Abstract
Retroviral vectors are commonly used in clinical gene therapy, but recent observations of insertional oncogene activation in preclinical and clinical settings have forced a discussion of their safety. Here we investigated the relationship between retroviral transduction efficiency in mass cultures and the actual number of integrated vector copies in single cells using K562 leukemia and primary CD34+ cells. We found an exponential increase of integration numbers correlated to gene transfer rates and a linear increase of expression levels with insertion frequency. On average we detected one vector insertion per transduced cell for a gene transfer of less than 30%, 3 for 60%, and approximately 9 for 90% (in K562). Clonal analysis revealed strikingly increased variations of both transgene copy numbers (more than 20-fold in primary cells) and expression levels associated with higher transduction. Therefore, limiting retroviral gene transfer to approximately 30% may be suggested to avoid generating clones containing multiple insertions.
Introduction
Retroviral vectors based on murine leukemia viruses (MLVs) are the most commonly used gene transfer system for stable transduction of various target cells,1 but their random genome insertion2 may lead to functional or structural alterations of cellular genes. This process, called insertional mutagenesis, has been considered a possible limitation of somatic gene therapy for more than 2 decades.3-5 However, until recently there were no reports of malignant transformation of normal somatic cells related to the insertion of replication-defective retroviral vectors. Indeed, based on the experience of numerous preclinical and clinical studies, the frequency of those events was predicted to be low.5-9
Last year we reported the first evidence that a retroviral vector insertion may directly lead to the activation of an oncogene, Evi-1, causing acute myeloid leukemia in a mouse gene-marking model.10 Later, a lymphoproliferative disease, probably causally linked to retroviral insertions into the oncogene LMO-2, developed in 2 patients in an otherwise highly successful gene therapy trial.11-13 These observations underscore that insertional mutagenesis represents a real risk factor for somatic gene therapy and that insertional alterations of crucial regulatory genes (eg, oncogenes) are more common than estimated.9
Given that problems related to insertional mutagenesis are likely dose dependent,9 dose finding requires careful investigations in powerful preclinical models. Here we investigated the correlation between retroviral transduction rates and numbers of vector insertions. To address this question we transduced primary hematopoietic CD34+ progenitors and cultured K562 cells with an enhanced green fluorescence protein (EGFP)-expressing vector at different multiplicities of infection (MOIs) and analyzed gene transfer (expression, copy numbers) in mass cultures and cell clones derived from them.
Study design
Cell culture, retroviral vector, and transduction
Primary CD34+ peripheral blood stem cells were isolated using magnetic cell sorting (MACS) technology (Miltenyi Biotec, Bergisch-Gladbach, Germany).14-16 CD34+, K562, and PG13 cells producing the retroviral vector SF11αEGFPrev17 were cultivated as described.14 For transduction, K562 or prestimulated CD34+ cells14 were transferred onto 6- or 24-well plates, preloaded up to 8 times with diluted (1:5, group A) or undiluted (groups B-F) supernatant.14 Twenty-four hours after the first round, cells from some groups were submitted to a second infection on plates preloaded accordingly. MOIs given in Table 1 were calculated based on titers determined as described.14 It was assumed that subsequent preloading steps are equally as efficient as the first one, although there might be a saturation effect (B.F. and K.K., unpublished data, April 2003). Transduced CD34+ and K562 cells were cloned in methyl cellulose (MC) or by limiting dilution.15,16
Retroviral transduction efficiency and vector copy numbers in mass cultures and cell clones derived thereof
Cell groups and clones . | MOI (× no. of preloading cycles) . | Gene transfer, % . | Vector copy no., real-time PCR . | Vectors/transduced cell, calculated . | MF, arbitrary units . | Average MF ± SD (variation factor)‡ . | Vector copies, Southern/qPCR . | Mean copy no./transduced cell ± SD (range)‡ . | MF/vector copy, arbitrary units (mean) . |
|---|---|---|---|---|---|---|---|---|---|
| K562 A | 0.5 | 28.6 | 0.24 | 0.9 | 33.3 ± 8.9 (1.9×) | 1.0 ± 0.0 (1) | (33.3) | ||
| 1 | 40.9 | 1 | 40.9 | ||||||
| 5 | 43.6 | 1* | 43.6 | ||||||
| 6 | 23.7 | 1* | 23.7 | ||||||
| 8 | 32.5 | 1 | 32.5 | ||||||
| 9 | 25.6 | 1 | 25.6 | ||||||
| K562 B | 2.5 | 60.9 | 1.78 | 2.9 | 43.5 ± 28.5 (6.5×) | 2.0 ± 0.8 (1-3) | (20.2) | ||
| 1 | 22.9 | 2* | 11.5 | ||||||
| 7 | 44.7 | 2 | 22.4 | ||||||
| 8 | 86.8 | 3 | 28.9 | ||||||
| 9 | 13.4 | 1 | 13.4 | ||||||
| 10 | 49.8 | 2 | 24.9 | ||||||
| K562 C | 5 | 78.0 | 2.89 | 3.7 | 162.0 ± 121.9 (14.2×) | 5.4 ± 3.7 (1-9) | (28.1) | ||
| 4 | 21.7 | 1 | 21.7 | ||||||
| 5 | 177.1 | 7* | 25.3 | ||||||
| 6 | 57.1 | 2* | 28.6 | ||||||
| 10 | 245.2 | 8 | 30.6 | ||||||
| 15 | 308.9 | 9 | 34.3 | ||||||
| K562 D | 20 | 70.3 | 6.27 | 8.9 | 162.9 ± 174.9 (29×) | 5.8 ± 6.0 (1-16) | (24.9) | ||
| 1 | 38.5 | 2 | 19.3 | ||||||
| 4 | 449.1 | 16 | 28.12 | ||||||
| 5 | 15.5 | 1 | 15.5 | ||||||
| 6 | 195.0 | 6 | 32.5 | ||||||
| 7 | 116.5 | 4* | 29.1 | ||||||
| K562 E | (2 ×) 5 | 93.5 | 8.90 | 9.5 | 251.2 ± 247.9 (19×) | 7.6 ± 5.5 (1-15) | (31.2) | ||
| 2 | 301.4 | 11 | 27.4 | ||||||
| 4 | 85.0 | 6 | 14.2 | ||||||
| 9 | 179.4 | 5* | 35.9 | ||||||
| 10 | 655.8 | 15* | 43.7 | ||||||
| 11 | 34.6 | 1 | 34.6 | ||||||
| K562 F | (2 ×) 20 | 89.6 | 9.07 | 10.1 | 752.9 ± 787.1 (17×) | > 16.0 ± 10.0 (4->31) | (< 37.1) | ||
| 1 | 860.3 | 19 | 35.8 | ||||||
| 5 | 367.5 | 14 | 26.2 | ||||||
| 8 | 339.1 | 13* | 26.1 | ||||||
| 10 | 2075.3 | > 31† | < 66.9 | ||||||
| 12 | 122.1 | 4 | 30.5 | ||||||
| CD34 A | 0.2 | 4.7 | 0.04 | 0.9 | ND | NA | ND | NA | NA |
| CD34 B | 1 | 31.6 | 0.39 | 1.2 | ND | NA | 1.6 ± 0.5 (1-2) | NA | |
| 6 | 2 | ||||||||
| 8 | 1 | ||||||||
| 10 | 1 | ||||||||
| 17 | 2 | ||||||||
| 18 | 2 | ||||||||
| CD34 C | 2 | 42.5 | 0.92 | 2.2 | ND | NA | 2.0 ± 1.2 (1-4) | NA | |
| 1 | 2 | ||||||||
| 6 | 2 | ||||||||
| 12 | 1 | ||||||||
| 13 | 1 | ||||||||
| 16 | 4 | ||||||||
| CD34 D | 4 | 58.7 | 1.01 | 1.7 | ND | NA | 2.0 ± 1.0 (1-3) | NA | |
| 3 | 2 | ||||||||
| 4 | 1 | ||||||||
| 6 | 1 | ||||||||
| 9 | 3 | ||||||||
| 12 | 3 | ||||||||
| CD34 E | (2 ×) 2 | 52.8 | 1.79 | 3.4 | ND | NA | 3.0 ± 1.2 (2-5) | NA | |
| 4 | 3 | ||||||||
| 5 | 3 | ||||||||
| 11 | 2 | ||||||||
| 16 | 5 | ||||||||
| 18 | 2 | ||||||||
| CD34 F | (2 ×) 4 | 67.2 | 2.79 | 4.2 | ND | NA | 6.0 ± 8.5 (1-21) | NA | |
| 1 | 4 | ||||||||
| 3 | 2 | ||||||||
| 6 | 2 | ||||||||
| 11 | 1 | ||||||||
| 12 | 21 |
Cell groups and clones . | MOI (× no. of preloading cycles) . | Gene transfer, % . | Vector copy no., real-time PCR . | Vectors/transduced cell, calculated . | MF, arbitrary units . | Average MF ± SD (variation factor)‡ . | Vector copies, Southern/qPCR . | Mean copy no./transduced cell ± SD (range)‡ . | MF/vector copy, arbitrary units (mean) . |
|---|---|---|---|---|---|---|---|---|---|
| K562 A | 0.5 | 28.6 | 0.24 | 0.9 | 33.3 ± 8.9 (1.9×) | 1.0 ± 0.0 (1) | (33.3) | ||
| 1 | 40.9 | 1 | 40.9 | ||||||
| 5 | 43.6 | 1* | 43.6 | ||||||
| 6 | 23.7 | 1* | 23.7 | ||||||
| 8 | 32.5 | 1 | 32.5 | ||||||
| 9 | 25.6 | 1 | 25.6 | ||||||
| K562 B | 2.5 | 60.9 | 1.78 | 2.9 | 43.5 ± 28.5 (6.5×) | 2.0 ± 0.8 (1-3) | (20.2) | ||
| 1 | 22.9 | 2* | 11.5 | ||||||
| 7 | 44.7 | 2 | 22.4 | ||||||
| 8 | 86.8 | 3 | 28.9 | ||||||
| 9 | 13.4 | 1 | 13.4 | ||||||
| 10 | 49.8 | 2 | 24.9 | ||||||
| K562 C | 5 | 78.0 | 2.89 | 3.7 | 162.0 ± 121.9 (14.2×) | 5.4 ± 3.7 (1-9) | (28.1) | ||
| 4 | 21.7 | 1 | 21.7 | ||||||
| 5 | 177.1 | 7* | 25.3 | ||||||
| 6 | 57.1 | 2* | 28.6 | ||||||
| 10 | 245.2 | 8 | 30.6 | ||||||
| 15 | 308.9 | 9 | 34.3 | ||||||
| K562 D | 20 | 70.3 | 6.27 | 8.9 | 162.9 ± 174.9 (29×) | 5.8 ± 6.0 (1-16) | (24.9) | ||
| 1 | 38.5 | 2 | 19.3 | ||||||
| 4 | 449.1 | 16 | 28.12 | ||||||
| 5 | 15.5 | 1 | 15.5 | ||||||
| 6 | 195.0 | 6 | 32.5 | ||||||
| 7 | 116.5 | 4* | 29.1 | ||||||
| K562 E | (2 ×) 5 | 93.5 | 8.90 | 9.5 | 251.2 ± 247.9 (19×) | 7.6 ± 5.5 (1-15) | (31.2) | ||
| 2 | 301.4 | 11 | 27.4 | ||||||
| 4 | 85.0 | 6 | 14.2 | ||||||
| 9 | 179.4 | 5* | 35.9 | ||||||
| 10 | 655.8 | 15* | 43.7 | ||||||
| 11 | 34.6 | 1 | 34.6 | ||||||
| K562 F | (2 ×) 20 | 89.6 | 9.07 | 10.1 | 752.9 ± 787.1 (17×) | > 16.0 ± 10.0 (4->31) | (< 37.1) | ||
| 1 | 860.3 | 19 | 35.8 | ||||||
| 5 | 367.5 | 14 | 26.2 | ||||||
| 8 | 339.1 | 13* | 26.1 | ||||||
| 10 | 2075.3 | > 31† | < 66.9 | ||||||
| 12 | 122.1 | 4 | 30.5 | ||||||
| CD34 A | 0.2 | 4.7 | 0.04 | 0.9 | ND | NA | ND | NA | NA |
| CD34 B | 1 | 31.6 | 0.39 | 1.2 | ND | NA | 1.6 ± 0.5 (1-2) | NA | |
| 6 | 2 | ||||||||
| 8 | 1 | ||||||||
| 10 | 1 | ||||||||
| 17 | 2 | ||||||||
| 18 | 2 | ||||||||
| CD34 C | 2 | 42.5 | 0.92 | 2.2 | ND | NA | 2.0 ± 1.2 (1-4) | NA | |
| 1 | 2 | ||||||||
| 6 | 2 | ||||||||
| 12 | 1 | ||||||||
| 13 | 1 | ||||||||
| 16 | 4 | ||||||||
| CD34 D | 4 | 58.7 | 1.01 | 1.7 | ND | NA | 2.0 ± 1.0 (1-3) | NA | |
| 3 | 2 | ||||||||
| 4 | 1 | ||||||||
| 6 | 1 | ||||||||
| 9 | 3 | ||||||||
| 12 | 3 | ||||||||
| CD34 E | (2 ×) 2 | 52.8 | 1.79 | 3.4 | ND | NA | 3.0 ± 1.2 (2-5) | NA | |
| 4 | 3 | ||||||||
| 5 | 3 | ||||||||
| 11 | 2 | ||||||||
| 16 | 5 | ||||||||
| 18 | 2 | ||||||||
| CD34 F | (2 ×) 4 | 67.2 | 2.79 | 4.2 | ND | NA | 6.0 ± 8.5 (1-21) | NA | |
| 1 | 4 | ||||||||
| 3 | 2 | ||||||||
| 6 | 2 | ||||||||
| 11 | 1 | ||||||||
| 12 | 21 |
CD34+ and K562 cells were transduced with SFα11eGFP17 at different MOIs (column 2). In columns 3 to 5, data for mass culture infections are given. Results represent 1 of 2 nearly identical duplicates. Five clones were derived from each mass culture (column 1) and subsequently analyzed for transgene expression (columns 6 and 7) and insertion (columns 8 and 9). See “Study design” regarding calculations of the given MOIs. The variation factors in the seventh column represent the variation factor between the lowest and the highest value in a given group. MF indicates mean fluorescence; qPCR, quantitative PCR; ND, not determined; and NA, not applicable.
For the 9 K562 clones, quantitative data were obtained by Southern blot analysis and real-time PCR, and a standard curve was generated allowing us to determine transgene copy numbers in the mass cultures
For clone 10 in K562 group F, the copy number was above the resolution limit of the Southern blot
Values in this column were calculated for the 5 clones shown in each cell group
Measuring gene transfer and vector copy numbers
Results and discussion
Data summarized in Table 1 show the expected linear relationship17,18 between MOI and vector copy number for primary CD34+ (groups A-D) and K562 cells (groups A-C). However, the increase in the number of transduced cells was linear (in relation to the MOI) only up to approximately 30% of gene transfer. Moreover, once maximal transduction was reached, further increasing the number of infectious vector particles per cell did not lead to an elevated number of transduced cells but only to much higher transgene copy numbers per cell (eg, K562 group C vs group D). As a consequence, raising the MOI under these circumstances would not only be useless, it would potentially be dangerous. Instead, to increase transduction rates (up to 68% for CD34+ and 93% for K562 cells) a second infection cycle had to be performed (groups E and F for both).
Nonlinear interdependence between transduction efficiency with MLV vectors and the applied MOI (Table 1) is based on an important feature of the original virus: provirus integration occurs during mitosis, after chromosomal duplication.2,19,20 In the presence of limiting amounts of infectious particles, only 1 of the 2 daughter cells will thus bear a virus copy, resulting in a theoretical maximal gene transfer of 50% with an MOI of 1. However, because the interaction of viral particles and target cells occurs by chance, the statistical use of 1 infectious vector copy per cell results in a gene transfer rate of approximately 30% to 35% with 1.4 to 1.7 vector copies per transduced cell and a Poisson-like distribution of insertion numbers (usually not exceeding 3). Consequently, any linear increase of gene transfer should be accompanied by an exponential increase of insertion numbers. This problem becomes even more complex if the target cells are not equally susceptible to infection at a given time. Indeed, we observed a mean of more than 9 vector copies per transduced K562 cells after 2 infection cycles in cell populations showing approximately 90% transgene expression (Table 1). Similarly, Brenner et al21 recently reported average copy numbers of approximately 8 for 90% gene transfer into primary CD34+ cells.
To estimate the number of insertions in single-cell-derived clones, transduced cells from all groups were subjected to limiting dilution (K562) or methyl cellulose (CD34+) cloning, and 5 randomly chosen EGFP-positive clones per group were further analyzed by transgene-specific, real-time PCR, Southern blot analysis, or both.
As evident from Table 1, data obtained with single-cell-derived clones are generally in good agreement with vector copy numbers in the corresponding mass culture. However, with increased transduction rates, the analysis of single-cell-derived clones revealed strong differences in copy numbers among clones (Table 1; Figure 1). Therefore, transduction at high MOI may strongly influence the outcome of functional studies by elevating transgene expression in mass cultures17 and creating clones with multiple transgene copies (K562 clone 10/F, more than 31 copies; CD34 clone 12/F, 21 copies), which might be far from normal. Recently, of CD34+ mass cultures transduced to 90% (average copy number, 8), only those CD34+ cells with limited vector copy number (mean, 2) were capable of repopulating NOD-SCID mice, which probably reflects receptor or postreceptor restrictions (eg, cell-cycle status) to gene transfer in true stem cells.21 However, a direct genotoxic impact of multiple vector insertions on cell survival (compare K562 groups C vs D, E vs F) cannot be excluded18 and requires urgent investigation, considering that preinsertion blocks will soon be overcome (as with lentiviral technology).22
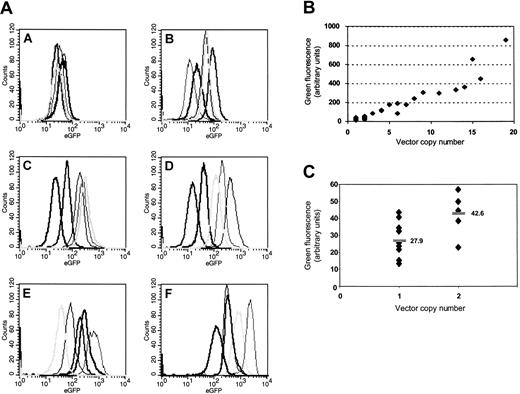
Striking increase of transgene expression and variability of gene expression and copy number depending on the MOI. Five clones selected by chance after limiting-dilution cloning of K562 cells (Table 1) were analyzed for each group (A-F). As shown in the histogram plots (panel A), both the level and the variability of transgene expression show a strong increase along with an elevated MOI. Analysis of mean fluorescence values reveals that (panel B) there is a clear positive correlation of insertion numbers and gene expression. Note that clone F10 has been excluded from this figure because the number of insertions above the resolution limit of the Southern blot analysis was obviously underestimated. A detailed analysis for all clones bearing 1 or 2 insertions (panel C) indicates that clonal variability is limited (approximately 3.2 ×) in clones with single insertions, and the mean expression level (horizontal bars) nearly doubles in the presence of 2 vector copies.
Striking increase of transgene expression and variability of gene expression and copy number depending on the MOI. Five clones selected by chance after limiting-dilution cloning of K562 cells (Table 1) were analyzed for each group (A-F). As shown in the histogram plots (panel A), both the level and the variability of transgene expression show a strong increase along with an elevated MOI. Analysis of mean fluorescence values reveals that (panel B) there is a clear positive correlation of insertion numbers and gene expression. Note that clone F10 has been excluded from this figure because the number of insertions above the resolution limit of the Southern blot analysis was obviously underestimated. A detailed analysis for all clones bearing 1 or 2 insertions (panel C) indicates that clonal variability is limited (approximately 3.2 ×) in clones with single insertions, and the mean expression level (horizontal bars) nearly doubles in the presence of 2 vector copies.
Finally, there was a clear positive correlation between transgene expression and insertion frequency in K562 clones (Figure 1), which is consistent with a limited variability of transgene expression depending on the individual insertion site. Indeed, variations in expression levels per single integration were in the range of less than 4 (groups A, B; Figure 1; Table 1). Thus, although dependent on insertion sites, retrovirus-mediated expression may be less variable than observed with many physicochemical transfection methods.23
In conclusion, generating cell clones with multiple vector insertions after high-efficiency retroviral gene transfer may lead to erroneous predictions of transgene efficiency and toxicity. Moreover, those clones might be at a higher risk for malignant transformation because the frequency of insertional mutagenesis may directly correlate with the number of integrated vector copies.6,7 Consequently, transduction protocols should be evaluated for transgene copy numbers obtained in single cell clones, even if that risk is lower in mature cells or with alternative, safety-improved vector systems. If a target population contains a fraction of cells that is more resistant to retroviral (including lentiviral)22 transduction, a further increase of the MOI cannot be recommended. Rather, procedures for posttransduction cell sorting and clonal selection in vivo must be elaborated with even stronger emphasis.
Prepublished online as Blood First Edition Paper, July 24, 2003; DOI 10.1182/blood-2003-05-1424.
Supported by the European Community (grant QLK3-2001-01265). A.W. is supported by the Deutsches Krebsforschungszentrum (grant Ca 97).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ”advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dung Nguyen for expert technical assistance and Francis A. Ayuk for critical reading.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal