Abstract
Interleukin-10 (IL-10) plays an important role in prevention of chronic inflammation in vivo. However, the molecular mechanism by which IL-10 exerts its anti-inflammatory response is poorly understood. Here, we performed a microarray analysis and identified Bcl-3 as an IL-10-inducible gene in macrophages. Lentiviral vector-mediated expression of Bcl-3 inhibited lipopolysaccharide (LPS)-induced production of tumor necrosis factor α (TNF-α), but not IL-6, in macrophages. In Bcl-3-transduced and IL-10-pretreated macrophages, LPS-induced nuclear translocation of nuclear factor κB (NF-κB) p65 was not impaired. However, DNA binding by NF-κB p50/p65 was profoundly inhibited. Nuclear localization of Bcl-3 was associated with inhibition of LPS-induced TNF-α production. Overexpression of Bcl-3 suppressed activation of the TNF-α promoter, but not the IL-6 promoter. Bcl-3 interacted with NF-κB p50 and was recruited to the TNF-α promoter, but not the IL-6 promoter, indicating that Bcl-3 facilitates p50-mediated inhibition of TNF-α expression. Furthermore, Bcl-3-deficient macrophages showed defective IL-10-mediated suppression of LPS induction of TNF-α, but not IL-6. These findings suggest that IL-10-induced Bcl-3 is required for suppression of TNF-α production in macrophages. (Blood. 2003; 102:4123-4129)
Introduction
Interleukin-10 (IL-10) is produced by activated macrophages and T cells, and plays an important role in anti-inflammatory responses.1 Indeed, mice lacking IL-10 or the IL-10 receptor developed chronic enterocolitis and showed enhanced inflammatory responses.2-4 Furthermore, transfer of the IL-10 gene reduced the incidence of colitis in mice, and treatment with IL-10 is now anticipated to be used clinically for patients with inflammatory bowel diseases.5,6 The anti-inflammatory activity of IL-10 is mainly elicited by inhibition of the activity of macrophages/monocytes. IL-10 inhibits lipopolysaccharide (LPS)-induced production of inflammatory cytokines, including tumor necrosis factor α (TNF-α), IL-6, and IL-12, by macrophages. Many attempts have been made to elucidate the mechanism by which IL-10 inhibits cytokine production by macrophages, and several models have been proposed. These include inhibition of nuclear factor κB (NF-κB) and mitogen-activated protein (MAP) kinase activity, and reduction of transcription and stability of mRNA for cytokine genes.7-10 However, the precise mechanism by which IL-10 suppresses macrophage activity remains unclear.
The IL-10 receptor consists of the ligand binding subunit, IL-10R1, and an accessory subunit, IL-10R2. Binding of IL-10 to the IL-10 receptor induces activation of the Janus kinase (JAK) family of tyrosine kinases, Jak1 and Tyk2, which are constitutively associated with IL-10R1 and IL-10R2, respectively.1 Jak1 and Tyk2 activation is followed by phosphorylation of the latent transcription factor signal transducer and activator of transcription 3 (Stat3). Studies with gene-targeted mice have revealed that Jak1 and Stat3 play a pivotal role in the IL-10 signaling pathway. Macrophages from Jak1-deficient mice did not show an anti-inflammatory response to IL-10.11,12 Mice in which Stat3 was specifically deleted in the myeloid lineage cells, including macrophages, developed chronic enterocolitis. Macrophages from these mutant mice were unresponsive to IL-10.13 Furthermore, a recent study with additional gene deletions demonstrated that overproduction of IL-12p40 from Stat3-deficient macrophages caused enhanced Th1 responses leading to the development of Th1-dependent chronic enterocolitis.14 These studies indicate that macrophages are the main target cells of IL-10, which exhibits anti-inflammatory responses in vivo. Several studies have indicated that de novo protein synthesis is required for IL-10 inhibition of LPS-induced cytokine production.9,15,16 Therefore, genes induced by IL-10-mediated Stat3 activation in macrophages are expected to be responsible for anti-inflammatory responses. In this context, several IL-10-inducible genes, such as SOCS3 and heme oxygenase-1, have been identified.17-19 However, it is still controversial how these gene products contribute to anti-inflammatory responses.
In this study, we addressed the mechanism by which IL-10 inhibits LPS-induced TNF-α production in macrophages. A gene array analysis led to the identification of several IL-10 target genes, including Bcl-3, in macrophages. Introduction of these genes into macrophages by lentiviral vectors revealed that Bcl-3 inhibited LPS-induced TNF-α production. Bcl-3 was associated with NF-κB p50 and preferentially recruited to the TNF-α promoter. Macrophages from Bcl-3-deficient mice were defective in IL-10-mediated suppression of TNF-α production. Thus, Bcl-3 induced by IL-10 is responsible for inhibition of LPS-induced TNF-α production in macrophages.
Materials and methods
Reagents and cell culture
LPS from Escherichia coli (O55:B5) was purchased from Sigma (St Louis, MO). The mouse macrophage cell line (RAW264.7) and human embryonic kidney cells (293T) were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin, and 10 U/mL penicillin G. Mouse peritoneal macrophages were collected by peritoneal lavage with Hanks balanced salt solution at 3 days after intraperitoneal injection of 2 mL of 4% sterile thioglycollate into 8- to 12-week-old mice. Peritoneal macrophages were cultured in RPMI 1640 medium with 10% FBS, 100 μg/mL streptomycin, and 10 U/mL penicillin G.
Mice
The generation of myeloid cell-specific Stat3-deficient mice was described previously.13 IL-10- and Bcl-3-deficient mice were purchased from The Jackson Laboratory (Bar Harbor, ME).
Affymetrix gene expression profiling
RAW264.7 and peritoneal macrophages were treated with IL-10 (10 ng/mL) for 2, 4, or 6 hours. Total RNA was extracted with an RNeasy kit (Qiagen, Hilden, Germany), followed by mRNA purification with an Oligotex mRNA Kit (Pharmacia, Piscataway, NJ). Double-stranded cDNA was synthesized from 1 μg mRNA with the SuperScript Choice System (Invitrogen, Carlsbad, CA) primed with T7-(dT) 24 primer. These cDNAs were used to prepare biotin-labeled cRNA by an in vitro transcription reaction performed using T7 RNA polymerase in the presence of biotinylated-ribonucleotides, according to the manufacturer's protocol (Enzo Diagnostics, Farmingdale, NY). The cRNA product was purified using an RNeasy kit (Qiagen), fragmented, and hybridized to Affymetrix Murine Genome U74Av2, Bv2, and Cv2 microarray chips, according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). The hybridized chips were stained, washed, and scanned with a GeneArray Scanner (Affymetrix).
Electrophoretic mobility shift assay (EMSA)
Macrophages were treated with 10 ng/mL IL-10 for 18 hours, and stimulated with 100 ng/mL LPS for 60 minutes. Then, nuclear proteins were extracted, and then incubated with an end-labeled, double-stranded oligonucleotide containing a NF-κB binding site of the TNF-α promoter in 25 μL binding buffer (10 mM HEPES-KOH [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid-potassium hydroxide, pH 7.8], 50 mM KCl, 1 mM EDTA [ethylenediaminetetraacetic acid, pH 8.0], 5 mM MgCl2, and 10% glycerol) for 20 minutes at room temperature and loaded on a native 5% polyacrylamide gel. The DNA-protein complexes were visualized by autoradiography. The specificities of the shifted bands were determined by adding antibodies specific for p65 and p50 (Santa Cruz Biotechnology, Santa Cruz, CA).
Production of lentiviral vector and infection
The lentiviral vector, CSII-EF-MCS-IRES-hrGFP (cPPT-containing SIN vector plasmid containing multicloning sites for cDNA insertion followed by the internal ribosomal entry site-green fluorescent protein [IRES-GFP] sequence under the control of the EF-1α promoter), was used to generate CSII-EF-Bcl-3 and CSII-EF-SOCS3. Woodchuck hepatitis virus posttranslational regulatory element (PRE) was ligated at the 3′ end of GFP. The lentiviral vectors were cotransfected into 293T cells with pMDLg/p RRE (packaging plasmid), pRSV-Rev (Rev expression plasmid), and pMD.G (VSV-G expression plasmid). Infectious lentiviruses in the culture supernatants were harvested at 48 hours after transfection. RAW cells (5 × 105) were cultured with the lentiviruses for 24 hours and then the culture medium was replaced. The transduction efficiency was monitored by GFP expression.
Flow cytometric analysis
Cells were stimulated with 100 ng/mL LPS, harvested, and incubated with 4% paraformaldehyde. The cells were then incubated with a phycoerythrin (PE)-conjugated anti-TNF-α antibody (Ab; PharMingen, San Diego, CA). Stained cells were analyzed in a fluorescence-activated cell sorter (FACS) Calibur using the Cell Quest software (Becton Dickinson, San Jose, CA).
Measurement of cytokines and NO in culture supernatants
RAW cells and peritoneal macrophages (5 × 104) were cultured in 96-well plates with 30 ng/mL interferon γ (IFNγ) and 10 ng/mL LPS for 24 hours. The concentrations of TNF-α and IL-6 in the culture supernatants were determined by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (Genzyme Techne, Minneapolis, MN). The concentration of nitric oxide (NO) was measured using Griess reagents according to the manufacturer's instructions (Dojindo, Kumamoto, Japan).
Luciferase assay
Mouse Bcl-3 expression constructs were cloned into the pEF-BOS mammalian expression vector. The TNFα and IL-6 promoters containing genomic fragments from -1260 to +140 and -1232 to +39, respectively, were cloned into the pGL3-basic vector (Promega, Madison, WI). RAW cells were transfected with expression constructs using the Superfect transfection reagent (Qiagen). The luciferase activities of total cell lysates were measured using the Dual-luciferase reporter assay system (Promega). The Renilla-luciferase reporter gene (50 ng) was used as an internal control. In all cases, the data were normalized for transfection efficiency by dividing the firefly luciferase by that of the Renilla luciferase.
Immunoprecipitation assays
For immunoprecipitation assay, Myc-tagged Bcl-3 and Flag-tagged p50, p52 were cloned into pEF-BOS and pcDNA3.1, respectively. These expression vectors were transfected into 293 cells using Lipofectoamine 2000 (Invitrogen). Transfected cells were solubilized with 1 mL radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.5% Na-deoxycholate, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 mg/mL each leupeptin and pepstatin). Whole cell lysates were incubated with antibodies to Myc-Tag 9B11 (Cell Signaling Technology, Beverly, MA), anti-Flag M2 antibody (Sigma). Immunoprecipitates were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride (PVDF) membrane, and incubated with anti-Myc tag antibody (A-14; Santa Cruz Biotechnology) or anti-Flag M2 antibody. Bound antibodies were visualized with an enhanced chemiluminescence system (Perkin Elmer, Boston, MA).
Immunofluorescence staining and confocal microscopy
Cells on 3.5-cm dishes were washed with Tris-buffered saline (TBS) and fixed with 3.7% formaldehyde in TBS for 15 minutes at room temperature. After permeabilization with 0.2% Triton X-100, cells were washed with TBS and incubated with a rabbit anti-Bcl-3 Ab (Santa Cruz Biotechnology) in TBS containing 1% bovine serum albumin at 10 ng/mL, followed by incubation with Alexa Fluor 594-conjugated goat anti-rabbit immunoglobulin G (IgG; Molecular Probes, Eugene, OR). To stain the nucleus, cells were cultured with 0.5 μg 4′,6-diamidino-2-phenylindole (DAPI; Wako, Osaka, Japan). Confocal microscopy was performed using an LSM510 model confocal microscope (Carl Zeiss, Oberkochem, Germany).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed essentially with a described protocol (Upstate Biotechnology, Lake Placid, NY). In brief, RAW cells were transfected with Myc-tagged Bcl-3 and Flag-tagged p50 expression vectors using Nucleofector (Amaxa, Germany). At 48 hours after transfection, cells were stimulated with 100 ng/mL LPS for 2 hours and then fixed with formaldehyde for 10 minutes. The cells were lysed, sheared by sonication, and incubated overnight with specific antibody followed by incubation with protein A-agarose saturated with salmon sperm DNA (Upstate Biotechnology). Precipitated DNAs were analyzed by quantitative polymerase chain reaction (PCR, 35 cycles) using primers 5′-ACTAGCCAGGAGGGAGAACAGAAACTC-3′ and 5′-CACAAGCAGGAATGAGAAGAGGCTGAG-3′ for the TNF-α promoter and 5′-TAGCAGCAGGTCCAACTGTGCTATCTG-3′ and 5′-AAGCCTCCGACTTGTGAAGTGGTATAG-3′ for the IL-6 promoter.
Results
Identification of IL-10-inducible genes in macrophages
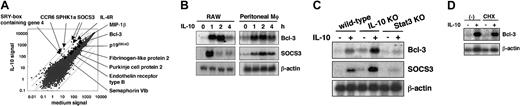
In order to identify IL-10-inducible genes that are responsible for the inhibition of LPS-induced cytokine production, a microarray analysis was performed. Thioglycollate-elicited mouse peritoneal macrophages and RAW264.7 cells were treated with 10 ng/mL IL-10 for 2, 4, and 6 hours, and gene induction was analyzed. This analysis resulted in the identification of several IL-10-inducible genes, including SOCS3, p19INK4D, and the IL-4 receptor α, which were all shown previously to be induced by IL-10 in macrophages.18-20 Among these IL-10-inducible genes, we found that expression of Bcl-3 was induced 3-fold in both RAW264.7 cells and peritoneal macrophages (Figure 1A, data not shown). We confirmed the IL-10-induced mRNA expression of Bcl-3 and SOCS3 in RAW264.7 cells and peritoneal macrophages by Northern blotting (Figure 1B). Both Bcl-3 and SOCS3 mRNA expression were rapidly induced within one hour of IL-10 treatment. In peritoneal macrophages from IL-10-deficient mice, IL-10-induced mRNA expression of Bcl-3 and SOCS3 was enhanced compared with wild-type macrophages. However, IL-10-induced Bcl-3 and SOCS3 expression was completely abolished in Stat3-deficient macrophages, indicating that the IL-10-induced expression of both genes is dependent on the Stat3-mediated pathway (Figure 1C). Addition of cycloheximide did not block the induction of either Bcl-3 or SOCS3 mRNA, indicating that protein synthesis is not required for the IL-10 induction of these genes (Figure 1D).
Microarray analysis of IL-10-inducible genes in macrophages. (A) Genes induced by IL-10 in peritoneal macrophages. Representative results in peritoneal macrophages treated with 10 ng/mL IL-10 for 4 hours are shown in the scatter plot. Similar results were observed in peritoneal macrophages treated with IL-10 for 2 hours, and RAW cells treated for 2 hours and 4 hours. (B) RAW cells and peritoneal macrophages were cultured in the absence or presence of 10 ng/mL IL-10 for the indicated periods. Total RNA was extracted and analyzed by a Northern blot using 32P-labeled Bcl-3 and SOCS3. Hybridization with a β-actin probe confirmed even loading of RNA in each lane. Data are representative of 3 independent experiments. (C) Peritoneal macrophages isolated from wild-type, IL-10-deficient, and Stat3-mutant mice were treated with 10 ng/mL IL-10 for 2 hours. Total RNA was extracted and analyzed for Bcl-3 and SOCS3 expression by Northern blotting. KO indicates knock-out. (D) Peritoneal macrophages isolated from wild-type mice were treated with 10 ng/mL IL-10 for 2 hours in the presence or absence of 1 μg/mL cycloheximide (CHX) for 0.5 hours. Samples were analyzed by Northern blotting as in panel B.
Microarray analysis of IL-10-inducible genes in macrophages. (A) Genes induced by IL-10 in peritoneal macrophages. Representative results in peritoneal macrophages treated with 10 ng/mL IL-10 for 4 hours are shown in the scatter plot. Similar results were observed in peritoneal macrophages treated with IL-10 for 2 hours, and RAW cells treated for 2 hours and 4 hours. (B) RAW cells and peritoneal macrophages were cultured in the absence or presence of 10 ng/mL IL-10 for the indicated periods. Total RNA was extracted and analyzed by a Northern blot using 32P-labeled Bcl-3 and SOCS3. Hybridization with a β-actin probe confirmed even loading of RNA in each lane. Data are representative of 3 independent experiments. (C) Peritoneal macrophages isolated from wild-type, IL-10-deficient, and Stat3-mutant mice were treated with 10 ng/mL IL-10 for 2 hours. Total RNA was extracted and analyzed for Bcl-3 and SOCS3 expression by Northern blotting. KO indicates knock-out. (D) Peritoneal macrophages isolated from wild-type mice were treated with 10 ng/mL IL-10 for 2 hours in the presence or absence of 1 μg/mL cycloheximide (CHX) for 0.5 hours. Samples were analyzed by Northern blotting as in panel B.
Lentiviral transduction of Bcl-3 into macrophages results in reduced LPS-induced TNF-α production
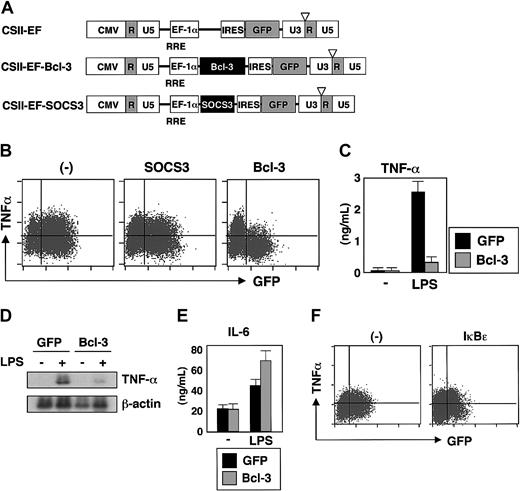
We next tried to transfer these IL-10-inducible genes into macrophages. Since macrophages are not susceptible to most gene-transfer methods, we tested lentiviral vectors that are capable of transducing nondividing cells.21-23 To assess the transduction efficiency, the CSII-EF lentiviral vector containing the green fluorescent protein (GFP) gene was used to transduce several types of macrophages and efficient transduction was achieved (GFP-positive cells represented more than 80% of RAW cells, 50% of bone marrow-derived macrophages, and 30% of peritoneal macrophages). We then constructed lentiviral vectors containing Bcl-3 or SOCS3 (Figure 2A) and transduced them into RAW264.7 cells and bone marrow-derived macrophages. The lentiviral vector containing only the GFP (CSII-EF) was used as a control in all experiments. RAW264.7 cells and bone marrow-derived macrophages were infected with lentivirus, and after 2 days of culture, cells were stimulated with LPS and analyzed for TNF-α production by intracellular staining. As shown in Figure 2B, neither lentiviral infection alone nor with SOCS3 caused any change in the LPS-induced TNF-α production in RAW264.7 cells. In contrast, expression of Bcl-3 resulted in a dramatic decrease in the number of cells producing TNF-α (Figure 2B). Similar findings were also observed in bone marrow macrophages (data not shown). Next, GFP-positive cells were isolated by FACS and stimulated with LPS, and the production of TNF-α in the culture supernatants was analyzed (Figure 2C). Cells expressing GFP alone secreted significant amounts of TNF-α in response to LPS. However, LPS-induced secretion of TNF-α was severely reduced in cells transduced with Bcl-3/GFP. We also analyzed the LPS-induced mRNA expression of TNF-α (Figure 2D). Introduction of Bcl-3/GFP severely impaired LPS induction of TNF-α mRNA. Thus, lentiviral expression of Bcl-3 in macrophages inhibited the LPS-induced production of TNF-α. However, in RAW264.7 cells transduced with Bcl-3/GFP, LPS-induced IL-6 production was not reduced, indicating that the expression of Bcl-3 did not result in the impairment of all LPS responses in macrophages (Figure 2E). We next lentivirally expressed IκBϵ, an inhibitor of κBϵ, that is structurally related to Bcl-3.24 Lentiviral expression of IκBϵ in RAW 264.7 cells did not inhibit LPS-induced TNF-α production, indicating that Bcl-3 is specifically involved in the inhibition of LPS-induced TNF-α production in macrophages (Figure 2F).
Lentiviral introduction of Bcl-3 results in reduced LPS-induced TNF-α production in macrophages. (A) Schematic drawings of the lentivirus vector containing a murine Bcl-3 or SOCS3 gene cassette. (B) RAW cells were infected with lentivirus expressing Bcl-3/GFP or SOCS3/GFP. After infection for 24 hours, RAW cells were washed and additionally incubated for 2 days. The cells were then stimulated with LPS (10 ng/mL) for 6 hours, intracellularly stained for TNF-α, and analyzed by flow cytometry. (C) GFP-positive cells were collected with FACS, and stimulated with IFN-γ (30 ng/mL) and LPS (10 ng/mL) for 24 hours. The concentration of TNF-α in the culture supernatants was determined by ELISA. Error bars indicate SD. (D) GFP-positive cells (GFP alone and Bcl-3/GFP) were purified and stimulated with LPS (100 ng/mL) for 4 hours, and then total RNA extracts were analyzed for the mRNA expression of TNF-α. Hybridization with the β-actin probe confirmed even loading of RNA in each lane. (E) GFP-positive cells were stimulated with IFN-γ (30 ng/mL) and LPS (10 ng/mL) for 24 hours. The IL-6 concentration in the culture supernatants was measured by ELISA. Error bars indicate SD. (F) RAW cells were infected with a lentivirus expressing IκBϵ/GFP. After infection for 24 hours, RAW cells were washed and additionally incubated for 2 days. The cells were then stimulated with LPS (10 ng/mL) for 6 hours and analyzed for intracellular production of TNF-α by flow cytometry.
Lentiviral introduction of Bcl-3 results in reduced LPS-induced TNF-α production in macrophages. (A) Schematic drawings of the lentivirus vector containing a murine Bcl-3 or SOCS3 gene cassette. (B) RAW cells were infected with lentivirus expressing Bcl-3/GFP or SOCS3/GFP. After infection for 24 hours, RAW cells were washed and additionally incubated for 2 days. The cells were then stimulated with LPS (10 ng/mL) for 6 hours, intracellularly stained for TNF-α, and analyzed by flow cytometry. (C) GFP-positive cells were collected with FACS, and stimulated with IFN-γ (30 ng/mL) and LPS (10 ng/mL) for 24 hours. The concentration of TNF-α in the culture supernatants was determined by ELISA. Error bars indicate SD. (D) GFP-positive cells (GFP alone and Bcl-3/GFP) were purified and stimulated with LPS (100 ng/mL) for 4 hours, and then total RNA extracts were analyzed for the mRNA expression of TNF-α. Hybridization with the β-actin probe confirmed even loading of RNA in each lane. (E) GFP-positive cells were stimulated with IFN-γ (30 ng/mL) and LPS (10 ng/mL) for 24 hours. The IL-6 concentration in the culture supernatants was measured by ELISA. Error bars indicate SD. (F) RAW cells were infected with a lentivirus expressing IκBϵ/GFP. After infection for 24 hours, RAW cells were washed and additionally incubated for 2 days. The cells were then stimulated with LPS (10 ng/mL) for 6 hours and analyzed for intracellular production of TNF-α by flow cytometry.
Nuclear expression of Bcl-3 is required for inhibition of LPS-induced TNF-α production
In accordance with previous reports, expression of Bcl-3 was predominantly observed in the nucleus in Bcl-3/GFP-transduced RAW cells25-27 (Figure 3B, upper). We addressed whether this nuclear localization of Bcl-3 was associated with the inhibition of LPS responses. Bcl-3 is composed of 7 central ankyrin repeats, which are essential for interaction with NF-κB p50 and p52 homodimers, and an N-terminal basic nuclear localization sequence (NLS).28,29 We generated deletion mutants that lacked the N-terminal NLS (ΔN) or the N-terminal 154 residues including the first ankyrin repeat (ΔANK) and lentivirally introduced these mutants into RAW cells (Figure 3A). Immunofluorescent staining with an anti-Bcl-3 Ab detecting the C-terminal portion of the Bcl-3 protein showed that ΔN and ΔANK were not localized in the nucleus but expressed in the cytoplasm (Figure 3B). When LPS-induced TNF-α production was analyzed, cells transduced with ΔN/GFP and ΔANK/GFP did not show any reduction in TNF-α production (Figure 3C). Thus, the nuclear localization of Bcl-3 was associated with its inhibitory effect on TNF-α production.
Nuclear-localized Bcl-3 inhibits the LPS response in macrophages. (A) Schematic structures of the deletion mutants of Bcl-3. ΔN lacks the N-terminal nuclear localization signal. ΔANK lacks the N-terminal 154 residues including the first ankyrin repeat. (B) Lentivirus-infected RAW cells (green) were stained with anti-Bcl-3 (red) and DAPI (blue), and analyzed by confocal microscopy. Scale bar: 2 μm. (C) Lentivirus-infected RAW cells were stimulated with LPS (10 ng/mL) for 6 hours, stained with anti-TNF-α, and analyzed by flow cytometry.
Nuclear-localized Bcl-3 inhibits the LPS response in macrophages. (A) Schematic structures of the deletion mutants of Bcl-3. ΔN lacks the N-terminal nuclear localization signal. ΔANK lacks the N-terminal 154 residues including the first ankyrin repeat. (B) Lentivirus-infected RAW cells (green) were stained with anti-Bcl-3 (red) and DAPI (blue), and analyzed by confocal microscopy. Scale bar: 2 μm. (C) Lentivirus-infected RAW cells were stimulated with LPS (10 ng/mL) for 6 hours, stained with anti-TNF-α, and analyzed by flow cytometry.
We next analyzed LPS-induced NF-κB activation in cells transduced with Bcl-3/GFP. GFP-positive cells were isolated by FACS and stimulated with LPS for 60 minutes, and p65 expression in the nucleus was analyzed by Western blotting (Figure 4A). LPS-induced nuclear translocation of p65 was observed in Bcl-3-transduced cells as well as in cells transduced with GFP alone. We next analyzed the DNA binding activity of NF-κB (Figure 4B). LPS stimulation led to enhanced DNA binding activity of p50/p50 homodimers and p50/p65 heterodimers in cells transduced with GFP alone. In Bcl-3-transduced cells, the DNA binding activity by p50/p50 homodimers was observed before LPS stimulation, and further LPS stimulation did not induce any activation of p50/p65 heterodimers. In IL-10-pretreated peritoneal macrophages and RAW cells, LPS-induced nuclear translocation of p65 was normally observed, but the DNA binding activity of p50/p65 heterodimers was severely reduced (Figure 4C-D). The specificity of the bands was confirmed by supershifts by treatment with anti-p50 and anti-p65 antibodies (Figure 4D). These findings are similar to those observed in Bcl-3-transduced macrophages, indicating that IL-10-inducible Bcl-3 is required for suppression of NF-κB activity in the nucleus.
Impaired NF-κB activity in Bcl-3-transduced and IL-10-pretreated macrophages. (A) Western blot showing the nuclear expression of p65 subunit of NF-κB after LPS stimulation (1 hour) in RAW cells transduced with GFP alone or Bcl-3/GFP. (B) RAW cells transduced with GFP alone or Bcl-3/GFP were stimulated with LPS and nuclear extracts were subjected to EMSA. There are 2 types of NF-κB binding to the probe, p50/p65 heterodimers and p50/p50 homodimers, indicated by the arrow and arrowhead, respectively. (C) Western blot showing the nuclear expression of p65 before and 60 minutes after LPS stimulation in nonpretreated (med) and IL-10-pretreated (IL-10) RAW macrophages. (D) RAW cells were pretreated with 10 ng/mL IL-10 for 18 hours and stimulated with 10 ng/mL LPS for 60 minutes. Nuclear extracts were subjected to EMSA using the NF-κB binding site of the TNF-α promoter as a probe. The specificities of the shifted bands were determined by adding specific Abs to p50 (arrow) and p65 (arrowhead). Similar results were observed in mouse peritoneal macrophages.
Impaired NF-κB activity in Bcl-3-transduced and IL-10-pretreated macrophages. (A) Western blot showing the nuclear expression of p65 subunit of NF-κB after LPS stimulation (1 hour) in RAW cells transduced with GFP alone or Bcl-3/GFP. (B) RAW cells transduced with GFP alone or Bcl-3/GFP were stimulated with LPS and nuclear extracts were subjected to EMSA. There are 2 types of NF-κB binding to the probe, p50/p65 heterodimers and p50/p50 homodimers, indicated by the arrow and arrowhead, respectively. (C) Western blot showing the nuclear expression of p65 before and 60 minutes after LPS stimulation in nonpretreated (med) and IL-10-pretreated (IL-10) RAW macrophages. (D) RAW cells were pretreated with 10 ng/mL IL-10 for 18 hours and stimulated with 10 ng/mL LPS for 60 minutes. Nuclear extracts were subjected to EMSA using the NF-κB binding site of the TNF-α promoter as a probe. The specificities of the shifted bands were determined by adding specific Abs to p50 (arrow) and p65 (arrowhead). Similar results were observed in mouse peritoneal macrophages.
Ectopic expression of Bcl-3 inhibits LPS-induced activation of the TNF-α promoter, but not the IL-6 promoter
We next examined the effect of transient overexpression of Bcl-3 on LPS-induced activation of the TNF-α and IL-6 promoters using a reporter gene assay. Bcl-3 overexpression suppressed LPS-induced transcriptional activity of the TNF-α promoter in RAW cells (Figure 5A). In contrast, Bcl-3 expression had no effect on LPS-induced transactivation of the IL-6 promoter in RAW cells (Figure 5B). Overexpression of Bcl-3ΔN, the mutant Bcl-3 lacking the N-terminal NLS, did not suppress the activity of the TNF-α promoter, indicating that the N-terminal NLS is required for suppression of the TNF-α promoter (Figure 5A). Thus, Bcl-3 has an inhibitory effect on the LPS-induced activation of the TNF-α promoter, but not the IL-6 promoter.
Overexpression of Bcl-3 inhibits LPS-induced transactivation of the TNF-α promoter. (A) RAW cells were transiently cotransfected with the TNF-α promoter-luciferase construct (10 ng) and the expression vectors for full-length Bcl-3 or ΔN (10 ng, 100 ng, or 500 ng), as indicated. After 24 hours of transfection, cells were treated with or without 1 μg/mL LPS for 8 hours and then the luciferase activities were measured. Data are representative of 3 independent experiments yielding similar results. Data are expressed as relative fold activation compared with the nonstimulated (-) set. (B) RAW cells were transiently cotransfected with the IL-6 promoter-luciferase construct (100 ng) and the expression vector for Bcl-3 (10 ng, 100 ng, or 500 ng), as indicated. The cells were treated with 1 μg/mL LPS for 8 hours and luciferase activity was detected. Representative results of 3 independent experiments are shown. (C) The 293 cells were transfected with various combinations of pEF-BOS-Myc-Bcl-3, pcDNA3.1-Flag-p50, and pcDNA3.1-Flag-p52 in a 6-well dish (each 1 μg/well). Total amount of transfected DNA was kept at 2 μg by adding pEF-BOS or pcDNA 3.1. p50 and p52 were immunoblotted by anti-Flag antibody (M2 monoclonal antibody). Bcl-3 was immunoblotted by rabbit polyclonal anti-c-Myc antibody. Plasmids transfected are indicated on top of the panel. (D) RAW cells expressing Myc-Bcl-3 and Flag-p50 were stimulated with 100 ng/mL LPS for 2 hours, and chromatin immunoprecipitation assays were performed with α-Myc or α-Flag antibodies. The detection of the immunoprecipitated TNF-α promoter (left panel) or IL-6 promoter (right panel) was analyzed by PCR with promoter-specific primers. Representative of 3 independent experiments. (E) RAW cells were transiently cotransfected with pELAM-1, Flag-p50 (0, 1, 10, 100 ng/well), and pEF-BOS Bcl-3 (100 ng/well) expression plasmids. After 24 hours of transfection, cells were treated with 1 μg/mL LPS for 8 hours and then luciferase activity was detected. Data are expressed as relative fold activation compared with the nontransfected set. Error bars indicate SD.
Overexpression of Bcl-3 inhibits LPS-induced transactivation of the TNF-α promoter. (A) RAW cells were transiently cotransfected with the TNF-α promoter-luciferase construct (10 ng) and the expression vectors for full-length Bcl-3 or ΔN (10 ng, 100 ng, or 500 ng), as indicated. After 24 hours of transfection, cells were treated with or without 1 μg/mL LPS for 8 hours and then the luciferase activities were measured. Data are representative of 3 independent experiments yielding similar results. Data are expressed as relative fold activation compared with the nonstimulated (-) set. (B) RAW cells were transiently cotransfected with the IL-6 promoter-luciferase construct (100 ng) and the expression vector for Bcl-3 (10 ng, 100 ng, or 500 ng), as indicated. The cells were treated with 1 μg/mL LPS for 8 hours and luciferase activity was detected. Representative results of 3 independent experiments are shown. (C) The 293 cells were transfected with various combinations of pEF-BOS-Myc-Bcl-3, pcDNA3.1-Flag-p50, and pcDNA3.1-Flag-p52 in a 6-well dish (each 1 μg/well). Total amount of transfected DNA was kept at 2 μg by adding pEF-BOS or pcDNA 3.1. p50 and p52 were immunoblotted by anti-Flag antibody (M2 monoclonal antibody). Bcl-3 was immunoblotted by rabbit polyclonal anti-c-Myc antibody. Plasmids transfected are indicated on top of the panel. (D) RAW cells expressing Myc-Bcl-3 and Flag-p50 were stimulated with 100 ng/mL LPS for 2 hours, and chromatin immunoprecipitation assays were performed with α-Myc or α-Flag antibodies. The detection of the immunoprecipitated TNF-α promoter (left panel) or IL-6 promoter (right panel) was analyzed by PCR with promoter-specific primers. Representative of 3 independent experiments. (E) RAW cells were transiently cotransfected with pELAM-1, Flag-p50 (0, 1, 10, 100 ng/well), and pEF-BOS Bcl-3 (100 ng/well) expression plasmids. After 24 hours of transfection, cells were treated with 1 μg/mL LPS for 8 hours and then luciferase activity was detected. Data are expressed as relative fold activation compared with the nontransfected set. Error bars indicate SD.
We further addressed the mechanism by which Bcl-3 inhibits the TNF-α promoter activity. Consistent with the previous reports,28,29 coimmunoprecipitation analysis showed that Bcl-3 interacted with NF-κB p50 and p52 (Figure 5C). We next performed chromatin immunoprecipitation (ChIP) assays to further investigate whether Bcl-3 regulates activity of the TNF-α promoter. Since the available anti-Bcl-3 antibody could not be used for immunoprecipitation, we expressed Myc-tagged Bcl-3 and Flag-tagged p50 in RAW cells and used them for the experiments. RAW cells were stimulated with LPS for 2 hours, and ChIP assays were performed using anti-Myc and anti-Flag Abs (Figure 5D). In RAW cells expressing Bcl-3 and p50, both Bcl-3 and p50 were recruited to the TNF-α promoter before LPS stimulation. LPS stimulation did not induce any change in the recruitment of Bcl-3 and p50. In contrast, Bcl-3 was not recruited to the IL-6 promoter. Furthermore, LPS-induced p50 recruitment was normally observed in the IL-6 promoter, indicating that Bcl-3 expression did not have any effect on the IL-6 promoter. We next analyzed the effect of p50 overexpression on activity of the TNF-α promoter (Figure 5E). Although expression of a small amount of p50 enhanced activity of the TNF-α promoter, increasing amounts of p50 suppressed the promoter activity. Coexpression of Bcl-3 further enhanced p50-mediated suppression of the TNF-α promoter activity. Taken together, these results suggest that Bcl-3 is specifically involved in suppression of the TNF-α promoter together with NF-κB p50.
Macrophages from Bcl-3-deficient mice are defective in IL-10-induced suppression of TNF-α production
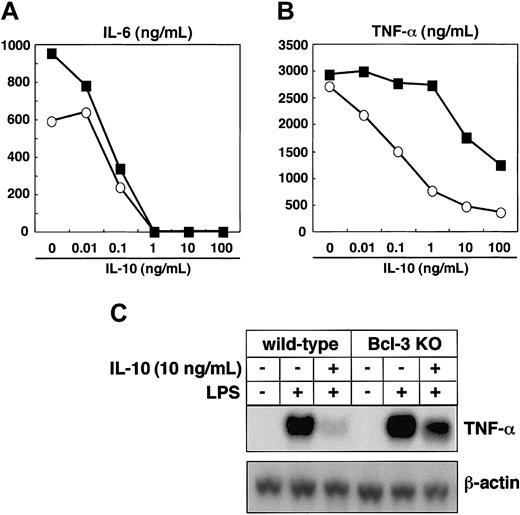
Finally, we analyzed whether Bcl-3 deficiency resulted in defective IL-10 responses. Peritoneal macrophages were isolated from wild-type and Bcl-3-deficient mice, pretreated with increasing amounts of IL-10, and stimulated with LPS. In wild-type mice, IL-10 pretreatment resulted in a dose-dependent suppression of LPS-induced production of TNF-α and IL-6 (Figure 6A-B). The IL-10-induced suppression of IL-6 production was similarly observed in Bcl-3-deficient macrophages (Figure 6A). However, when Bcl-3-deficient macrophages were pretreated with less than 1 ng/mL IL-10, suppression of LPS-induced TNF-α production was scarcely detected, although higher concentrations of IL-10 partially decreased TNF-α production (Figure 6B). When macrophages from wild-type mice were pretreated with 10 ng/mL IL-10, LPS-induced TNF-α mRNA expression was severely inhibited, but only partial impairment was observed in Bcl-3-deficient macrophages (Figure 6C). Thus, Bcl-3-deficient macrophages showed defective IL-10-mediated suppression of LPS-induced TNF-α production, demonstrating that IL-10-induced Bcl-3 is critical for the inhibition of LPS-induced TNF-α production.
Bcl-3-deficient macrophages are defective in IL-10-mediated suppression of LPS-induced TNF-α production. (A-B) Peritoneal macrophages from wild-type and Bcl-3-deficient mice were pretreated with IL-10 at the indicated concentrations for 18 hours and stimulated with 100 ng/mL LPS for an additional 24 hours. Concentrations of IL-6 (A) and TNF-α (B) in the culture supernatants were measured by ELISA. The experiments were repeated with 3 different animals and produced similar results. Open circles indicate wild-type mice; closed squares, Bcl-3-deficient mice. (C) Peritoneal macrophages from wild-type and Bcl-3-deficient mice were pretreated with 10 ng/mL IL-10 for 18 hours and stimulated with 100 ng/mL LPS for 2 hours. TNF-α mRNA expression was analyzed by Northern blotting.
Bcl-3-deficient macrophages are defective in IL-10-mediated suppression of LPS-induced TNF-α production. (A-B) Peritoneal macrophages from wild-type and Bcl-3-deficient mice were pretreated with IL-10 at the indicated concentrations for 18 hours and stimulated with 100 ng/mL LPS for an additional 24 hours. Concentrations of IL-6 (A) and TNF-α (B) in the culture supernatants were measured by ELISA. The experiments were repeated with 3 different animals and produced similar results. Open circles indicate wild-type mice; closed squares, Bcl-3-deficient mice. (C) Peritoneal macrophages from wild-type and Bcl-3-deficient mice were pretreated with 10 ng/mL IL-10 for 18 hours and stimulated with 100 ng/mL LPS for 2 hours. TNF-α mRNA expression was analyzed by Northern blotting.
Discussion
In this study, we analyzed the mechanism by which IL-10 exhibits negative regulatory effects on macrophages. A microarray analysis led to the identification of IL-10-inducible genes in macrophages. In the course of analyzing the mechanism underlying the anti-inflammatory effect of IL-10, several IL-10-inducible genes have so far been identified.17-20,30 Among these gene products, heme oxygenase-1 (HO-1) and SOCS3 have been proposed to be involved in IL-10-mediated suppression of macrophage activity.17,18 SOCS3 is a member of the family of suppressors of cytokine signaling (SOCS) proteins, which play important roles in the negative regulation of cytokine signaling pathways.31 A member of this family, SOCS1, has recently been shown to negatively regulate LPS signaling, as well as cytokine signaling, since SOCS1-deficient mice were highly sensitive to LPS.32,33 Similarly, in vitro studies indicated that IL-10-induced SOCS3 negatively regulates the LPS induction of TNF-α and IL-6 in macrophages.18 In this study, however, lentiviral expression of SOCS3 did not cause any reduction in LPS-induced TNF-α production in macrophages (Figure 2).
In addition to SOCS3, we identified Bcl-3 as an IL-10-inducible gene in macrophages. Bcl-3 is a member of the IκB protein family, harboring ankyrin repeat domains. However, unlike other IκB proteins, which are associated with NF-κB transcription factors and sequester them in the cytoplasm, Bcl-3 is localized in the nucleus.25,34 The role of Bcl-3 in the regulation of NF-κB activity is still controversial. Although several studies have indicated that Bcl-3 is a negative regulator of NF-κB,28,35-37 Bcl-3 has been shown to act as a transcriptional activator in some cases.25,34 Studies with Bcl-3-deficient mice have demonstrated an important role of Bcl-3 in B-cell and T-cell functions, but how Bcl-3 functions in macrophages remains unclear.38,39 Lentiviral expression of Bcl-3 inhibited LPS-induced production of TNF-α, but not IL-6, in macrophages (Figure 2). Overexpression of Bcl-3 also inhibited LPS-induced activation of the TNF-α promoter, but not the IL-6 promoter, in macrophages (Figure 5). Furthermore, Bcl-3-deficient macrophages showed defective IL-10-mediated suppression in the production of TNF-α, but not IL-6 (Figure 6). Thus, the present study clearly demonstrates that Bcl-3 functions as a negative regulator of LPS-induced TNF-α production in macrophages. Bcl-3 interacted preferentially with homodimers of p50 and p52, neither of which harbors the transactivation domains25,34,40 (Figure 5C). Furthermore, Bcl-3 was preferentially recruited to the TNF-α promoter and enhanced p50-mediated inhibition of the TNF-α promoter activity (Figure 5D-E). Therefore, although the molecular mechanism by which Bcl-3 inhibits TNF-α production remains unclear, Bcl-3 may interfere with the transcriptional activity of NF-κB in the TNF-α promoter through association with p50 or p52.
In Bcl-3-transduced macrophages, the LPS-induced DNA binding activity of NF-κB p50/p65 heterodimers was impaired, and this was also observed in IL-10-pretreated macrophages. However, LPS-induced IL-6 production was not impaired in Bcl-3-transduced cells. From these observations, we hypothesize 2 possibilities. First, although the inductions of both TNF-α and IL-6 are dependent on NF-κB, individual NF-κB subunits differentially regulate the induction of inflammatory cytokine genes. For instance, the IL-12p40 gene is selectively regulated by the c-Rel subunit,41 and the IL-6 gene is mainly regulated by the p50 subunit, since macrophages from mice lacking the p50 subunit were defective in LPS-induced production of IL-6, but not TNF-α.42 Introduction of increasing amounts of p50 into RAW cells resulted in a dose-dependent inhibition of transactivation of the TNF-α promoter, but not the IL-6 promoter (Figure 5E; H.K., unpublished data, February 2003). These results indicate that p50 subunits, especially p50 homodimers, may have an inhibitory effect on the TNF-α promoter, but not the IL-6 promoter, and may enable LPS induction of the IL-6 gene in IL-10-pretreated macrophages. Alternatively, since the IL-6 gene has been shown to be regulated by several transcription factors, such as NF-IL6 and AP-1 family members in addition to NF-κB, IL-6 may be induced by activation of these transcription factors. Thus, IL-10-inducible Bcl-3 is responsible for part of the IL-10-mediated suppression of macrophage activity, and additional molecules must be involved in the IL-10-mediated macrophage suppression. Indeed, although Stat3 mutant mice produced tremendously high levels of inflammatory cytokines in response to LPS, Bcl-3-deficient mice did not show increased production (Takeda et al13 ; and H.K., unpublished data, March 2003). A novel molecule harboring ankyrin repeats and showing high similarity with Bcl-3 has been identified as IκBζ/IL-1-inducible nuclear ankyrin-repeat protein (INAP)/molecule possessing ankyrin repeats induced by lipopolysaccharide (MAIL).43-45 IκBζ/INAP/MAIL has been shown to be induced in macrophages by inflammatory stimuli such as IL-1 and Toll-like receptor (TLR) ligands, and is preferentially expressed in the nucleus. Thus, although lentiviral expression of IκBϵ had no effect on the LPS response, other Bcl-3-related molecules may have regulatory roles in NF-κB activation.
In this study, we identified Bcl-3 as an IL-10-inducible molecule responsible for the suppression of LPS-induced TNF-α production. However, as-yet-unknown mechanisms underlying IL-10-mediated suppression of macrophages must exist. Further studies will be required to fully understand the inhibitory function of IL-10 in macrophages, which will certainly be very beneficial for the clinical treatment of inflammatory bowel diseases.
Prepublished online as Blood First Edition Paper, August 7, 2003; DOI 10.1182/blood-2003-04-1228.
Supported by grants from Special Coordination Funds, the Ministry of Education, Culture, Sports, Science, and Technology, and the Japan Research Foundation for Clinical Pharmacology.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank N. Okita for technical assistance, P. Lee for critical reading of the manuscript, and E. Horita and M. Hashimoto for secretarial assistance.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal