Abstract
Plasminogen activator inhibitor 1 (PAI-1) is the main fibrinolysis inhibitor, and high plasma levels are associated with an increased risk for vascular diseases. Inflammatory cytokines regulate PAI-1 through a hitherto unclear mechanism. Using reporter gene analysis, we could identify a region in the PAI-1 promoter that contributes to basal expression as well as to tumor necrosis factor α (TNFα) induction of PAI-1 in endothelial cells. Using this region as bait in a genetic screen, we could identify Nur77 (NAK-1, TR3, NR4A1) as an inducible DNA-binding protein that binds specifically to the PAI-1 promoter. Nur77 drives transcription of PAI-1 through direct binding to an NGFI-B responsive element (NBRE), indicating monomeric binding and a ligand-independent mechanism. Nur77, itself, is transcriptionally up-regulated by TNFα. High expression levels of Nur77 and its colocalization with PAI-1 in atherosclerotic tissues indicate that the described mechanism for PAI-1 regulation may also be operative in vivo.
Introduction
Plasminogen activator inhibitor 1 (PAI-1) is the main inhibitor of tissue-type plasminogen activators (t-PAs) and urokinase-type plasminogen activators (u-PAs) (reviewed in Binder et al1 and Chapman2). It functions as a serine protease inhibitor (SERPIN) by forming stable 1:1 stochiometric complexes3 with its target proteases that are removed by scavenger receptors.4 PAI-1 regulates intravascular fibrinolysis and tissue proteolysis, and thereby controls thrombus dissolution as well as invasion and migration of cells through the extracellular matrix. PAI-1 is secreted by several tissues, including the liver, adipose tissue, smooth muscle cells (SMCs), and endothelial cells (ECs). ECs produce low amounts of PAI-1 under normal resting conditions. PAI-1 is present only in low concentrations (6-80 ng/mL) in normal human plasma. Elevated levels of plasma PAI-1 found in several pathologic conditions are thought to increase the risk for vascular complications such as thrombosis5,6 or myocardial infarction.7Indeed, PAI-1 mRNA levels are increased in atherosclerotic vessels.8-10 Such local up-regulation of PAI-1 in vascular tissues might be caused by several mechanisms, including hypoxia,11 lipid mediators,12 and growth factors,13,14 as well as inflammatory cytokines. Atherosclerosis is indeed regarded as a disease with a significant inflammatory component (reviewed in Libby et al15), and PAI-1 expression is up-regulated by inflammatory mediators such as tumor necrosis factor α (TNFα),16interleukin-1 β (IL-1β),17,18 or lipopolysaccharide (LPS)19 in human endothelial cells and in the model cell line HepG2.17,18,20 In fact, PAI-1 levels are increased during sepsis,21 in which endothelial cells are a major source of this protein. Thus, PAI-1 is regarded as an “inflammatory response gene” that can alter the fibrinolytic potential of the vessel22 and might lead to a thrombotic tendency.
The PAI-1 promoter has been analyzed for the presence of transcription factor consensus binding sites possibly responsible for its inducible expression: best studied is the strong induction by TGF-β mediated by binding of SMAD 3 and 4 proteins to consensus sites in the PAI-1 promoter.23 SP1 elements in the proximal promoter region have been shown to mediate the response of PAI-1 to glucose24 and angiotensin II25; in addition, an AP-1–like binding site mediates the PAI-1 response to protein kinase C (PKC) and protein kinase A (PKA) signals26,27; and furthermore, a hypoxia response element (HRE) mediates the binding of HIF-1 and the response to hypoxia.11 The regulation of PAI-1 by inflammatory mediators is, however, still unclear. Nuclear factor–κB (NF-κB) is the major transcription factor translocated to the nucleus in response to inflammatory stimuli where it directly drives transcription of responsive genes such as E-selectin, VCAM-1, IL-8, tissue factor, and others.18 However, no NF-κB binding site could be identified in the PAI-1 promoter. Others, and we, have shown that the early growth response gene 1 (EGR-1) is another important transcription factor within the TNFα signaling cascade28,29 leading to increased PAI-1 synthesis.30 However, as in the case of NF-κB, no EGR-1 consensus site is present in the PAI-1 promoter, and it therefore remains unclear how PAI-1 is regulated during inflammation.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs, different batches nos. 91, 92, 181, 182, 183; the reactivity of the different batches of HUVECs toward stimulation by inflammatory cytokines varied as described previously31) were cultured as described.31 32 Cells were used for experiments up to passage 5. MCF-7 cells and JURKAT were cultured as recommended by American Type Culture Collection (ATCC, Manassas, VA).
Relative quantitative reverse transcriptase–polymerase chain reaction (Q-PCR)
RNA was isolated using Trizol (Invitrogen, Carlsbad, CA) reagent. Total RNA (900 ng) was reverse transcribed with MuLV-reverse transcriptase using the Gene Amp RNA PCR kit (Applied Biosystems, Foster City, CA) and oligo dT16 primers. The mRNA sequences of the investigated genes were obtained from GenBank. The primers for β-2 microglobulin were used as described by Wellman et al.33 The other primers were designed using the PRIMER3 software from the Whitehead Institute for Biomedical Research (Cambridge, MA). The following forward (F) and reverse (R) primers were used for PAI-1: F, 5′-CAGACCAAGAGCCTCTCCAC-3′; R, 5′-ATCACTTGGCCCATGAAAAG-3′; and for Nur77: F, 5′-CACCCACTTCTCCACACCTT-3′; R, 5′-ACTTGGCGTTTTTCTGCACT-3′. Q-PCR was performed by LightCycler technology using the Fast Start SYBR Green I kit for amplification and detection (Roche Diagnostics, Basel, Switzerland). In all assays, cDNA was amplified using a standardized program (10′ denaturing step and 55 cycles of 5′ at 95°C; 15′ at 65°C, and 15′ at 72°C; melting point analysis in 0.1°C steps; final cooling step). Each LightCycler capillary was loaded with l.5 μL DNA Master Mix; l.8 μL MgCl2 (25 mM); 10.1 μL H2O; and 0.4 μL of each primer (10 μM). The final amount of cDNA per reaction corresponded to 2.5 ng total RNA used for reverse transcription. Relative quantification of target gene expression was performed using a mathematical model by Pfaffl.34 The expression of the target molecule was normalized to the expression of β-2 microglobulin.
Nuclear run-on assay
Nuclear run-on assays were performed as described.35 Nuclei were isolated from HUVECs that were either untreated or treated for 4 hours with 100 U/mL TNFα. The run-on reaction contained 1.5 × 107 isolated nuclei and 250 μCi (9.25 MBq) [α32P] UTP for incorporation into the nascent pre-mRNA chains. PAI-1 and glyceraldehyde phosphate dehydrogenase (GAPDH) PCR product (1 μg of each) was applied to Hybond-N (Amersham Biosciences, Piscataway, NJ) nylon membranes using a slot blot apparatus followed by ultraviolet (UV) crosslinking. The labeled RNA was isolated from the nuclei using Trizol reagent, washed, and resuspended in DEPC H2O. Of each labeled RNA, 1.5 million cpm was used for hybridization, and the membranes were exposed on Phosphoimager plates (Amersham) for 3 days.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays were performed as described.32 Confluent HUVECs were stimulated with 100 U/mL TNFα for 2 hours. JURKAT were induced with phorbol myristate acetate (PMA, 10 ng/mL) and ionomycin (0.5 μg/mL) for 2 hours, controls were induced with vehicle (1.1% dimethyl sulfoxide [DMSO]). For the electrophoretic mobility shift assay, 5 μg nuclear extracts of HUVECs were treated as indicated and incubated at room temperature for 30 minutes, with 5 × 106 cpm/mL 32P (Amersham) of labeled oligonucleotides representing the nucleotide (nt) −270 to −250 of the PAI-1 promoter, its single base mutations or the consensus NGFI-B responsive element (NBRE). As a control, a competition assay was performed by adding a 100-fold molar excess of unlabeled oligonucleotide to the reaction prior to the addition of the labeled probe. The rabbit anti Nur77 antibody E-20 (Santa Cruz Biotechnology, Santa Cruz, CA) that recognizes the C-terminal parts of Nur77 and Nurr1 was used for supershifting. The samples were separated on a 5% polyacrylamide gel. The gel was then dried and exposed to Phosphoimager plates (Amersham).
One-hybrid screen
Yeast one-hybrid screening was performed with the Matchmaker 1-Hybrid system (Clontech Laboratories, Palo Alto, CA) according to the manufacturer's recommendations. The “bait” was prepared by synthesizing 3 tandem copies of the nt −270 to −250 region that was subcloned into pHISi (Clontech). The resultant plasmids were sequenced for verification, transformed into the yeast strain YM4271, and selected for integration in the his3 locus. Using a library from activated lymphocytes (Clontech), 2 million transformants were obtained. There were 5 clearly HIS-positive colonies, and 3 of these contained inserts coding for Nur77; the other 2 were false positives.
Transfection of HUVECs
Fragments of the human PAI-1 promoter (deletion series from nt −1520 to −80 ending at +20 and a −850 construct with deletion of the bases from −250 to −270 ΔNBRE]) were cloned into a luciferase expression vector, pUBT-luc.36 At 24 hours before transfection, HUVECs were seeded in 6-well tissue-culture plates to reach 80% to 90% confluence the next morning. Transient transfections were performed by using the Lipofectamine Plus reagent (Invitrogen) according to the protocol. Cells were incubated with transfection mixture containing 1.5 μg DNA (including a cytomegalovirus [CMV]–beta-gal or CMV-renilla construct as internal control), 6 μL Plus reagent and 4 μL Lipofectamine in a total volume of 1 mL medium M-199 per well for 130 minutes. Induction with TNFα (100 U/mL) was performed the next day for 6 hours. Luciferase and beta-galactosidase assays were performed with cellular lysates of transfected cells as previously described.37 All experimental values were determined from duplicate wells and were performed at least twice.
The constructs for overexpression of Nur77 contained the coding sequence for amino acids 1 to 580 for the full-length (wild-type [wt]) clone and 249 to 598 for the truncated (dominant negative) clone in the vector pCDNA3.1. The dominant-negative construct for immunocytochemistry contained the coding sequence for amino acids 248 to 598 in the vector pEGFP-C1 (Clontech), resulting in a fusion protein with enhanced green fluorescent protein (EGFP) on its N-terminus. As transfection control we used the empty pEGFP-C1.
Immunohistochemistry
For detection of PAI-1, mouse anti–PAI-1 (3PAI5; Technoclone GmbH, Vienna, Austria) diluted 1:50 was used, for Nur77 rabbit anti-Nur77 (M-210 Santa Cruz Biotechnology), diluted 1:100 was used. Secondary antibodies rabbit antimouse Alexa 488, goat antirabbit Alexa 488, and goat antimouse Alexa 568, all from Molecular Probes (Eugene, OR), were used diluted 1:2000 for immunocytochemistry and 1:300 for immunohistochemistry. Tissues were snap frozen and embedded in optimal cutting temperature compound (OCT; Miles Laboratories, Elkhart, IN). An anti-CD31 antibody (WM59 mouse monoclonal, TCS Cellworks, Botolph Claydon, Great Britain) was used to identify endothelial cells. Cryosections were fixed in acetone at −20°C and used for standard immunodetection immediately.
Results
TNFα induces PAI-1 expression
To verify that TNFα induces PAI-1 in endothelial cells, we followed PAI-1 expression in HUVECs stimulated with TNFα using Q-PCR. PAI-1 mRNA levels increased after 1 hour and reached 10-fold higher levels after 4 hours (Figure1A). We performed nuclear run-on assays to ensure that this induced mRNA increase was due to de novo synthesis. As shown in the inset to Figure 1A, treatment with TNFα induced de novo synthesis of PAI-1 mRNA, consistent with results reported earlier.35 To analyze which part of the PAI-1 promoter might be responsible for TNFα-dependent induction, we constructed a series of deletion mutants of the PAI-1 promoter (between nucleotides [nt]−1520 and +20 relative to the transcription start) driving transcription of a luciferase reporter gene. When HUVECs were transfected with these reporter gene constructs a 40-bp stretch between nt −280 to −240 of the PAI-1 promoter could be identified as critical for the small38 but consistent TNFα-induced transcriptional activation in ECs (Figure 1B).
Expression of PAI-1 mRNA and PAI-1 reporter studies.
(A) Relative quantitative PCR (Q-PCR) analysis of PAI-1 mRNA expression. HUVECs were stimulated with TNFα for the indicated periods and harvested. Relative expression of PAI-1 was normalized to the expression of β-2 microglobulin. The inset shows nuclear run-on data. HUVECs were either untreated or treated for 4 hours with TNFα, and the RNA isolated after the run-on reaction was hybridized to PAI-1 and GAPDH probes that were immobilized on membranes. (B) Reporter gene analysis. A series of deletions in the PAI-1 promoter were fused to a luciferase reporter gene, transfected into HUVECs, and measured by luminometry. HUVECs expressing the reporter gene constructs were induced with TNFα. (C) EMSAs of overlapping parts of the PAI-1 promoter. Nuclear extracts were obtained from HUVECs and incubated with labeled double-stranded oligonucleotides representing the nt −280 to −260, −270 to −250, and −260 to −240 of the PAI-1 promoter, respectively. Specific binding activity of nuclear protein complexes is indicated by black arrows. (D) Overview of the region of the PAI-1 promoter containing the NBRE. Oligonucleotide sequences that were used for EMSAs and for the yeast screens are shown. Light gray indicates care consensus sequence; dark gray, 2 additional adenines required for binding; and black, nucleotides exchanged.
Expression of PAI-1 mRNA and PAI-1 reporter studies.
(A) Relative quantitative PCR (Q-PCR) analysis of PAI-1 mRNA expression. HUVECs were stimulated with TNFα for the indicated periods and harvested. Relative expression of PAI-1 was normalized to the expression of β-2 microglobulin. The inset shows nuclear run-on data. HUVECs were either untreated or treated for 4 hours with TNFα, and the RNA isolated after the run-on reaction was hybridized to PAI-1 and GAPDH probes that were immobilized on membranes. (B) Reporter gene analysis. A series of deletions in the PAI-1 promoter were fused to a luciferase reporter gene, transfected into HUVECs, and measured by luminometry. HUVECs expressing the reporter gene constructs were induced with TNFα. (C) EMSAs of overlapping parts of the PAI-1 promoter. Nuclear extracts were obtained from HUVECs and incubated with labeled double-stranded oligonucleotides representing the nt −280 to −260, −270 to −250, and −260 to −240 of the PAI-1 promoter, respectively. Specific binding activity of nuclear protein complexes is indicated by black arrows. (D) Overview of the region of the PAI-1 promoter containing the NBRE. Oligonucleotide sequences that were used for EMSAs and for the yeast screens are shown. Light gray indicates care consensus sequence; dark gray, 2 additional adenines required for binding; and black, nucleotides exchanged.
A nuclear protein complex binds from nt −270 to −250 in the PAI-1 promoter
To define the binding site in this part of the PAI-1 promoter for the putative nuclear proteins, we performed serial electrophoretic mobility shift assays (EMSAs) with overlapping oligonucleotide probes spanning the region from nt −280 to −240. The central region (nt −270 to −250) showed specific binding of a protein complex(es) when incubated with nuclear extracts from HUVECs (Figure 1C). We therefore used this 20-bp stretch (Figure 1D) of the PAI-1 promoter as bait in a yeast one-hybrid screen using an activated lymphocyte cDNA fusion library. We identified 3 positive clones, all containing the DNA binding domain of the orphan nuclear receptor Nur77.
Nur77 binds to an NBRE in the PAI-1 promoter
Upon analysis, we found that the PAI-1 promoter contains a putative NBRE (NGFI-B responsive element) from nt −261 to −254 also present in the oligonucleotides used for EMSA and in the bait sequence used for the yeast one-hybrid screen (Figure 1D). The NBRE is a well-described binding site for Nur77 monomers, and binding of Nur77 to this element has been shown to strongly activate transcription of other genes.39 40
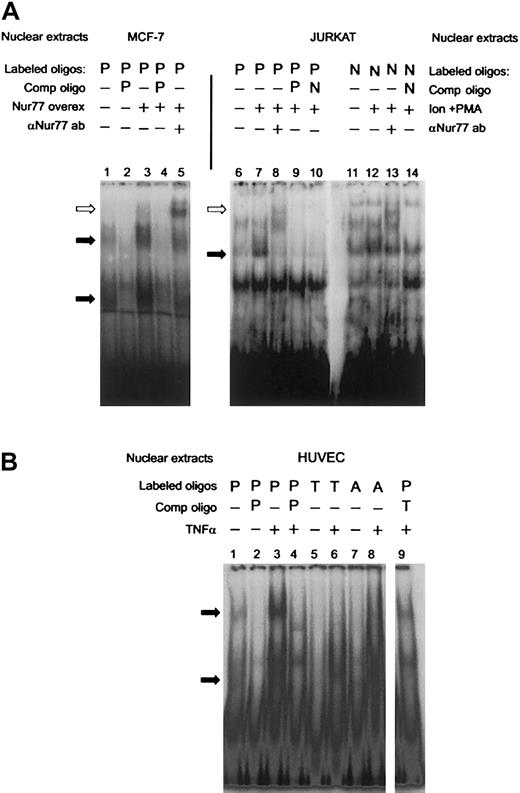
To establish that Nur77 binds to that site in the PAI-1 promoter, we performed supershift assays after overexpressing a truncated Nur77 (amino acids 249-598, containing the DNA and ligand-binding domains) in MCF-7 cells, which express low endogenous amounts of Nur77. Lane 3 in Figure 2A shows increased specific binding to the labeled PAI-1 oligonucleotide (P) in transfected cells. This indicates that overexpression of Nur77 is sufficient to induce binding. Upon addition of an antibody recognizing Nur77, the specific bands were retarded in the gel, identifying the presence of Nur77 in the complex(es) bound to the labeled oligonucleotide (Figure 2A, lane 5). The human T-cell leukemia cell line JURKAT is known to express significant levels of Nur77 only after stimulation,41 and indeed nuclear extracts of PMA- and ionomycin-stimulated JURKAT cells exhibited specific binding to the PAI-1 oligonucleotide (P, Figure 2A, lane 7), which was also supershifted by the anti-Nur77 antibody (Figure 2A, lane 8). The differing positions of the antibody-supershifted bands in lane 5 compared with lane 8 can be explained by the fact that in MCF-7 cells a shorter, truncated form of Nur77 was expressed, which still exhibited full DNA binding activity,42 while JURKAT expressed a wild-type Nur77. Binding of JURKAT nuclear extracts to the PAI-1 oligonucleotide was comparable with that of an oligonucleotide containing a canonical NBRE consensus site (5′-AAAAGGTCAAG-3′ [Consensus NBRE, N], Figure 2A, lanes 11-14). Mutation of the NBRE in the PAI-1 oligonucleotide from 5′-AGGTCA-3′ (PAI-NBRE) to 5′-AGGACA-3′ (PAI-T15A, T, Figure 1D), thereby destroying the most conserved base of the consensus sequence, resulted in loss of binding of the nuclear protein complexes to the oligonucleotide (Figure 2B, lane 6). A part of the CTE (C-terminal extension) of Nur77, called the A box, confers specificity for binding of Nur77 monomer to the NBRE. This interaction is mediated by a stretch of 2 or more adenines located 5′ from the NBRE.43 44 In order to analyze whether binding of Nur77 to the NBRE in the PAI-1 promoter involves interaction with the 5′ adenines, we designed an oligonucleotide in which the original site in the PAI-1 promoter was changed from 5′-GAAAGGTCA-3′ (PAI-NBRE) to 5′-GACAGGTCA-3′ (PAI-A11C, A, Figure 1D). This mutation strongly reduced binding of Nur77 to the oligonucleotide (Figure 2B, lane 8). Taken together these data indicate that the PAI-1 promoter contains an NBRE site that is a target for monomeric binding of Nur77 and that Nur77 by itself is sufficient to induce specific binding.
PAI-1 promoter: EMSAs and supershifts of Nur77.
(A) Overexpression of a truncated clone of Nur77 (amino acids 248 to 580 that have full DNA-binding activity) in MCF-7 cells (lanes 1-4) and addition of the E-20 antibody, which recognizes the C-terminus of the Nur77 family of proteins (lane 5). JURKAT cells were induced with ionomycin and PMA (Ion+PMA) and incubated with a radio-labeled NBRE found in the PAI-1 promoter (P, lanes 6-10) and a canonical consensus NBRE (N, lanes 11-14). The extracts were also incubated with E-20 antibody (lanes 8,13). Binding was competed with a 100-fold molar excess of unlabeled oligonucleotides as indicated (PAI-NBRE, P; Con NBRE, N). Specific binding activity of nuclear protein complexes is indicated by black arrows. A supershift resulting from binding of the Nur77 antibody is indicated by white arrows. (B) HUVECs were stimulated with TNFα, and nuclear extracts were incubated with radioactively labeled oligonucleotides containing the NBRE of the PAI-1 promoter (P). The same extracts were incubated with radioactively labeled oligonucleotides containing the NBRE of the PAI-1 promoter that was mutated in the positions indicated in Figure 1D (PAI-T15A, T; PAI-A11C, A). Specific binding activity of nuclear protein complexes is indicated by black arrows.
PAI-1 promoter: EMSAs and supershifts of Nur77.
(A) Overexpression of a truncated clone of Nur77 (amino acids 248 to 580 that have full DNA-binding activity) in MCF-7 cells (lanes 1-4) and addition of the E-20 antibody, which recognizes the C-terminus of the Nur77 family of proteins (lane 5). JURKAT cells were induced with ionomycin and PMA (Ion+PMA) and incubated with a radio-labeled NBRE found in the PAI-1 promoter (P, lanes 6-10) and a canonical consensus NBRE (N, lanes 11-14). The extracts were also incubated with E-20 antibody (lanes 8,13). Binding was competed with a 100-fold molar excess of unlabeled oligonucleotides as indicated (PAI-NBRE, P; Con NBRE, N). Specific binding activity of nuclear protein complexes is indicated by black arrows. A supershift resulting from binding of the Nur77 antibody is indicated by white arrows. (B) HUVECs were stimulated with TNFα, and nuclear extracts were incubated with radioactively labeled oligonucleotides containing the NBRE of the PAI-1 promoter (P). The same extracts were incubated with radioactively labeled oligonucleotides containing the NBRE of the PAI-1 promoter that was mutated in the positions indicated in Figure 1D (PAI-T15A, T; PAI-A11C, A). Specific binding activity of nuclear protein complexes is indicated by black arrows.
TNFα, LPS, and IL-1 induce expression of Nur77
The specific binding activity to the PAI-1 oligonucleotide was strongly increased when nuclear extracts from TNFα-stimulated cells were used, indicating regulation of Nur77 by TNFα (Figure 2B, lane 3); such increased binding activity resembles the results seen in Figure 2A (lane 3) when Nur77 was overexpressed.
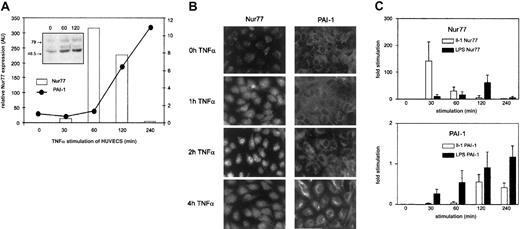
To assess whether in fact TNFα regulates Nur77 at the level of expression, we followed Nur77 mRNA levels by Q-PCR. TNFα induced a rapid burst of Nur77 detectable after 30 minutes that peaked after one hour of stimulation and declined to unstimulated levels after 4 hours (Figure 3A); this change in Nur77 mRNA was followed by an increase in Nur77 protein (Figure 3A, inset). PAI-1 mRNA levels increased with a delay and then continued to rise steadily until 4 hours after stimulation as shown in Figure 1A. Immunofluorescence data of corresponding experiments in cultured cells are shown in Figure 3B: upon stimulation of endothelial cells by TNFα, Nur77 protein increased after one hour and accumulated in the nucleus after 2 hours; PAI-1 protein synthesis started in the cells after 4 hours. These data indicate that TNFα leads to induction of Nur77 transcription; Nur77 protein then accumulates in the nucleus followed by PAI-1 mRNA and protein expression suggesting that TNFα regulates PAI-1 via Nur77.
Expression of Nur77 and PAI-1 in HUVECs induced with TNFα.
(A) Q-PCR analysis of Nur77 and PAI-1 mRNA expression. HUVECs were induced with TNFα for the indicated minutes; relative expression was normalized to β-2 microglobulin. The inset shows a Western blot of HUVECs stimulated for the indicated number of seconds with TNFα and stained using an antibody recognizing the N-terminus of Nur77 (representative experiment). (B) Immunocytochemistry. HUVECs were stimulated by TNFα for indicated periods and stained with antibodies recognizing Nur77 or PAI-1. (C) Q-PCR analysis of Nur77 and PAI-1 mRNA expression. HUVECs were induced with LPS and IL-1 for the indicated time periods (±SD of triplicates).
Expression of Nur77 and PAI-1 in HUVECs induced with TNFα.
(A) Q-PCR analysis of Nur77 and PAI-1 mRNA expression. HUVECs were induced with TNFα for the indicated minutes; relative expression was normalized to β-2 microglobulin. The inset shows a Western blot of HUVECs stimulated for the indicated number of seconds with TNFα and stained using an antibody recognizing the N-terminus of Nur77 (representative experiment). (B) Immunocytochemistry. HUVECs were stimulated by TNFα for indicated periods and stained with antibodies recognizing Nur77 or PAI-1. (C) Q-PCR analysis of Nur77 and PAI-1 mRNA expression. HUVECs were induced with LPS and IL-1 for the indicated time periods (±SD of triplicates).
To test whether other inflammatory mediators also regulate Nur77 and PAI-1 in a similar way, we performed experiments as shown in Figure 3A using LPS (lipopolysaccharide) or IL-1β (interleukin 1). As shown in Figure 3C, IL-1β induced up-regulation of Nur77 and PAI-1 mRNA in a comparable way. This indicates that Nur77-mediated PAI-1 regulation might not be restricted to TNFα. Also, LPS induced up-regulation of Nur77 and PAI-1; however, the different time course seen in response to LPS indicates overall differences in the signal transduction pathway between LPS, IL-1β, and TNFα. Such different kinetics of the induction of inflammatory response genes have been described previously.45 The early increase in PA1-1 mRNA further indicates additional regulatory mechanisms including changes in mRNA stability as indicated by previous studies.46
TNFα induction of PAI-1 depends on Nur77 and the presence of the NBRE in the PAI-1 promoter
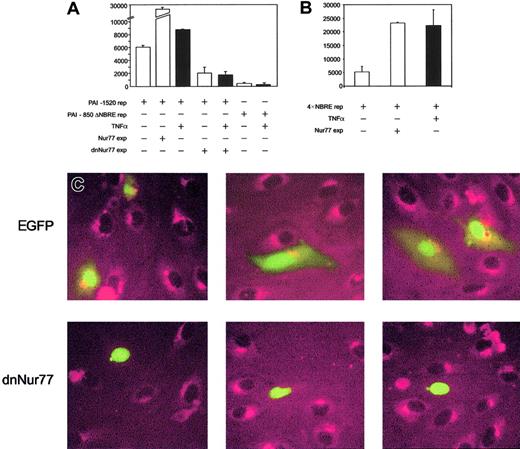
To prove that increased expression of Nur77 directly activates the PAI-1 promoter, we performed reporter gene assays. PAI-1 reporter constructs containing the NBRE of the PAI-1 promoter were strongly induced when Nur77 was coexpressed (Figure4A), whereas PAI-1 promoter constructs with an NBRE deletion did not respond to Nur77 overexpression (not shown) or to TNFα (Figure 4A). Consistently, overexpression of Nur77 as well as TNFα stimulation strongly induced a 4 ×NBRE luciferase reporter gene construct (Figure 4B). Coexpression of a dominant-negative mutant42 of Nur77 (dnNur77) reduced basal reporter gene activity and completely abolished the up-regulation of the reporter gene by TNFα (Figure 4A). When a dnNur77 fused to EGFP (dnNur77-EGFP) was transfected into HUVECs, followed by stimulation with TNFα for 4 hours, no PAI-1 expression was seen in the transfected cells expressing the construct. Cells transfected with EGFP alone or untransfected cells responded normally with PAI-1 expression to TNFα stimulation (Figure 4C).
Analysis of PAI-1 reporter gene activity and protein expression upon coexpression of full-length and dominant-negative Nur77 constructs.
(A) PAI-1 reporter gene activity. Bars 1 to 3 represent overexpression of Nur77 as well as TNFα induction in HUVECs; bars 4 and 5 represent overexpression of a dominant-negative mutant of Nur77 lacking its transactivation domain in untreated or TNFα-stimulated HUVECs; bars 6 and 7 represent the activity of a PAI-1 reporter gene construct lacking the 20 bp containing the NBRE with and without TNFα stimulation. Black bars are TNFα+(±SD of triplicates). (B) 4 × NBRE reporter gene activity. Effect of TNFα stimulation and overexpression of Nur77 on activity of a reporter gene construct containing a 4 × tandem repeat of an NBRE fused to a minimal promoter. Black bars are TNFα+ (±SD of triplicates). (C) Immunocytochemistry. HUVECs were transiently transfected to express EGFP or dnNur77-EGFP. After treatment with TNFα for 4 hours, cells expressing EGFP alone (green nuclear and cytoplasmatic signal) as well as untransfected cells produced large amounts of PAI-1 protein (red), whereas cells expressing dnNur77 (green, nuclear localization) showed no expression of PAI-1. Original magnification × 100 (C).
Analysis of PAI-1 reporter gene activity and protein expression upon coexpression of full-length and dominant-negative Nur77 constructs.
(A) PAI-1 reporter gene activity. Bars 1 to 3 represent overexpression of Nur77 as well as TNFα induction in HUVECs; bars 4 and 5 represent overexpression of a dominant-negative mutant of Nur77 lacking its transactivation domain in untreated or TNFα-stimulated HUVECs; bars 6 and 7 represent the activity of a PAI-1 reporter gene construct lacking the 20 bp containing the NBRE with and without TNFα stimulation. Black bars are TNFα+(±SD of triplicates). (B) 4 × NBRE reporter gene activity. Effect of TNFα stimulation and overexpression of Nur77 on activity of a reporter gene construct containing a 4 × tandem repeat of an NBRE fused to a minimal promoter. Black bars are TNFα+ (±SD of triplicates). (C) Immunocytochemistry. HUVECs were transiently transfected to express EGFP or dnNur77-EGFP. After treatment with TNFα for 4 hours, cells expressing EGFP alone (green nuclear and cytoplasmatic signal) as well as untransfected cells produced large amounts of PAI-1 protein (red), whereas cells expressing dnNur77 (green, nuclear localization) showed no expression of PAI-1. Original magnification × 100 (C).
These data further indicate that the regulation of the PAI-1 promoter as well as of PAI-1 protein by TNFα is Nur77 dependent and requires the NBRE.
Nur77 is present in atherosclerotic vessels and colocalizes with PAI-1
Having shown that TNFα up-regulates Nur77 and in turn PAI-1 expression in ECs, we were interested to see whether Nur77 is also up-regulated in atherosclerotic vessels in vivo. When normal or atherosclerotic coronary arteries were stained for Nur77 and PAI-1, a strong Nur77 signal was seen only in SMCs, macrophages, and ECs (stained with antibodies against CD-31, Figure5G) of the plaque area and of the neointima and vessels of the adventitia. In these areas, PAI-1 staining colocalized with Nur77 staining (Figure 5A-J). PAI-1 was additionally seen in the extracellular matrix especially of the plaque (Figure5D,F). Nur77 was also strongly up-regulated in the neointima (Figure5L), while in the media only a scattered staining in SMCs was seen (Figure 5M). This pattern of Nur77 staining is consistent with a model in which inflammatory cells in the neointima release inflammatory cytokines, which in turn induces Nur77 in SMC of the neointima and the media.47 Our finding that inflammatory cytokines induce Nur77 mRNA in cultured SMCs (not shown) supports this model.
Nur77 and PAI-1 expression in normal and atherosclerotic vascular tissues.
(A-D) Minimal expression for Nur77 (A) and PAI-1 (B) in normal vessels; high expression of both proteins in the plaque area in endothelial cells and smooth muscle cells in an atherosclerotic coronary artery (C-D). The 50-μm scale bar in panel D applies to panels A-D. (E-G) Coimmunostaining of Nur77 and PAI-1 in a human atherosclerotic lesion. The vessel was stained for Nur77 (green, E), PAI-1 (red, F), or CD31 (blue, G). The 100-μm scale bar in panel G applies to panels E-G. (H-J) Endothelial cells of vasa vasora in human atherosclerotic coronary arteries express both Nur77 (H) and PAI-1 (J), which colocalize (I). The 20-μm scale bar in panel J applies to panels H-J. (K-M) Expression of Nur77 in a human atherosclerotic vessel (K) with higher expression in the neointima (L) and lower expression in the media (M). Panels L and M show further magnification of the indicated areas of panel K.
Nur77 and PAI-1 expression in normal and atherosclerotic vascular tissues.
(A-D) Minimal expression for Nur77 (A) and PAI-1 (B) in normal vessels; high expression of both proteins in the plaque area in endothelial cells and smooth muscle cells in an atherosclerotic coronary artery (C-D). The 50-μm scale bar in panel D applies to panels A-D. (E-G) Coimmunostaining of Nur77 and PAI-1 in a human atherosclerotic lesion. The vessel was stained for Nur77 (green, E), PAI-1 (red, F), or CD31 (blue, G). The 100-μm scale bar in panel G applies to panels E-G. (H-J) Endothelial cells of vasa vasora in human atherosclerotic coronary arteries express both Nur77 (H) and PAI-1 (J), which colocalize (I). The 20-μm scale bar in panel J applies to panels H-J. (K-M) Expression of Nur77 in a human atherosclerotic vessel (K) with higher expression in the neointima (L) and lower expression in the media (M). Panels L and M show further magnification of the indicated areas of panel K.
Discussion
Up-regulation of PAI-1 during inflammation has long since been known to occur, and in fact, the deleterious outcome of sepsis was for some time thought to be caused by the extremely high levels of PAI-1 found under these conditions.48 However, the PAI-1 promoter is lacking consensus sequences for the inflammatory transcription factor NF-κB,49 and even binding sites for other transcription factors implicated in the inflammatory pathways such as EGR-128,29 are missing. Therefore, several indirect mechanisms have been implicated in PAI-1 induction during inflammation.38,50,51 TNFα efficiently induced de novo synthesis of PAI-1 mRNA in endothelial cells and induced an approximately 10-fold increase in PAI-1 mRNA levels after 4 hours of treatment. Stimulation of the PAI-1 promoter reporter gene construct used here by the same stimulus was only weak. A similar weak stimulation of PAI-1 promoter constructs was also reported earlier by others,38 and the differences between transcriptional activity on the PAI-1 promoter construct and induced PAI-1 mRNA levels might be inherent to the methodologies used. However, a contribution of a regulatory element present upstream from the promoter constructs used,38 as well as additional PAI-1 mRNA stability induced by the inflammatory mediators, cannot be excluded. Independent of such additional mechanisms, we here present a novel regulatory element by which TNFα and other inflammatory stimuli can activate PAI-1 expression in endothelial cells.
We defined a consensus sequence in the proximal part of the PAI-1 promoter for the orphan nuclear receptor Nur77 that drives basal transcription and is crucial for up-regulation of PAI-1 in ECs in response to the inflammatory cytokine TNFα. This consensus sequence mediates monomeric binding of Nur77. Such monomeric binding is compatible with a ligand-independent mechanism and consistent with regulation of PAI-1 expression by changes in the levels of Nur77 expression, which then functions as a “constitutive orphan steroid receptor.”52 Consistent with such a mechanism, we found that TNFα and other inflammatory stimuli induce transcriptional up-regulation of Nur77: upon stimulation with TNFα, Nur77 mRNA and, in turn, protein are induced within 30 and 60 minutes, respectively; Nur77 is then translocated to the nucleus followed by PAI-1 mRNA and protein expression. Our data, showing that the PAI-1 response to TNFα is abolished in ECs transfected with a dominant-negative form of Nur77, point toward a pivotal role of Nur77 for the PAI-1 response to TNFα and possibly other inflammatory stimuli that concomitantly up-regulate Nur77 and PAI-1.
Our data demonstrating that Nur77 expression is increased in atherosclerotic vessels and colocalizes there with PAI-1 indicate the importance of this mechanism also in vivo. Recent work by deVries et al,53 who showed that Nur77 is found induced in a screen for genes up-regulated by proatherogenic cytokines in smooth muscle cells, supports our finding, as well as a report by Arkenbout et al,54 demonstrating the involvement of the nuclear receptor subfamily 4 in neointima formation in a mouse model. Nur77 (NAK-1, TR3) is a member of this nuclear receptor subfamily and the human homolog55,56 of mouse Nur77 and rat NGFI-B. Nur77 was shown to be involved in T-cell receptor–induced apoptosis42,57 and to be induced during B-cell differentiation58 in response to growth factors,59 mechanical stress,60 and dietary fatty acids.61 We here show for the first time that the transcription factor Nur77 is part of the TNFα response and mediates TNFα-induced PAI-1 up-regulation.
In conclusion, we have delineated the mechanism by which TNFα induces PAI-1 expression in endothelial cells and identified Nur77 as the responsible transcription factor. Increased expression of Nur77 and PAI-1 in vascular cells of atherosclerotic tissues indicates that Nur77 might be a pivotal response gene for inflamed vascular cells and a possible target for therapeutic intervention.
Prepublished online as Blood First Edition Paper, December 27, 2002; DOI 10.1182/blood-2002-07-2331.
Supported in part by grants from the Austrian Science Foundation within the program project grant F005 (projects no. 509 to B.R.B. and no. 512 to R.d.M.); by the Interdisciplinary Cooperation Project Program of the Austrian Federal Ministry for Education, Science, and Culture; and the competence center for Bio Molecular Therapeutics (BMT). Earlier work within this project was funded by grants from the Anton Dreher Foundation no. 234/93 and the Herzfeld-Foundation (P.H.).
F.G. and P.H. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Bernd R. Binder, Department of Vascular Biology and Thrombosis Research, Schwarzspanierstr 17, A-1090 Vienna, Austria; e-mail:bernd.binder@univie.ac.at.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal