Activation-induced cytidine deaminase (AID) induces somatic hypermutation (SHM), class switch recombination (CSR), and immunoglobulin gene conversion in B-lymphocytes. Here we report for the first time the expression of AID in healthy human B-lymphocytes and in B-cell non-Hodgkin lymphomas (B-NHL). AID mRNA expression in humans is restricted to the CD19+CD38+IgD−germinal center cells, namely the CD19+CD38+CD44− centroblasts. After in vitro stimulation of naive human B cells by CD40-L and IL-4, AID mRNA is strongly induced for only 48 hours. In a survey of human B-NHL AID was found to be constitutively expressed in follicular lymphoma and in diffuse large B-cell lymphoma but to be absent in B-precursor lymphoblastic leukemia, in mantle cell lymphoma, and in plasma cell myeloma. In B-cell chronic lymphatic leukemia, in immunocytoma, and in extranodal marginal zone B-cell lymphoma of MALT, AID mRNA was expressed only in some samples. In follicular lymphoma and diffuse large B-cell lymphoma, the expression of AID mRNA was coincident with the presence of SHM in the variable region exons of the immunoglobulin heavy-chain gene. In human B-NHL, the AID mRNA is spliced into 4 different variants but does not contain point mutations. Thus AID, which is highly regulated during healthy B-cell development, is constitutively expressed in human germinal center B-NHL and in subsets of nongerminal center B-NHL. This constitutive expression of AID may promote illegitimate DNA recombinations and somatic mutations in B-NHL.
Introduction
Specific chromosomal translocations are a genetic hallmark of lymphoid malignancies in non-Hodgkin lymphoma (NHL). Immunoglobulin gene loci are involved in most of these translocations in B-cell NHL, suggesting that mistaken maturation of the immunoglobulin genes may contribute to lymphoma formation.1,2 During B-cell development the immunoglobulin genes are altered by 3 distinct mechanisms: In B lymphoblasts, V(D)J joining rearranges the immunoglobulin-gene variable (V), diversity (D), and joining (J) segments to produce the primary antibody repertoire; this well defined process is mediated by the recombination activating genes 1 and 2.3,4 After antigen encounter, an effective secondary antibody response is achieved by somatic hypermutation (SHM) of the variable region exons and by class switch recombination (CSR) that replaces the μ constant region with one of the downstream-located constant regions.5,6 Activation-induced cytidine deaminase (AID) is indispensable for SHM, CSR, and immunoglobulin gene conversion in B cells.7-10 AID is a close homologue of APOBEC-1, the catalytic subunit of the apo-B mRNA editing-complex; AID and APOBEC-1 are located in tandem on chromosome 12 and apparently have arisen by gene duplication.11-14APOBEC-1 is an RNA-dependent cytidine deaminase that specifically deaminates C6666 in the apo-B mRNA together with the APOBEC-1–stimulating protein.12,15-18 In AID-deficient mice, SHM and CSR are completely abolished, leading to a severe defect of the humoral immune response.8 In human patients with the autosomal recessive form of the hyper-immunoglobulin M syndrome (HIGM2), various mutations in the human AID gene have been found that cripple AID function.7 In these patients not only is CSR absent but SHM is strongly reduced.7 In fibroblasts, the expression of AID is sufficient to mediate CSR of an artificial switch substrate and SHM in an actively transcribed artificial gene substrate.19,20 Expression of AID inEscherichia coli leads to a hypermutator phenotype with nucleotide transitions at dC/dG in a context-dependent manner.21 Moreover, expression of a bacteriophage-derived inhibitor protein of uracil-DNA glycosylase in chicken DT40 cells that lack the repair gene XRCC2 shifts the pattern of SHM from predominantly transversions to transitions.22 These recent results indicate that AID may simply act by deamination of cytidines in the DNA.21 22
Because APOBEC-1 transgenic animals develop hepatocellular carcinomas,23 we reasoned that AID might be a potential oncogene. Here we demonstrate that AID is constitutively expressed in follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL), whereas in healthy B-cell development the expression of AID is highly regulated and restricted to germinal center B cells. This constitutive expression of AID may contribute to NHL formation.
Patients, materials, and methods
Isolation of B-cell populations from human tonsils
Human nonmalignant tonsils were obtained with written consent from adult patients undergoing tonsillectomy. The tonsil specimens were minced, and the single-cell suspension was filtered through a 100-μm filter (Filcon; DAKO, Copenhagen, Denmark). Lymphocytes were isolated by gradient centrifugation using Ficoll-Paque medium (Amersham Pharmacia Biotech, Uppsala, Sweden). Total B cells were isolated with anti-CD19 using immunomagnetic beads (Dynal Biotech, Oslo, Norway) and subsequently were fractionated into the following: naive B-cells (CD19+CD38−IgD+), naive germinal center cells (CD19+CD38+IgD+), germinal center cells (CD19+CD38+IgD−), and memory cells (CD19+CD38−IgD−). The CD19+CD38+cells were fractionated into CD19+CD38+CD44− centroblasts and CD19+CD38+CD44+ centrocytes using anti-CD44. The purity of the cell fractions was generally greater than 95%. Total RNA was isolated from 1 × 108 cells with Trizol (Invitrogen, Carlsbad, CA). Single-stranded cDNA was synthesized using 500 μg total RNA and a commercially available random priming synthesis kit (Superscript First Strand Synthesis Kit; Invitrogen).
Isolation of naive human B cells by negative selection and in vitro stimulation with CD40L
Naive IgM+ B cells (5-10 × 108) were prepared by negative selection. Briefly, single-cell suspensions of B cells prepared from human tonsils were resuspended in Hanks buffered salt solution (HBSS) with 5% fetal bovine serum (FBS) and were centrifuged for 6 minutes at 1600 rpm at 4°C. The cells were resuspended in HBSS/5% fetal bovine serum (FBS) (5 × 107cells/mL), mixed with biotin-conjugated antibodies against CD2, CD3, CD14, CD16, CD33, CD56 glycophorin A, immunoglobulin G (IgG), and IgA (Stem Cell Technologies, Vancouver, BC, Canada) (100 μL cocktail/mL), and incubated on ice for 45 minutes. Magnetic colloid coupled to streptavidin (60 μL/mL) was added, and the incubation was continued for another 60 minutes. Approximately 5 × 108 cells in antibody cocktail plus magnetic colloid were applied to a 0.9-cm preparative metallic matrix-assisted cell sorting (MACS) column, and the IgM/IgD+ cells were eluted with HBSS/5% FBS. Cells were pelleted for 6 minutes at 1600 rpm and resuspended in RPMI 1640 plus 10% FBS at a density of approximately 1 × 106 cells/mL. These human naive B cells were stimulated for various times with 2.7 mg/mL plasma membranes prepared from Sf21 insect cells stably infected with a human CD40L-expressing recombinant baculovirus (CD40L-BV) in the absence or presence of 40 ng/mL interleukin (IL)–4 (PharMingen, Franklin Lakes, NJ).
Lymphoma tissue specimens
For all lymphomas investigated, fresh and paraffin-embedded samples were available. Fresh tissue specimens were obtained during surgery, immediately shock frozen in liquid nitrogen, and kept at −80°C until use. Diagnoses of the lymphoma subtypes were established by the reference pathologists of the Lymph Node Registry Kiel on the basis of standard histopathology with additional immunhistochemical stainings with mouse monoclonal antibodies using the alkaline phosphatase–anti-alkaline phosphatase (APAAP) technique. Informed consent was obtained from all patients analyzed in this study. All immunoreagents were purchased from DAKO (Hamburg, Germany), except for CD20, Ki-B3, and Ki-S5, which had been generated in the Lymphoma Registry, Kiel, Germany.
B-precursor lymphoblastic leukemia/lymphoma (cALL) was diagnosed when blasts expressed terminal deoxyribonucleotidyl transferase (TdT), CD79a, CD19 or CD20, and CD10. Diffuse large B-cell lymphoma (DLBCL) was diagnosed when centroblasts or immunoblasts expressed B-cell marker–like CD20 and showed a high proliferation index determined by Ki-S5 staining. Follicular lymphoma (FL) was diagnosed on the basis of a follicular growth pattern with the expression of CD20, CD10, and bcl-2 in the absence of CD5. B-chronic lymphocytic leukemia (B-CLL) and mantle cell lymphoma (MCL) showed a predominance of small or medium-sized lymphocytes with coexpression of CD20, CD23, and CD5 in B-CLL or coexpression of CD20, CD5, and cyclin D1 in the absence of CD23 in MCL. Extranodal marginal zone B-cell lymphoma of MALT (mucosa-associated lymphatic tissue) (MALT-lymphoma) was diagnosed on the basis of the diagnostic lymphoepithelial lesions with expression of CD20 in the absence of Ki-B3 or DBA44. The diagnosis of plasma cell multiple myeloma (MM) was established by the presence of plasma cell infiltrates expressing monotypic light chains. Immunocytoma (lymphoplasmacytic lymphoma) (IC) was diagnosed when lymphoplasmacytoid cells expressed CD20 and IgM with intracytoplasmic monotypic light chains, and the lymphoma lacked features of other entities such as follicular growth or marginal zone features. Based on the Kiel classification, lymphoplasmacytic lymphoma included clonal plasma cells determined by clonal expression of one immunoglobulin light chain. Lymphocytoid immunocytoma contained polyclonal plasma cells. This group of lymphomas is classified as B-CLL in the World Health Organization (WHO) classification. All samples were completely composed of lymphoma tissue and contained no portions of adjacent tissue. RNA was prepared from 5 to 7 slices of 7 μm using Trizol (Invitrogen). cDNA pools of the individual lymphoma specimens were synthesized using 1 μg total RNA and a commercially available kit with random primers (SMART RACE cDNA Amplification Kit; Clontech, Palo Alto, CA).
Oligonucleotides
The following oligonucleotides were used: AID 1 (sense, nucleotide [nt] 48-78, GAGGCAAGAAGA CAC TCT GGA CAC CAC TAT); AID 2 (antisense, nt 408-381, GCGGTGAAGATCCTCAGGCTGAGG TTG); AID 3 (sense, nt 328-358, AGCCCCTGCTACGACTGTGCC CGACATGTG); AID 4 (antisense, nt 681-654, AGTTGCTAT CAA AGT CCCAAA GTA CGA); AID 5 (sense, nt 11-40, TAATTGAAG TGAGAT TTTTCT GGCCTGAGA); AID 6 (antisense, nt 720-690, CTTCAGCAGAGATAT TTC ATC GTGTGTGAC); AID 7 (antisense, nt 520-490, CCAGCAGTAAAA ATA ATC TTTGAA GGTCAT); AID 13 (antisense, 232-203, CTTATT GCGAAGATA ACC AAA GTCCAGTGA); AID 14 (sense, intron 8201-8230, GGA GCC GAA ATT AAA GAT TAG AAG CAG AGA); AID s-1 (sense, nt 200-232, TTTTCACTGGAC TTTGGTTAT CTTCGC); AID as-1 (antisense, nt 520-485, CCAGCAGTAAAA ATA ATC TTTGAAGGT); AID as-2 (antisense, nt 484-461, TATTTGCAC CCC GGCGCGGTGCAG); β-actin s-2 (sense, nt 391-420, CCCCCTGAACCC CAAGGCCAACCGCGA GAA); β-actin (as-2) (antisense, nt 660-631, TAGCCGCGCTCGGTGAGGATCTTCATGAGG); β-actin as-4 (nt 630-601, TAGTCAGTCAGGTCC CGGCCAGCCAGGTCC); Blimp-1 s-1 (sense, nt 421-444), GAGTAAAGAATA CAT ACC AAA GGG); Blimp-1 as-1 (antisense, nt 824-801, CATTTTTCTCAG TGCTCGGTTGCT); Blimp 1 as-2 (antisense, nt 730-701, TGCAAAGTCCCGACA ATA CCA CACAAG); c-myc s-1(sense, nt 594-617, TCTCAACGACAGCAG CTCGCCCAA); c-myc as-1 (antisense, 893-870, TTGAGGACCAGTGGGCTGTGAGGA); c-myc s-2 (sense, nt 618-642, GTCCTGCGCCTCGCAAGACTCCAG); Bcl-6 s-1 (sense, nt 248-271, CAA GAA GTT TCT AGG AAA GGC CGG); Bcl-6 as-1 (antisense, nt 547-524, GATTGATCA CACTAA GGTTGCATT); Bcl-6 as (antisense, nt 520-497, ACTGGT CTGTAA AGATGC TATAGA).
RT-PCR analysis
One microliter cDNA synthesis mix was used for polymerase chain reaction (PCR) amplification. PCR reactions were performed as described24,25 using the following conditions—AID s-1 and AID as-2: 30 cycles at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds; AID 3 and AID 7: 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; AID 5 and AID 6: 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; nested PCR with AID 14 and AID 6 and 30 cycles using the same PCR conditions; β-actin s-1 and β-actin as-1: 25 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; Bcl-6 s-1 and Bcl-6 as-1: 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; blimp-1 s-a and blimp-1 as-1: 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds; c-myc s-1 and c-myc as-1: 30 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. PCR products were hybridized with32P-labeled internal oligonucleotides.25
Quantitative PCR was performed with a Light Cycler (Roche, Basel, Switzerland) using standard software for quantification as recommended by the manufacturer. Expression levels were normalized to the expression level of β-actin mRNA. The following conditions were used: AID 3 and AID 7: 45 cycles at 95° for 1 second, 55°C for 5 seconds, and 72°C for 6 seconds; β-actin s-1 and β-actin as-2: 45 cycles at 95°C for 1 second, 58°C for 5 seconds, and 72°C for 6 seconds; Bcl-6 s-1 and bcl-6 as-1: 45 cycles at 95°C for 1 second, 58°C for 5 seconds, and 72°C for 6 seconds; blimp-1 s-a and blimp-1 as-1: 45 cycles at 95°C for 1 second, 58°C for 5 seconds, and 72°C 6 seconds. Expression levels were expressed in relation to the expression level of β-actin mRNA that was separately determined in each sample.
Ribonuclease protection assay
Radiolabeled antisense AID RNA was synthesized as described spanning 222 bases of the AID mRNA sequence (nt 232-10).25Total RNA from human lymphoma tissues (10 μg) was coprecipitated with 2 × 104 cpm α-32P–labeled AID antisense RNA probe. Hybridization and RNA digestion with the Ribonuclease Protection Assay II Kit (Ambion, Austin, TX) and analysis of the protected RNAs on denaturing polyacrylamide sequencing gels was performed as described.25 26
Analysis of somatic hypermutation of the variable region of the immunoglobulin heavy-chain gene
Recombined V(D)J segments of the IgH transcripts of the lymphoma specimens were amplified from the respective cDNA pools by PCR using a set of degenerate oligonucleotides exactly as described.27
Determination of immunoglobulin heavy- and light-chain expression
Immunglobulin heavy-chain and light-chain expression was determined by standard immunohistochemistry using antibodies against κ, λ, IgM, IgG, IgD, IgE, and IgA as described.28
Results
Expression of AID mRNA in human germinal center B cells
AID mRNA was amplified by reverse transcription (RT)–PCR from naive B cells (CD19+CD38−IgD+), naive germinal center cells (CD19+CD38+IgD+), germinal center cells (CD19+CD38+IgD−), and memory cells (CD19+CD38−IgD−) (Figure1). In germinal center B cells, a strong AID PCR product was generated, and a faint band was detectable in the memory cell fraction (Figure 1). No RT-PCR products for AID mRNA were detectable in naive B cells or in naive germinal center B cells (Figure1). CD19+CD38+ germinal center cells were further fractionated with anti-CD44 monoclonal antibodies into centroblasts (CD19+CD38+CD44−) and centrocytes (CD19+CD38+CD44+). AID mRNA was strongly detected in the CD44− centroblasts but was hardly detectable in CD44+ centrocytes (Figure1).
Expression of AID mRNA in human germinal center B lymphocytes.
Total human B cells were fractionated by immunoaffinity purification into naive B cells (CD19+CD38−IgD+), naive germinal center cells (CD19+CD38+IgD+), germinal center cells (CD19+CD38+IgD−), memory cells (CD19+CD38−IgD−), centroblasts (CD19+CD38+CD44−), and centrocytes (CD19+CD38+CD44+). cDNA pools were synthesized from total RNA, and the cDNAs of AID (nt 328-520) and β-actin (nt 391-660) were amplified by PCR. The PCR products were separated by agarose gel electrophoresis and hybridized with internal radiolabeled oligonucleotides.
Expression of AID mRNA in human germinal center B lymphocytes.
Total human B cells were fractionated by immunoaffinity purification into naive B cells (CD19+CD38−IgD+), naive germinal center cells (CD19+CD38+IgD+), germinal center cells (CD19+CD38+IgD−), memory cells (CD19+CD38−IgD−), centroblasts (CD19+CD38+CD44−), and centrocytes (CD19+CD38+CD44+). cDNA pools were synthesized from total RNA, and the cDNAs of AID (nt 328-520) and β-actin (nt 391-660) were amplified by PCR. The PCR products were separated by agarose gel electrophoresis and hybridized with internal radiolabeled oligonucleotides.
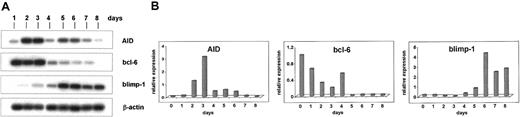
Naive human IgM+ B cells isolated from human tonsils were cultivated for up to 8 days in the presence of IL-4 and plasma membranes of Sf21 cells that had been infected with a CD40-L expressing baculovirus. AID mRNA was strongly induced between days 2 and 3 (Figure2A). At day 4, the levels of AID mRNA strongly decreased (Figure 2A). The mRNA of bcl-6 decreased at approximately day 4 and was hardly detectable from day 5 to day 8 (Figure 2A). In contrast, the mRNA of blimp-1 was strongly induced at approximately day 5 (Figure 2A). The mRNA expression of AID, bcl-6, and blimp-1 in these cells was quantitated by real-time PCR (Figure 2B). The increase of AID mRNA at day 3 was approximately 40-fold in comparison with the AID mRNA levels of naive B-lymphocytes at day 1 (Figure 2B). At day 0, AID mRNA could not be detected by real-time RT-PCR. From day 3 to day 4, the AID mRNA level decreased by more than 80% (Figure 2B). The mRNA of bcl-6 steeply decreased from day 4 to day 5, and the mRNA of blimp-1 increased approximately 50-fold between day 4 and day 6 (Figure 2B). When the naive human IgM+ B cells were cultivated in the presence of CD40-L without IL-4, the mRNA of AID was induced much later, at day 4 instead of day 2, and this induction was far less pronounced (data not shown).
Induction of AID mRNA in human naive B cells by CD40-L and IL-4.
Human naive B cells were cultivated in the presence of CD40-L and IL-4 for up to 8 days. The mRNA of AID (nt 200-520), bcl-6 (nt 248-547), blimp-1 (421-824), and β-actin (391-660) was amplified by PCR from cDNA pools, and the PCR products were separated by agarose gel electrophoresis and hybridized with an internal radiolabeled oligonucleotide (A). The mRNA of AID, bcl-6, and blimp-1 was measured by real-time PCR using a Light Cycler (Roche) with the primer pairs as above and normalized to the expression of β-actin mRNA (B).
Induction of AID mRNA in human naive B cells by CD40-L and IL-4.
Human naive B cells were cultivated in the presence of CD40-L and IL-4 for up to 8 days. The mRNA of AID (nt 200-520), bcl-6 (nt 248-547), blimp-1 (421-824), and β-actin (391-660) was amplified by PCR from cDNA pools, and the PCR products were separated by agarose gel electrophoresis and hybridized with an internal radiolabeled oligonucleotide (A). The mRNA of AID, bcl-6, and blimp-1 was measured by real-time PCR using a Light Cycler (Roche) with the primer pairs as above and normalized to the expression of β-actin mRNA (B).
Expression of AID mRNA in human B-cell non-Hodgkin lymphomas
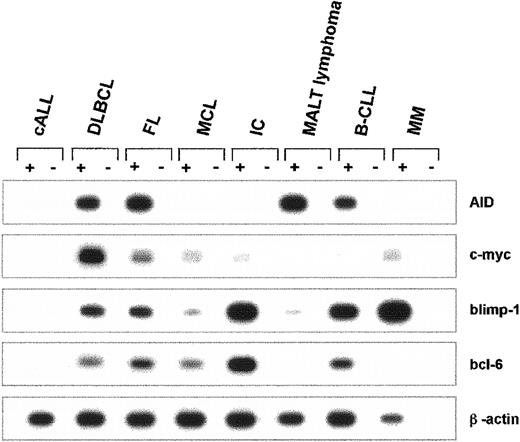
The mRNA expression of AID and of c-myc, blimp-1, bcl-6, and β-actin was studied in B-precursor lymphoblastic leukemia/lymphoma (cALL), diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), mantle cell lymphoma (MCL), immunocytoma (IC), extranodal marginal zone B-cell lymphoma of MALT (MALT lymphoma), B-cell chronic lymphatic leukemia (B-CLL), and multiple myeloma (MM). Individual mRNA expression profiles for each entity are depicted in Figure3. Within this series, AID mRNA was not detected in any of the patients with cALL (n = 3), MCL (n = 4), or MM (n = 3). Strong RT-PCR products of AID mRNA were generated in all patients with FL (n = 5) and in all patients with DLBCL (n = 13). In patients with MALT lymphoma (n = 8), IC (n = 8), and B-CLL (n = 5), a heterogeneous expression pattern was found for AID mRNA, with strong expression in some and absence of expression in other patients. Although blimp-1 was most abundantly expressed in IC, MM, and, to a lesser extent, in B-CLL (Figure 3), c-myc mRNA was strongly amplified in DLBCL, and Bcl-6 was strongly expressed in IC (Figure 3). In healthy human lymph nodes (n = 4), AID mRNA could only be detected by nested RT-PCR but not by the conventional RT-PCR, indicating that AID is expressed in healthy lymphatic tissues at lower levels than in germinal center B-NHL (data not shown).
Expression of AID mRNA in human non-Hodgkin lymphomas.
cDNA pools were prepared from cALL, DLBCL, FL, MCL, IC, MALT lymphoma, B-CLL, and MM. The cDNA of AID (nt 200-520), c-myc (nt 594-893), blimp-1 (nt 421-824), bcl-6 (nt 248-547), and β-actin (nt 391-660) was amplified by PCR. PCR products were separated by agarose gel electrophoresis and hybridized with an internal radiolabeled oligonucleotide. A representative autoradiograph with the PCR products of one individual tissue sample for each NHL entity is shown.
Expression of AID mRNA in human non-Hodgkin lymphomas.
cDNA pools were prepared from cALL, DLBCL, FL, MCL, IC, MALT lymphoma, B-CLL, and MM. The cDNA of AID (nt 200-520), c-myc (nt 594-893), blimp-1 (nt 421-824), bcl-6 (nt 248-547), and β-actin (nt 391-660) was amplified by PCR. PCR products were separated by agarose gel electrophoresis and hybridized with an internal radiolabeled oligonucleotide. A representative autoradiograph with the PCR products of one individual tissue sample for each NHL entity is shown.
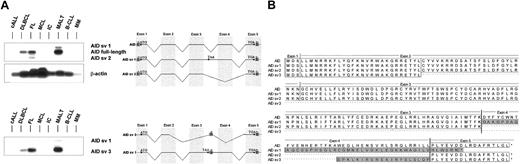
In all AID-expressing lymphomas, full-length AID mRNA is the most abundant splice form (Figure 4). Alternative splicing generates 3 additional AID mRNA variants. In splice variant 1 (sv 1), the intron between exon 3 and exon 4 is not spliced, extending the open-reading frame for an additional 45 amino acid residues up to the first stop codon in intron 3 (Figure 4). In splice variant 2 (sv 2), exon 4 is spliced, leading to an internal deletion of 39 amino acid residues. In splice variant 3 (sv 3), a short neo-exon located in intron 3 is used, but exon 3 and exon 4 are excluded (Figure 4). This spliced variant encodes a truncated peptide consisting of the amino terminus and the carboxy terminus of AID linked by 20 amino acid residues encoded by the neo-exon 3 (Figure 4). In FL, sv 2 was the second minor splice variant, whereas in MALT lymphoma, sv 1 was the second minor variant (Figure 4A). In MALT lymphoma, both sv 1 and sv 3 are expressed, whereas in FL and in DLBCL, only sv 3 could be found (Figure 4). Sequencing did not detect point mutations in AID full-length cDNA from patients with FL (n = 3), DLBCL (n = 3), and MALT lymphoma (n = 3).
Alternative splicing of AID mRNA in human non-Hodgkin lymphoma.
Full-length AID mRNA (nt 11-720) was amplified by PCR from cDNA pools of human NHL, and the PCR products were hybridized with an internal oligonucleotide (A, upper panel). AID splice variants sv1 and sv2 were amplified by half-nested PCR using a specific oligonucleotide (AID14) complementary to the neo-exon in intron 3 (A, lower panel). The primers used for the RT-PCR are indicated with their respective numbers. The amino acid sequence of the 4 different splice variants of the human AID gene is shown (B). The shaded boxes mark the amino acid residues aberrant to the full-length sequence; the asterisks indicate the deleted amino acid residues of the splice variants.
Alternative splicing of AID mRNA in human non-Hodgkin lymphoma.
Full-length AID mRNA (nt 11-720) was amplified by PCR from cDNA pools of human NHL, and the PCR products were hybridized with an internal oligonucleotide (A, upper panel). AID splice variants sv1 and sv2 were amplified by half-nested PCR using a specific oligonucleotide (AID14) complementary to the neo-exon in intron 3 (A, lower panel). The primers used for the RT-PCR are indicated with their respective numbers. The amino acid sequence of the 4 different splice variants of the human AID gene is shown (B). The shaded boxes mark the amino acid residues aberrant to the full-length sequence; the asterisks indicate the deleted amino acid residues of the splice variants.
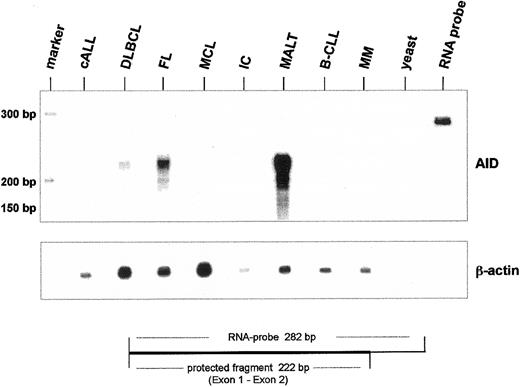
The expression of AID mRNA in human B-cell non-Hodgkin lymphoma was directly quantitated by ribonuclease protection analysis (RPA). For these experiments an AID antisense RNA probe complementary to exons 1 and 2 was used. Therefore, full-length AID and the mRNA variants sv 1 and sv 2 protect an identical fragment of 222 nucleotides. The expected protected fragment of 222 nucleotides was observed with the highest intensity in MALT, followed by FC and DLBCL (Figure5). Quantification of AID mRNA based on the expression of β-actin mRNA revealed that in this particular MALT lymphoma, the AID mRNA levels were approximately 10-fold higher than in FL. In the samples of B-CLL and of immunocytoma used in this experiment, no AID mRNA was detectable by RPA (Figure 5).
Ribonuclease protection for AID mRNA in human non-Hodgkin lymphoma.
Total RNA (10 μg) from human NHL was hybridized with α–32P-labeled AID antisense RNA probe (nt 232-10). After RNA digestion the protected AID antisense RNA was analyzed on a denaturing polyacrylamide sequencing gel. As an internal standard, ribonuclease protection was performed for β-actin mRNA (lower panel). Yeast RNA (yeast) was used as a negative control, and undigested antisense RNA probe was run aside.
Ribonuclease protection for AID mRNA in human non-Hodgkin lymphoma.
Total RNA (10 μg) from human NHL was hybridized with α–32P-labeled AID antisense RNA probe (nt 232-10). After RNA digestion the protected AID antisense RNA was analyzed on a denaturing polyacrylamide sequencing gel. As an internal standard, ribonuclease protection was performed for β-actin mRNA (lower panel). Yeast RNA (yeast) was used as a negative control, and undigested antisense RNA probe was run aside.
AID mRNA expression, somatic hypermutation, and class switch recombination in human B-cell non-Hodgkin lymphomas
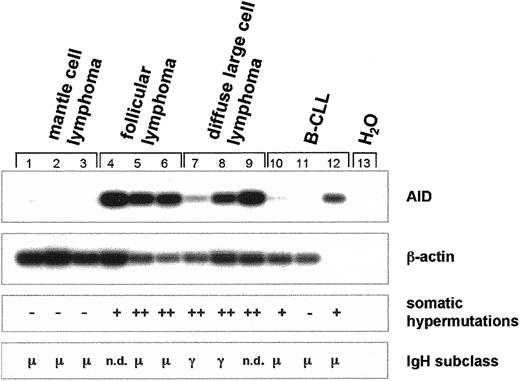
Next we studied somatic hypermutation (SHM) of the recombined V(D)J segment and determined the subtype of the constant region of the immunoglobulin heavy-chain (IgH) gene in MCL (n = 3), FL (n = 3), DLBCL (n = 3), and B-CLL (n = 3) in correlation to the expression of AID mRNA (Figure 6). In MCL, no AID mRNA is expressed, no somatic hypermutation of the variable region is detectable, and the μ constant region of the IgHgene is expressed (Figure 6). In contrast, in FL and DLBCL, both of which express AID mRNA, the variable region of the IgH gene is hypermutated (Figure 6). In most patients with FL, theIgH gene locus has not undergone isotype class switch recombination (CSR), and mainly IgM is expressed (Figure 6). In contrast, in most patients with DLBCL, CSR of the IgH gene has occurred and IgG is expressed (Figure 6). In the B-CLLs investigated, CSR had not occurred and somatic hypermutation was either absent or low (Figure 6).
Expression of AID mRNA, somatic hypermutation, and class switch recombination of the
IgH gene in germinal center non-Hodgkin lymphoma.cDNA pools were synthesized from total RNA of MCL (n = 3), FL (n = 3), DLBCL (n = 3), and B-CLL (n = 3). AID (nt 328-520) and β-actin cDNA (nt 391-660) were amplified by PCR and hybridized with radiolabeled internal oligonucleotides. Recombined V(D)J segments of the IgH transcripts were amplified using a set of degenerate primers and were subsequently sequenced. Point mutations of less than 3% in comparison with the published sequences were regarded as somatic hypermutation negative (−), point mutations of more than 3% but less than 6% were regarded as somatic hypermutation positive (+), and point mutations of more than 6% were regarded as strong positive for somatic hypermutation (++). Immunoglobulin heavy-chain expression was determined by standard immunohistochemistry, and the constant region of the IgH locus is indicated. n.d. denotes nondetectable IgH gene expression.
Expression of AID mRNA, somatic hypermutation, and class switch recombination of the
IgH gene in germinal center non-Hodgkin lymphoma.cDNA pools were synthesized from total RNA of MCL (n = 3), FL (n = 3), DLBCL (n = 3), and B-CLL (n = 3). AID (nt 328-520) and β-actin cDNA (nt 391-660) were amplified by PCR and hybridized with radiolabeled internal oligonucleotides. Recombined V(D)J segments of the IgH transcripts were amplified using a set of degenerate primers and were subsequently sequenced. Point mutations of less than 3% in comparison with the published sequences were regarded as somatic hypermutation negative (−), point mutations of more than 3% but less than 6% were regarded as somatic hypermutation positive (+), and point mutations of more than 6% were regarded as strong positive for somatic hypermutation (++). Immunoglobulin heavy-chain expression was determined by standard immunohistochemistry, and the constant region of the IgH locus is indicated. n.d. denotes nondetectable IgH gene expression.
Because not all DLBCLs are thought to be derived from germinal center cells, we analyzed 13 patients with DLBCL and stratified them according to their expression of CD10 as detected by immunohistochemistry, which can be used to discriminate the germinal center B-cell–like type of DLBCL from the activated B-cell–like type.29 Five of these DLBCLs were positive for CD10, and 8 were negative (Table1). AID mRNA was detected in all 5 CD10+ and all 8 CD10− DLBCLs (Table1). Three of the CD10+ DLBCLs had developed from confirmed FLs, but none of the CD10− cases had progressed from FL (Table 1). SHM was detected in 3 of 3 patients with the CD10+ DLBCL, in whom we sequenced the variable region of the IgH gene, and in 5 of 5 of the CD10− DLBCLs (Table 1).
AID expression in DLBCL
| . | n . | AIDpositive . | Preceded by FL . | SHM greater than 6% . |
|---|---|---|---|---|
| CD10+ | 5 | 5 of 5 | 3 of 5 | 3 of 3 |
| CD10− | 8 | 8 of 8 | 0 of 8 | 5 of 5 |
| . | n . | AIDpositive . | Preceded by FL . | SHM greater than 6% . |
|---|---|---|---|---|
| CD10+ | 5 | 5 of 5 | 3 of 5 | 3 of 3 |
| CD10− | 8 | 8 of 8 | 0 of 8 | 5 of 5 |
CD10 protein, AID mRNA, and somatic hypermutation of theIgH gene in DLBCL. In 13 patients with DLBCL, CD10 protein expression was analyzed by immunohistochemistry. AID mRNA expression was analyzed by RT-PCR. Three of the CD10+ patients showed progression from documented FL. The variable region exons of the IgH gene were sequenced in 3 of the CD10+ and in 5 of the CD10− patients.
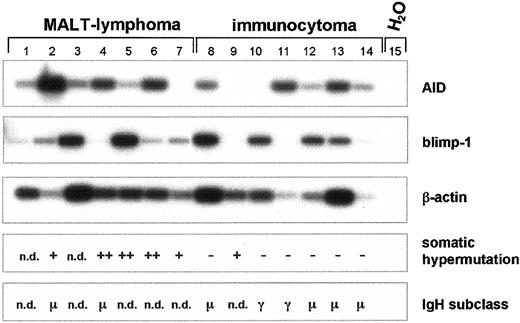
The expression of AID and blimp-1 was also studied in 7 MALT lymphomas and in 7 immunocytomas (Figure 7). In the MALT lymphomas, a great variation of AID mRNA expression was observed with relatively high AID expression in 3 patients and low or no expression of AID mRNA in 5 patients (Figure 7). The expression of blimp-1 is inversely correlated to the expression of AID in MALT lymphomas, and to lesser extent, in immunocytomas (Figure 7). Thus, the lymphomas with high-level expression of blimp-1 demonstrated less AID mRNA, and high-level expression of AID mRNA was linked to low amounts of blimp-1 (Figure 7). In the MALT lymphomas, SHM was present in all patients except for 2 in whom no IgH transcripts could be detected. Interestingly, the MALT lymphoma with the high-level expression of AID mRNA (Figure 7, lane 2, and Figure 5) demonstrated only low levels of SHM (3.2% base changes in the variable region) and expressed IgM (Figure 7). Although AID mRNA was absent in 2 and was expressed in 5 ICs, SHM and CSR could not be detected in the ICs with the exception of one sample (lane 9) (Figure 7). Notably, this IC with low-level SHM did not express AID, whereas the IC with AID expression did not demonstrate SHM (Figure 7). Therefore, in MALT lymphoma and in IC, no correlation of AID expression with the presence of SHM in the IgH variable regions is observed. In addition, AID expression was studied in the blood lymphocytes of 10 additional patients with B-CLL. In 7 of these patients, AID mRNA was detected in blood lymphocytes, and in the other 3 patients AID mRNA was undetectable (data not shown). As in IC, no correlation of AID expression and SHM was found in the blood lymphocytes of these 10 patients with B-CLL.
Expression of AID mRNA, somatic hypermutation, and class switch recombination of the
IgH gene in MALT lymphoma and immunocytoma. cDNA pools were synthesized from total RNA of MALT lymphomas (n = 7) and immunocytoma (n = 7). AID (nt 200-520), blimp-1 (421-824), and β-actin cDNA (nt 391-660) were amplified by PCR and hybridized with a radiolabeled internal oligonucleotide. Recombined V(D)J segments of the IgH transcripts were amplified using a set of degenerate oligonucleotides as primers and subsequently entirely sequenced. Point mutations of less than 3% in comparison to the published sequences are defined as somatic hypermutation negative (−), point mutations of more than 3% but less than 6% are defined as somatic hypermutation positive (+), and point mutations of more than 6% are defined as strong positive for somatic hypermutation (++). Immunoglobulin heavy-chain expression was determined by standard immunohistochemistry, and the constant region of the IgH locus is indicated. n.d. denotes nondetectable IgH gene expression.
Expression of AID mRNA, somatic hypermutation, and class switch recombination of the
IgH gene in MALT lymphoma and immunocytoma. cDNA pools were synthesized from total RNA of MALT lymphomas (n = 7) and immunocytoma (n = 7). AID (nt 200-520), blimp-1 (421-824), and β-actin cDNA (nt 391-660) were amplified by PCR and hybridized with a radiolabeled internal oligonucleotide. Recombined V(D)J segments of the IgH transcripts were amplified using a set of degenerate oligonucleotides as primers and subsequently entirely sequenced. Point mutations of less than 3% in comparison to the published sequences are defined as somatic hypermutation negative (−), point mutations of more than 3% but less than 6% are defined as somatic hypermutation positive (+), and point mutations of more than 6% are defined as strong positive for somatic hypermutation (++). Immunoglobulin heavy-chain expression was determined by standard immunohistochemistry, and the constant region of the IgH locus is indicated. n.d. denotes nondetectable IgH gene expression.
Discussion
This investigation demonstrates for the first time that AID is highly regulated in healthy human germinal center B cells, namely the CD44− centroblasts, but is constitutively expressed in FL and DLBCL. AID mRNA is absent in cALL, MCL, and MM. In MALT lymphoma, IC, B-CLL, AID mRNA can be detected only in a subset of patients. Further studies with more patients will be required to investigate whether the expression of AID in these cases is associated with a different growth phenotype, a different response to treatment, or even a different clinical outcome.
Recent studies have demonstrated the pivotal role of AID for the induction of SHM and CSR that mark the transition of the primary humoral immune response to the secondary phase of mature, highly specific antibody production.10,30,31 The identification of AID enables studies of these events at a molecular level. We started to investigate AID expression in healthy human germinal center B cells and B-cell NHL because AID might be a pacemaker for lymphoma development and malignant progression. This assumption is supported by the finding of AID-induced mutations in E coli,demonstrating that AID alone is sufficient to confer a mutator phenotype.21 Moreover, aberrant hepatic overexpression of the AID homologue APOBEC-1 induces hepatocellular carcinomas, indicating the oncogenic potential of these genes.23Previous studies in healthy human B cells and in B-NHL have shown that SHM occurs not only occur in the variable region of the IgHgene locus but also in other genes specifically expressed in B cells such as bcl-6.32-35 In DLBCL, a loss of specificity for SHM and DNA recombination events is observed with respect to the healthy B cell that could well be caused by the observed constitutive expression of AID.35
During healthy B-cell maturation, AID expression is highly restricted to only a short period of time. The constitutive expression of AID in B-NHL must be attributed to an autocrine or a paracrine secretion of the required growth factors by the neoplastic B cells or by lymphoma-infiltrating T cells or, alternatively, by a constitutive activation of the AID promoter. Current experiments in our laboratories try to address this important issue. If indeed AID is a pacemaker of B-NHL formation, measures to reverse the expression of AID would be an obvious option for the treatment or prevention of germinal center B NHL.
Alternative splicing generates 3 AID variants in addition to the full-length protein. Whether these splice variants have a biologic role remains to be investigated. The regularly spliced mRNA of AID is the most prevalent AID transcript in healthy and neoplastic B cells, and AID mRNA remains unmutated in human B-NHL. Thus, in B-NHL the expression of the AID gene is altered, but the function of the encoded AID protein is unchanged. With respect to the recent results on AID function in E coli, it is possible that the AID protein acts as a mutator in neoplastic human B cells.21 35
The expression of AID in DLBCL and FL was coincident with the occurrence of SHM in the variable region exons of the IgH locus. On the contrary, the absence of AID expression in MCL was associated with a lack of CSR and SHM. In DLBCL, the expression of AID is predictable for SHM and CSR and may become a molecular marker for these processes. AID expression was found in all patients with DLBCL studied regardless of whether CD10 protein was expressed or whether the DLBCL developed from a previous FL. We are currently generating a monoclonal antibody directed against AID that will allow a much easier determination of AID protein expression in human B-NHL.
In MALT lymphoma, IC, and B-CLL a great variation of AID expression was observed, and no correlation of AID expression with SHM and CSR could be demonstrated. A striking negative correlation of AID mRNA to blimp-1 mRNA was observed in MALT lymphomas. With respect to these markers, 2 entities of MALT lymphomas can be defined that are characterized by the expression of AID and the absence of blimp-1 or the reverse. This assumption is in accordance with recent results on blimp-1 action in human B-cell lines and mouse splenic B cells.36 Blimp-1 extinguishes the gene expression for germinal center B-cell function by repressing several transcription factors such as Spi-B and Id3, and it induces plasma cell genes such as XBP-1.36 The results of our investigation indicate that certain subsets within MALT lymphoma, IC, and B-CLL can be defined solely based on the expression of AID. Further studies with more patients are required to analyze whether the expression of AID in these lymphomas correlates to a different clinical outcome in patients. Taken together, the results of our study suggest that AID indeed may be a potent oncogene for B-cell NHL development. In addition to conducting more extended clinical studies, this hypothesis should now be tested experimentally using transgenic animals.
We thank Anna Sotnikova for her help in purifying B-lymphocytes from human tonsils.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/blood-2002-08-2424.
Supported by Deutsche Forschungsgemeinschaft grant SFB 545, Teilprojekt A6 (J.G.) and Bundesministerium für Bildung und Forschung, Förderkennzeichen 01KV95090 (J.G.), and supported in part by National Institutes of Health grant GM26939 (K.B.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jobst Greeve, Klinik für Allgemeine Innere Medizin, Inselspital-Universitätsspital Bern, CH-3010 Bern Switzerland; e-mail: jobst.greeve@insel.ch.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal