We read with great interest the recent article by Miller et al1 that examined the transcobalamin (TC) genetic polymorphism encoding proline or arginine at codon 259 in theTC gene (Pro259Arg) and its possible influence on indices of vitamin B12 status in healthy older adults. Based on serum levels of holoTC (the B12-TC complex) that were significantly higher in individuals who were homozygous for the wild-type gene encoding the proline form of TC compared with heterozygotes and homozygotes for the arginine-encoding gene variant, Miller et al concluded that the TC Pro259Arg polymorphism may influence the intracellular availability of vitamin B12 and thus influence susceptibility to B12 deficiency.1 There are several ways of describing human genetic sequence variations, and sometimes the inconsistent use of genetic terms may cause confusion. In international guidelines for genetic nomenclature it is preferable to give the nucleotide position of a polymorphism rather than giving the codon number only.2 Miller et al mention both nucleotide position and codon number, but unfortunately the nucleotide substitution that is the basis for the TC phenotypic variability on codon 259 is assigned to the wrong nucleotide compared with the full-length complementary DNA sequence of the TCgene (GenBank accession number M60396 for the proline-encoding sequence and NM000355 for the arginine-encoding sequence; Figure1). The polymorphism is a C-to-G substitution at position 776 in relation to the first nucleotide (+1) of the ATG-translation initiation codon, not a G-to-C substitution at nucleotide 775. This mistake, however, does not interfere with their genotyping method, and all results reported in the investigation seem valid for the TC 776C>G polymorphism. Nevertheless, we would like to make the following comment regarding the method. The presence of a G nucleotide at position 776, which encodes arginine at codon 259, removes a recognition site forMvaI. The polymerase chain reaction (PCR) product remains uncleaved if the individual is homozygous for the 776G allele and partially cleaved if the individual is heterozygous. This method is sensitive to insufficient cleavage due to excess of PCR product or poor cleavage conditions in a specific tube and is for this reason not suitable for routine diagnostic laboratory work. We suggest the use of alternative methods not based on restriction enzyme digestion for determining the TC 776C>G polymorphism. Recently, we presented a method for TC genotyping based on the minisequencing technique.3 4 This technique does not depend on the presence of restriction enzyme recognition sites and also is less sensitive to silent polymorphisms.
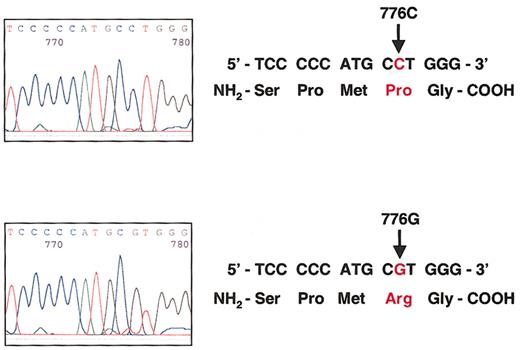
The TC Pro259Arg polymorphism is a C-to-G substitution at nucleotide 776.
(Top) Sequence analysis of the TC gene in one individual homozygous for the wild-type 776CC genotype that encodes proline at codon 259. (Bottom) Sequence analysis of the TC gene in one individual homozygous for the 776GG genotype that encodes arginine at codon 259.
The TC Pro259Arg polymorphism is a C-to-G substitution at nucleotide 776.
(Top) Sequence analysis of the TC gene in one individual homozygous for the wild-type 776CC genotype that encodes proline at codon 259. (Bottom) Sequence analysis of the TC gene in one individual homozygous for the 776GG genotype that encodes arginine at codon 259.
Transcobalamin 776C>G: significance and determination
We thank Dr Zetterberg and his colleagues for their comments regarding our paper1-1 and for pointing out that the correct designation for the transcobalamin codon 259 polymorphism is 776C>G. On the issue of which isoform of the protein (proline or arginine) should be considered the wild type, we acknowledge that convention dictates that the more commonly occurring isoform (proline) should be given this designation. It is noteworthy, however, that the arginine isoform does have a relatively high prevalence (∼ 20% homozygosity among whites),1-1-1-5 despite the fact that it is associated with poorer vitamin B12 status compared with the proline isoform.1-1-1-6 This observation implies that the mutation does not afford any obvious selective advantage, at least as far as standard methods of B12 status assessment are concerned. Therefore, the relatively high prevalence of the arginine isoform remains unexplained.
Concerning methodology, the minisequencing technique for genotyping transcobalamin does have the advantage of not requiring restriction enzyme digestion. However, the minisequencing method may not be readily available in all laboratories. In developing our method, which utilizes the restriction enzyme MvaI, we validated it by direct sequencing of the polymerase chain reaction (PCR) products prior to digestion. We concluded that our method was robust and accurate, and thus is a relatively inexpensive method for determining transcobalamin genotype.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal