδ | ND | 2P | D2-D3 | D2-D3 | Neg | Neg | Neg | Pos | Neg | Neg | ND | Pos | Pos | Neg | Pos | Neg | Neg | Pos |
| 3188 | IMγ | RR | 2P | D2-J1 | D2-J1 | Vf1-J1/2 | Vf1-JP1/2 | Neg | Neg | Neg | Neg | ND | Neg | Pos | Neg | Neg | Neg | Neg | Pos |
| 1439 | IMγ | RRr | 2P | V2-D3 | D2-J1 | Vf1-J1/2 | V11-J1/2 | Neg | Pos | Neg | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 1488 | IMγ | GR | 1P | D2-J1 | GL | V11-J1/2 | Neg | Neg | Pos | Neg | Pos | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Neg |
| 4161 | IMγ | ND | 1P | D2-J1 | ND | Vf1-J1/2 | V10-J1/2 | Neg | Neg | Neg | Neg | ND | Pos | Pos | Neg | Neg | Neg | Neg | ND |
| 634 | IMγ | RR | 2P | V2-D3 | D2-D3 | Vf1-JP | Neg | Neg | Neg | Neg | Neg | Neg | Neg | Pos | Neg | Neg | Neg | Pos | Pos |
| 1666 | IMγ | RR | 2P | V1-J1 | D2-J1 | V10-J1/2 | V10-J1/2 | 1 DJ | Neg | Pos | Pos | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Pos |
| 160 | IMγ | ND | 1P | D2-J1 | ND | V10-J1/2 | Vf1-J1/2 | Neg | Pos | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 4158 | IMγ | ND | 2P | V2-D3 | D2-J1 | V9-J1/2 | V9-J1/2 | Neg | Neg | Neg | Neg | ND | Neg | Neg | Neg | Neg | Neg | Neg | ND |
| 3973 | IMγ | RR | 2P | D2-J1 | V2-D3 | V10-JP1/2 | V11-JP1/2 | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Neg | Neg | Neg | Neg |
| 226 | IMγ | RR | 1P | V2-D3 | V2-D3 | V10-JP1/2 | V9-J1/2 | 1 DJ | Pos | Neg | Pos | Neg | Pos | Pos | Neg | Neg | Neg | Pos | Pos |
| 1434 | IMγ | DD | 1P | V2-V3 | Del | Vf1-J1/2 | Vf1-J1/2 | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Neg | Pos | Neg | Pos |
| 3891 | IMγ | ND | 1P | D2-J1 | ND | V11-J1/2 | Neg | 2 DJ | Neg | Neg | Pos | Neg | Neg | Pos | Neg | Neg | Neg | Neg | Neg |
| 2289 | IMγ | RR | 2P | V1-J1 | V2-D3 | V10-J1/2 | V9-J1/2 | 1 DJ | Pos | Neg | Pos | Pos | Pos | Pos | Neg | Neg | Neg | Neg | Pos |
| 844 | IMβ | RD | Neg | NI | Del | Vf1-J1/2 | V11-J1/2 | 1 V(D)J | Pos | Neg | Neg | Pos | Neg | Pos | Neg | Neg | Neg | Pos | Pos |
| 3790 | IMβ | RD | 1P | V2-J1 | Del | Vf1-J1/2 | V10-J1/2 | 1 V(D)J | Neg | Neg | Neg | Neg | Pos | Pos | Pos | Pos | Pos | Neg | Pos |
| 1029 | IMβ | RR | 2P | V2-D3 | D2-J1 | Vf1-J1/2 | V11-JP1/2 | V(D)J+2 DJ | Pos | Neg | Neg | Pos | Neg | Pos | Neg | Neg | ND | Pos | Neg |
| 3603 | IMβ | ND | 1P | D2-J1 | ND | Vf1-J1/2 | V10-J1/2 | 2 V(D)J | Pos | Pos | Neg | ND | Pos | Pos | Pos | Neg | Pos | Pos | ND |
| 1769 | IMβ | GR | 1P | V1-J1 | GL | Vf1-J1/2 | Vf1-J1/2 | 2 V(D)J | Neg* | Neg | Neg | Neg | Pos | Pos | Pos | Neg | Pos | Pos | Pos |
| 1456 | IMβ | RR g | 2P | V6-J1 | D2-J1 | Vf1-J1/2 | Vf1-J1/2 | 2 V(D)J | Pos | Pos | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Pos |
| 1435 | IMβ | DD | Neg | Del | Del | Vf1-J1/2 | Vf1-J1/2 | 1 V(D)J | Pos | Neg | Neg | Neg | Pos | Pos | Neg | Pos | Neg | Pos | Pos |
| 1021 | IMβ | RR g | 1P | V5-J1 | NI | Vf1-J1/2 | Vf1-J1/2 | V(D)J+1 DJ | Pos | Neg | Neg | Neg | Pos | Pos | Neg | Neg | Neg | Pos | Pos |
CD117+.
Full details of non-IM T-ALLs are available on request toelizabeth.macintyre@nck.ap-hop-paris.fr.
δ, γ, and β refer to TCR rearrangements. ND indicates not done; NI, not informative; Del, deleted; Stn, Southern.
RQ-PCR analysis of pTα and RAG-1 transcripts.
RAG-1 (shaded triangles) and pTα (shaded circles) expression in T-ALLs, acute myeloid leukemia (AML), healthy circulating PBMCs, and bone marrow (BM) controls. The HPB-ALL cell line expressed high-level pTα and RAG-1 and was arbitrarily attributed a value of 100%; all results were expressed relative to this value. Linearity of HPB-ALL quantitation for both transcripts extended over 5 logs. Low levels of pTα expression, 2 to 3 logs below HPB-ALL, were seen in normal PBMCs and most normal bone marrow samples, with a mean ± SD for all 15 samples of 0.55% ± 0.5%. AML samples expressed lower levels. RAG-1 transcripts were virtually undetectable in PBMCs and were at least 3 logs lower than HPB-ALL in the AML samples tested. Variable low-level expression (mean, 3.5% ± 4.5%) was seen in normal bone marrow because of the presence of lymphoid lineage precursors. pTα values at least 2 logs lower than HPB-ALL were considered negative. RAG1 values at least 2 logs lower than HPB-ALL were considered low/negative, bone marrow values between 1% and 15% were considered uninterpretable, and PB T-ALL values greater than 1% or BM T-ALLs greater than 15% (1.5 × 10−1) were considered positive. All results, including those considered uninterpretable, are shown. Mean value for each category is indicated as a horizontal bar.
RQ-PCR analysis of pTα and RAG-1 transcripts.
RAG-1 (shaded triangles) and pTα (shaded circles) expression in T-ALLs, acute myeloid leukemia (AML), healthy circulating PBMCs, and bone marrow (BM) controls. The HPB-ALL cell line expressed high-level pTα and RAG-1 and was arbitrarily attributed a value of 100%; all results were expressed relative to this value. Linearity of HPB-ALL quantitation for both transcripts extended over 5 logs. Low levels of pTα expression, 2 to 3 logs below HPB-ALL, were seen in normal PBMCs and most normal bone marrow samples, with a mean ± SD for all 15 samples of 0.55% ± 0.5%. AML samples expressed lower levels. RAG-1 transcripts were virtually undetectable in PBMCs and were at least 3 logs lower than HPB-ALL in the AML samples tested. Variable low-level expression (mean, 3.5% ± 4.5%) was seen in normal bone marrow because of the presence of lymphoid lineage precursors. pTα values at least 2 logs lower than HPB-ALL were considered negative. RAG1 values at least 2 logs lower than HPB-ALL were considered low/negative, bone marrow values between 1% and 15% were considered uninterpretable, and PB T-ALL values greater than 1% or BM T-ALLs greater than 15% (1.5 × 10−1) were considered positive. All results, including those considered uninterpretable, are shown. Mean value for each category is indicated as a horizontal bar.
Taken together, these data suggest that the classification of cTCRβ− T-ALLs on the basis of TCR configuration allows identification of progressively T-restricted precursors. They confirm that the order of human TCR rearrangements is TCRδ followed by TCRγ, just before TCRβ, and suggest that TCRδ rearrangement starts with Dδ2-Dδ3, followed by Dδ2-Jδ1. Jδ1 rearrangements initiate at the same stage as the earliest TCRγ rearrangements, whereas the switch from TCRβ DJ to V(D)J occurs at the same time as the transition from immature to end-stage TCRγ rearrangements, as pTα and RAG-1 transcripts appear.
Pre-αβT-ALL resemble pre-TCR-expressing precursors, and mature TCRαβ-expressing T-ALLs demonstrate a transitional DP/SP phenotype
The phenotype of pre-αβ T-ALL correlated with a cortical thymic precursor, insofar as most were CD4/8 DP and expressed CD1a. Expression of nonlineage-restricted markers such as CD34, CD13, CD33, and CD56 became rare or absent, whereas CD2 and CD5 were virtually universal, in keeping with a T-restricted stage of maturation arrest. In contrast to IM T-ALLs, no pre-αβ T-ALL was germline for δ, γ, or β TCR. Identified TCRδ rearrangements were V(D)J, and end-stage TCRγ Vf1-Jγ1/2 were frequent. Virtually all TCRαβ expressed CD5 and CD2, but CD1a expression was less frequent. All had deleted at least one TCRδ allele, and two thirds deleted both alleles. Biallelic TCRδ deletion was rare in all other categories. All pre-αβ cases were strongly positive for both RAG-1 and pTα transcripts (Figure 3). Both transcripts decreased in TCRαβ T-ALLs, particularly in SP/DN cases, where the only negative cases were observed.
Human pTα and RAG-1 transcripts therefore undergo closely coordinated regulation. They appear as immature T-ALLs complete TCRβ rearrangement, concurrent with the expression of CD4, CD8, or both, just before cTCRβ expression. Each is expressed by virtually all TCRαβ-lineage T-ALLs, other than a proportion of mature SP/DN cases. The highest levels of expression correspond to those cases undergoing or having just undergone β selection.
pTα− and pTα+TCRγδ-lineage T-ALLs are different
Surprisingly, 8 (38%) of 21 TCRγδ cases expressed pTα at levels only slightly lower than those observed in TCRαβ T-ALLs (Figure 3). High-level RAG-1 expression was observed in all pTα+ cases, compared with none of the pTα− cases (Table 2). cTCRβ expression was detected in 2 of 5 pTα+ cases but 0 of 6 pTα− cases and was restricted to the 2 cases with the highest levels of pTα. pTα+ cases were predominantly CD4/CD8 DP, CD1a+, CD10+, and TdT+, whereas pTα− cases were less frequently positive for these markers. CD34 and CD13/33 were also commoner in pTα+cases. pTα TCRγδ T-ALLs therefore resemble IMβ and pre-αβ cases.
All 8 pTα+ TCRγδ T-ALLs demonstrated TCRβ V(D)J rearrangements; these were often biallelic and were associated with a third DJ rearrangement in 2 cases. In contrast, pTα− cases demonstrated predominantly monoallelic DJ rearrangements. TCRγ JP1/2 and monoallelic rearrangements were restricted to pTα− TCRγδ+ cases, which were also more likely to demonstrate incomplete TCRδ VD, DD, or DJ, as if they had shut down TCRγ and TCRδ rearrangement. In contrast, pTα+ cases were more likely to have undergone downstream Jδ2-4 rearrangement or TCRδ deletion and end-stage TCRγ VfI-Jγ1/2, suggestive of continuing rearrangement.
TCRγδT-ALLs can therefore be divided into 2 distinct categories: (1) pTα RAG-1, and often TdT− cases that have undergone a form of TCRγ and TCRδ allelic exclusion and only partial TCRβ DJ rearrangement and (2) pTα+ cases that express a cortical phenotype and demonstrate ongoing TCRγ, TCRδ, and TCRβ rearrangements, associated with cTCRβ protein expression in a proportion. It is likely that the former correspond to malignant counterparts of classical TCRγδ lineage cells. The latter may well represent expansions of TCRαβ lineage cells that are selected on the basis of TCRγδ rather than pre-TCR expression.
Discussion
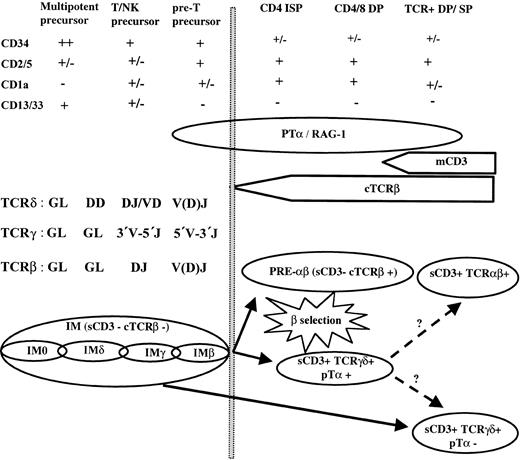
In this manuscript we have used TCR, pTα, and RAG-1 analysis to show that T-ALLs can further our understanding of the early stages of human T-lymphoid development (Figure4).
Human thymic T-lymphoid development.
Schematic representation of TCRαβ and TCRγδ lineage development and their leukemic equivalents.
Human thymic T-lymphoid development.
Schematic representation of TCRαβ and TCRγδ lineage development and their leukemic equivalents.
Half of TCRαβ lineage T-ALLs express pre-TCR
Cases belonging to the TCRαβ lineage were equally divided into those expressing TCRαβ and those expressing pre-αβ, which expressed cTCRβ in the absence of a classical TCR. Both demonstrated a CD2+, CD5+, CD1a+, CD4/8 DP > SP > DN phenotype, complete TCRβ and end-stage TCRγ rearrangements, and high-level expression of full-length pTα and RAG-1 transcripts in all but a proportion of TCRαβ SP/DN-negative cases. As such they correspond to the early ISP, DP, and SP stages of thymic development,32 when differentiation potential is restricted to the T lymphoid, and probably only the TCRαβ lineage. Several features of pre-αβ T-ALLs suggest that they correspond to cases that express the pre-TCR and are undergoing β selection. Surface pre-TCR expression is seen at the CD25+, CD44−, DN3 stage of murine development and at the CD4 ISP-to-DP transition during human development, when maximal levels of pTα transcripts are found. Flow cytometric detection of pre-TCR–associated sCD3 is, however, difficult because of the limited number of complexes expressed at the cell surface. Pre-TCR–expressing T-ALLs should therefore express high-level pTα, cTCRβ, and CD4 and/or CD8 but low- level or undetectable sCD3. They should have undergone TCRγ, TCRδ, and TCRβ but not TCRα rearrangement. Pre-αβ T-ALLs correspond exactly to this phenotype. Their main distinguishing feature from TCRαβ T-ALLs was a switch from TCRδ rearrangement to TCRδ deletion, reflecting TCRα rearrangement. One fourth of the TCRδ rearrangements detected by Southern blot analysis in pre-αβ T-ALLs were negative by TCRδ PCR (Table 2) and may thus include Vα-Jδ rearrangements. δRec rearrangements were also virtually restricted to this category (data not shown). Formal demonstration that pre-αβ T-ALLs express the pre-TCR will require analysis with pTα-specific antibodies. The frequency of pre-TCR–expressing T-ALLs is consistent with a population undergoing massive expansion. It is also possible that these precursors are particularly prone to leukemic transformation. In keeping with this, pTα expression is necessary for Notch-3-mediated33 and potentially also Notch-1–meditated34 leukemic transformation. Our data demonstrate that this is likely to result from the physiologic proliferative signal mediated by the pre-TCR in combination with a differentiation block related to Notch deregulation rather than from a specific oncogenic effect of pTα. T-ALLs that expressed sCD3 and cTCRβ but no detectable TCR could also potentially correspond to pre-TCR–expressing cases. Alternatively, they may express a TCRβδ35 or an unusual CD3-TCRαβ complex, thus preventing presentation of the CD3 epitope detected by BMA031. Full details of these cases will be published elsewhere.
Unlike murine development, in which cTCRβ and pTα expression have been identified in DN3 and DN4 thymocytes,36 we could not identify a significant number of pTα-expressing CD4/CD8 DN T-ALLs, consistent with previous evidence that pTα appears at the ISP/DP stage of human development.37 The immediate precursors of pre-TCR–expressing T-ALLs were the IMβ T-ALLs, most of which expressed CD4, CD8, or both. Human pTα in cCD3+ cells appears as the TCRβ rearrangement is completed and at the onset of CD4 or CD8 expression, immediately before cTCRβ expression. This discrepancy is likely to result from differences in the appearance of CD4 or CD8 and cTCRβ in healthy murine and human development.6 Our data demonstrate that human cTCRβ expression appears at the DP stage and identifies human T-lymphoid restriction.
TCRγδ-lineage T-ALLs
Given that healthy TCRγδ lymphocytes are not thought to express a pre-TCR or to undergo β selection, expression of pTα transcripts is not necessary for their maturation.38 We demonstrate that TCRγδ T-ALLs can be divided into 2 distinct categories based on their pTα expression; 60% are pTα− and resemble classical TCRγδ lymphocytes, whereas 40% are pTα+ and are likely to correspond to precursors that retain TCRαβ and TCRγδ potential. TCRγδ pTα− T-ALLs express few T-restricted markers apart from TCRγδ and CD5 (Table 2). TCR profiles show relatively frequent use of immature 3′ V segments and 5′ J segments. This may represent a form of allelic exclusion, whereby recombinase competence is shut down at an early stage, following the expression of functional TCRγδ. Hence, these T-ALLs are RAG-1 and often TdT negative, and they include the only TCRγδ T-ALLs with monoallelic TCRγ and incomplete TCRδ rearrangements. TCRβ rearrangements are restricted to predominantly monoallelic DJ, and none express cTCRβ protein. As such, they resemble healthy TCRγδ lymphocytes.13 Their denomination as immature acute leukemias is based on their clinical presentation and the paucity of recognized mature T-cell markers. In contrast, pTα+ TCRγδ T-ALLs are predominantly DP or CD4 ISP, CD1a+, and CD10+, and all express RAG-1 and TdT. In keeping with an active recombinase, all TCRγ are biallelic and predominantly end-stage, and all TCRδ rearrangements are complete, with relatively frequent use of downstream Jδ segments. TCRβ rearrangements are complete, biallelic, and unusually extensive. Maturation of murine DN precursors to the DP stage requires the expression of a pre-TCR. In the absence of either pTα or TCRβ, some, albeit inefficient, maturation to the DP stage is possible by replacement of the pre-TCR by TCRγδ15 It is possible that the pTα+ TCRγδ T-ALLs described here correspond to these cells, demonstrating for the first time that this pathway is also operational in at least leukemic human T-lymphoid development. The frequency of this category suggests either that these precursors are particularly susceptible to leukemic transformation or that they represent a relatively common developmental pathway. Genotypic comparison of pTα+ and pTα- TCRγδ T-ALL will help determine whether the former result from leukemic deregulation. Until recently, analysis of various TCR- and pTα-deficient mice suggested that the maturation of αβ lineage cells by TCRγδ expression was only operational in the absence of a pre-TCR.18,19 If this is also true for human T-ALLs, it is likely that it is the absence of TCRβ that prevents pre-TCR expression in pTα+ TCRγδ ALLs. Because all our cases expressed pTα+ transcripts at levels similar to DP TCRαβ-lineage cases, it is not the level of transcription that is the limiting factor. We cannot exclude abnormalities in pTα protein expression without further analysis, particularly the use of pTα-specific antibodies. In contrast, most did not express detectable cTCRβ protein, despite extensive ongoing TCRβ rearrangements. Absence of TCRβ is not, however, the limiting step in all cases, because the 2 TCRγδ T-ALLs with the highest level of pTα transcripts expressed cTCRβ protein on 56% and 77% of blasts. Gounari et al39 have recently demonstrated the expression of a pTα-driven reporter in TCRγδ thymocytes. They interpreted this as evidence of early TCRγδ cells that have recently derived from a pTα+ common αβ/γδ precursor. pTα+ TCRγδ T-ALLs were more frequently CD34, CD13/33, TdT, and RAG-1 positive. However, they demonstrated more frequent CD4/CD8, CD1a, and CD10 expression and more extensive TCR rearrangement. Our data suggest that pTα+ TCRγδ cells are intermediate between TCRαβ- and TCRγδ-lineage precursors and lead us to postulate that pTα+ TCRγδ precursors can differentiate into TCRαβ or TCRγδ mature lymphocytes (Figure 4). TCRγδ expression could precede or replace pre-TCR expression, even in the absence of any abnormalities of pTα or TCRβ. TCRγ and TCRδ analysis of pTα+ murine TCRγδ cells will clarify whether these cells are also closer to TCRαβ lineage precursors. The pTα+ TCRγδ T-ALLs described here will facilitate analysis of human TCRαβ versus TCRγδ lineage commitment, including analysis of their transcriptional profiles. Determination of their physiological relevance will require analysis of their healthy human counterparts.
Immature T-ALLs
We used the expression of CD7 and cCD3 in the absence of cTCRβ to identify approximately 30% of T-ALLs as immature. These are likely to include cases arrested at a non–T-restricted stage, because most were DN, pTα, and RAG-1 negative and expressed immature markers such as CD34, CD13, and CD33. The earliest human thymic precursor has an identical phenotype.40,41 CD117/c-kit expression is an important marker in murine DN42 and has been described in 11% of T-ALLs, particularly in immature cases that express myeloid antigens or isolated CD7.43 We found a lower level of CD117 expression, possibly because of differences in the evaluation of cCD3 expression, suggesting that CD117 plays a different role in the early stages of human and murine T lymphopoiesis.
Analysis of IM T-ALL TCR configurations allowed identification of progressively more T-restricted leukemic precursors. The order of TCR rearrangement was clearly (1) TCRδ Dδ2-Dδ3 (IMδ), (2) Dδ2-Jδ1 or Vδ2-Dδ3, immature TCRγ, and TCRβ DJ (IMγ), (3) end-stage TCRγ, TCRβ V(D)J, and TCRδ V(D)J or deletion (IMβ). This confirms previous analyses of human and murine early thymic development7 and suggests that the order of TCR rearrangement is not significantly disrupted by leukemic transformation. Complete TCRδ rearrangements were rare in all IM T-ALLs other than IMβ, implying that complete TCRδ rearrangement occurs immediately before TCRγδ or pre-TCR expression. IMβ T-ALLs were clearly the immediate precursors of pre-αβ T-ALLs and were virtually the only IM cases to express RAG-1 and pTα. If a pTα+ common αβ/γδ T-lymphoid–restricted sCD3-precursor exists,39 its leukemic equivalent is likely to be found in this subset. Only 4 T-ALLs were TCR germline (IM0), demonstrating that TCRδ rearrangements occur almost immediately after cCD3ε expression. Despite their immaturity, IM0 T-ALLs were TdT positive, demonstrating that TdT expression precedes TCR rearrangement, RAG-1, and pTα expression. Within murine development, cCD3ε expression does not occur until the DN2-to-DN3 transition, when TCR rearrangement starts.36 This implies that the human leukemic equivalent of DN1 is cCD3− and is likely to be classified as an immature acute myeloid leukemia (M0 AML).21 It is, of course, also possible that cCD3 appears earlier in human than in murine thymic development. Human, but not murine, NK precursors express cCD3.44-46
Two IM0 T-ALLs (UPN 281 and UPN 2586) were potentially DC precursors, based on a unique CD34+, CD13/33 DP, CD1a+, CD4/8 DN profile, as described for myeloid DC1 precursors.47 This population had recently been identified in the human postnatal thymus.48 Interestingly, one expressed pTα+, as previously described for DC2, but not DC1 precursors.49 A leukemic DC2 equivalent has been identified by CD56 and CD4 expression in the absence of cCD3. These leukemias express pTα50 By definition, no such cases were included in the present series, and all our CD56+cases were CD4−.
CD56 is often used to indicate NK potential, as may be a CD5−/CD2+ phenotype and absence of TCR rearrangement.51,52 Most (6 of 9) CD56+ IM T-ALLs, however, expressed CD5, and all but one CD2+/CD5+ IM0 (UPN 1147) had undergone TCR rearrangement. Only 1 IMδ was CD5−/CD2+ (UPN 1334). CD56 expression may be absent on early NK precursors.53,54 Our data suggest that CD56 is likely to identify a nonlineage-restricted lymphoid precursor that down-regulates CD56 during TCRαβ and TCRγδ but not NK maturation. It is also possible that CD56+ TCR-rearranged T-ALLs correspond to expansions of T/NK cells.55
Progressive restriction of a human multipotent (T/DC/NK) precursor with a CD5−, CD1a− phenotype, followed by a CD5+, CD1a− T/NK precursor and, finally, T-restricted potential at the CD5+, CD1a+ stage of CD34+, DN thymocyte development has been described.6,32 Six IM T-ALLs were CD5−, CD1a−, CD34/CD117+ and demonstrated an extremely immature phenotype, consistent with multipotent precursors. All were TdT−; only 2 expressed CD2, but 3 were CD56+, and 4 were CD13/33+. In contrast to previous suggestions that CD5−, CD1a− thymic precursors have not yet started TCR rearrangement,7 all but one had undergone at least partial TCRδ rearrangement. Six IM T-ALLs were CD1a+, but only the 2 potential DC precursors were CD34/117+, CD5+, DN and, as such, did not correspond to a T-restricted precursor. We were unable to identify any CD5+, CD1a+, TCR-rearranged DN T-ALLs. These data suggest that, within cTCRβ− T-ALLs, CD1a expression identifies either a non–T-lymphoid precursor or the previously described ISP immediate precursor of TCRαβ lineage DP cells about to undergo β selection.
A human common lymphoid precursor (CLP) has been identified in adult bone marrow by its CD34+, CD45RA+, CD10+, Lin− phenotype.56 Eight IM T-ALLs expressed CD10, but all had undergone TCRγ and TCRδ rearrangement, and most demonstrated TCRβ VJ, in keeping with T-restricted precursors. Conversely, no CD10+ IM0 or IMδ were identified, demonstrating that, if a CLP acute leukemia exists, it must be cCD3−. This is in keeping with the fact that the aforementioned CD56+, CD4+ CLP leukemias are cCD3−, although 60% express CD7.50 In contrast, evidence for T-myeloid potential within cTCRβ− IM T-ALLs was clear. More than 50% expressed CD33, CD13, or CD117. Their classification as T-ALL rather than AML is justified by MPO negativity and cCD3 positivity in all cases, CD2 or CD5 positivity in most cases, and extensive TCR rearrangement. Our data confirm the separation of B- and T-lineage development before the loss of myeloid potential, as recently proposed,57 rather than the initial separation of a CLP from a myeloid precursor.
Twenty-three immature T-ALLs were RAG-1lo/neg, including all but one IMδ and IMγ (Table 2). None had undergone end-stage TCRγ or complete TCRβ V(D)J, and only 2 of 30 TCRδ rearrangements were complete, demonstrating that relatively high levels of RAG-1 are necessary for TCRβ and TCRδ V(D)J rearrangements and complete opening of the TCRγ locus. Alternatively, other regulatory elements may be missing in these T-ALLs, as suggested by the fact that RAG-1 and RAG-2, even in combination with E2A, are insufficient for the induction of TCRδ V(D)J, and indeed DJ, in nonlymphoid cells58 and that Jδ1 rearrangement requires TCR-specific transcription factors.59 These regulatory elements are more likely to be expressed by IMγ than IMδ T-ALLs because only the former undergo Jδ1 rearrangement. Comparison of their transcriptional profiles is therefore likely to provide useful information regarding the transcription factors implicated in the regulation of TCRδ rearrangement.
Conversely, immature rearrangements, predominantly of TCRδ and TCRγ, must have occurred during an earlier stage of RAG-1 expression or must be able to occur in the presence of very low levels, considered as RAG-1lo/neg in the present study (Figure 3). Differential dependence on RAG-1 expression for TCRγ and TCRδ compared with TCRβ has already been described.60High-level RAG-1 expression was not, however, seen in immunoglobulin (Ig)– or TCR-rearranged AML,61 and only 2 waves of RAG-1 expression for TCRβ and TCRα rearrangement have been described.62 We have not yet analyzed RAG-2 transcripts in T-ALL, but they are thought to be coordinately expressed to RAG-1, with the possible exception of murine early DN3.14It is also possible, although unlikely, that RAG-1 and pTα are (coordinately) down-regulated in IMδ and IMγ T-ALLs as part of the oncogenic process. pTα expression is inhibited by TAL1/SCL expression,63 a known T-ALL oncogene. TAL1/SCL deregulation, however, occurs virtually exclusively in pre-αβ and TCRαβ T-ALLs.28 64
We have therefore shown that cCD3+ T-ALLs reflect all stages of human T-lymphoid development and can provide useful homogeneous populations arrested at different stages of development. Not all features of T-ALL can be explained on this basis—for example, the expression of CD34 by 25% to 50% of mature TCRαβ/γδ cases. This is not because of delayed down-regulation, given that only 7% of pre-αβ T-ALLs are CD34+. Appropriate classification, as proposed here, will facilitate the separation of physiological and leukemogenic profiles. Analysis of the genotype and the transcriptional profile of T-ALLs classified on this basis will clarify the mechanisms leading to leukemic transformation and our understanding of normal lymphopoiesis.65
We thank C. Bayle (Institut Gustave-Roussy, Villejuif), F-X. Mahon and C. Bilhou-Nabera (Bordeaux), X. Troussard (Caen), M. Dupont (Montpellier), R. Garand (Nantes), E. Kuhlein and N. Dastugue (Toulouse) and their clinical colleagues for providing T-ALL samples, Jean-Pierre de Villartay and Orly Azogui for constructive criticism, and Dorothée Menage for secretarial assistance. We also thank all members of the Biomed-2 Concerted Action who contributed to the design of the TCRδ and TCRβ multiplex PCR, particularly Louise Lavender and John Smith (Southampton), Monika Bruggeman and Michael Kneba (Kiel), and Ton Langerak and Jacques van Dongen (Rotterdam).
Prepublished online as Blood First Edition Paper, November 21, 2002; DOI 10.1182/blood-2002-08-2438.
Supported by the Fondation Contre la Leucémie de la Fondation de France, l'Association de la Recherche sur le Cancer (ARC), the Direction de Recherche Clinique de l'Assistance Publique-Hôpitaux de Paris (PHRC 97-106), and the Biomed-2 BMH4-CT98-3936 Concerted Action.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Elizabeth Macintyre, Laboratoire d'Hématologie, Tour Pasteur, Hôpital Necker, 149-161, rue de Sèvres, 75743 Paris cedex 15, France; e-mail:elizabeth.macintyre@nck.ap-hop-paris.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal