Abstract
High-level α-globin expression depends on cis-acting regulatory sequences located far upstream of the α-globin cluster. Sequences that contain the α-globin positive regulatory element (PRE) activate α-globin expression in transgenic mice. The α-globin PRE contains a pair of composite binding sites for the transcription factors activating protein 1 and nuclear factor erythroid 2 (AP1/NFE2). To determine the role of these binding sites in α-globin gene transcription, we mutated the AP1/NFE2 sites in the α-globin PRE in mice. We replaced the AP1/NFE2 sites with a neomycin resistance gene (neo) that is flanked by LoxP sites (floxed). Mice with this mutation exhibited increased embryonic death and α-thalassemia intermedia. Next, we removed the neo gene by Cre-mediated recombination, leaving a single LoxP site in place of the AP1/NFE2 sites. These mice were phenotypically normal. However, α-globin expression, measured by allele-specific RNA polymerase chain reaction (PCR), was decreased 25%. We examined the role of the hematopoietic-restricted transcription factor p45Nfe2 in activating expression through these sites and found that it is not required. Thus, we have demonstrated that AP1/NFE2 binding sites in the murine α-globin PRE contribute to long-range α-globin gene activation. The proteins that mediate this effect remain to be determined. (Blood. 2003;102:4223-4228)
Introduction
High-level expression of the α- and β-like globin genes depends on cis-acting DNA sequences located far upstream of the globin gene clusters. The locus control region (LCR), which is upstream of the β-globin cluster, confers position-independent expression on the β-like globin genes in transgenic mice.1 The LCR encompasses 4 erythroid-specific DNase I hypersensitive sites, 5′HS1 to 5′HS4, which mark the location of discrete enhancer elements.2-4 Individually, these enhancers are not required for high-level globin gene expression.5-8 Collectively, however, they account for much of the activity conferred by this region.7,9
Sequences upstream of the α-globin cluster also support expression of the α-like globin genes. In contrast to the LCR, these sequences do not confer full activity, or position-independent expression, on linked globin genes.10-12 However, naturally occurring deletions of the α-globin cluster that remove these sequences are associated with α-thalassemia, suggesting that they are essential for α-globin expression.13-16 The region defined by the minimum overlap between these deletions contains 2 erythroid-specific hypersensitive sites (HS-33 and HS-40). HS-40, also known as the α-globin PRE, activates globin gene expression in stably transfected murine erythroleukemia (MEL) cells and transgenic mice.10 Targeted disruption of HS-40 inactivates the α-globin genes in stable interspecific hybrids.17 Recently, the murine equivalent of HS-40 (HS-26) has been inactivated in mice. A 1.3-kb deletion, which removes the entire enhancer, is associated with approximately a 50% decrease in α-globin expression.18
The mechanism whereby enhancers in the β-globin LCR and the α-globin PRE activate high-level β- and α-globin gene expression remains to be determined. The minimum size of the enhancers, defined in transgenic mice, is several hundred nucleotides.19-21 These enhancer cores contain a high density of GATA, AP1/NFE2, and CACCC transcription factor binding sites, which have been conserved through evolution.22-24 Indeed, tandem AP1/NFE2 binding sites associated with 5′HS2 are one of the most highly conserved elements in the LCR.22,25 GATA and CACCC binding sites are present in the globin promoters and local enhancers and are important for the function of those elements.26-28 In contrast, in globin regulatory elements, AP1/NFE2 binding sites are restricted to the β-globin LCR and the α-globin PRE.11,19-21,25,29 This distribution suggests that AP1/NFE2 binding sites may have a role in long-range gene activation.
The consensus sequence for the AP1/NFE2 binding site (c/t)gctga(g/c)tca(c/t) is a composite binding site for the transcription factors Nfe2 and AP-1 (underlined).30 Nfe2 is a heterodimeric member of the basic region-leucine zipper (BZIP) family, which is composed of p18Nfe2 and p45Nfe2.30,31 p18Nfe2 is a member of the small Maf family (MafG or MafK).32 p45Nfe2 is a member of the cap'n collar family of BZIP proteins.30 In the human and mouse, there are 4 members of the cap'n collar family—p45Nfe2 and the Nfe2-related factors 1, 2, and 3 (Nrf1, Nrf2, and Nrf3).33-37 Expression of p45Nfe2 is hematopoietic restricted.30,31 Deficiency of p45Nfe2 is associated with a defect of megakaryocyte maturation, severe thrombocytopenia, and perinatal lethality in mice.38 p45Nfe2-deficient mice have an erythroid defect characterized by mild anemia and increased splenic erythropoiesis.39 Studies of erythroid cells from p45Nfe2-deficient mice failed to reveal specific changes in the total RNA levels of α- or β-globin.39 This lack of effect has been attributed to redundancy, both of cis-acting regulatory sequences and of proteins that bind AP1/NFE2 sites.
To gain insight into the role of AP1/NFE2 binding sites in globin gene regulation, we inactivated the AP1/NFE2 sites in the α-globin PRE (HS-26) in mice. This element contains a pair of closely spaced AP1/NFE2 binding sites evolutionarily conserved since pufferfish.40-42 Here we show that the AP1/NFE2 binding sites in HS-26 contribute to long-range α-globin gene activation. Further, we show that the transcription factor p45Nfe2 is not required for activity of these sites.
Materials and methods
Targeted inactivation of the AP1/NFE2 sites in HS-26
A P1 clone that included HS-26 was obtained from a 129-strain embryonic stem (ES) cell library (Genome Systems, St Louis, MO). Genomic fragments used in constructing the targeting vector were (GenBank accession number AY016021): 5′ arm, StuI 87239-HincII 90966; 3′ arm, KpnI 91137-BglI 94272. The AP1/NFE2 binding sites, located between the HincII 90966 and KpnI 91137 sites, were replaced by an oligonucleotide containing 2 LoxP sites. A neo gene was subcloned between the LoxP sites to provide positive selection for transfectants, and a Diphtheria A toxin gene was subcloned downstream of the 3′ arm to provide negative selection against nonhomologous integrants. The targeting vector was transfected into 2 different ES cell lines (RW4 and E14). Transfectants were screened for homologous recombination with a BglI 94272-HindIII 94694 probe that was downstream of the 3′ arm of the targeting vector. Chimeric mice were produced as previously described.43 Five chimeras provided germline transmission. Progeny from the F1 generation were bred with CMV-Cre transgenic mice to excise the neo gene.44 The Cre transgene was removed by breeding.
Analysis of mice
Peripheral blood was obtained from anesthetized mice by retro-orbital puncture with heparinized microhematocrit tubes. The hematocrit (Hct) was determined by centrifugation in a micro-MB centrifuge (International Equipment, Needham, MA). The red blood cell (RBC) count was determined using an automated Multisizer II cell counter (Coulter, Miami, FL). The hemoglobin (Hb) concentration was determined by the Drabkin method45 (Sigma, St Louis, MO). For the floxed strain of mice, it was necessary to pellet excess RBC membranes by centrifugation before measuring the absorbance to prevent interference from sample turbidity. Mean corpuscular volume (MCV) and mean corpuscular hemoglobin (MCH) were derived from the Hct, RBC, and Hb values. Reticulocytes were determined by fluorescence-activated cell sorter (FACS) analysis after staining with thiazole orange, as previously described,46 except that the stock solution of thiazole orange (1 mg/mL in methanol) was diluted 1:500 in phosphate-buffered saline (PBS) and the erythrocytes were washed 3 times with PBS after staining. Peripheral blood smears were stained with Wright-Giemsa, examined by light microscopy (100 ×, oil objective), and digitally imaged with a DXM1200 camera (Nikon, Tokyo, Japan).
Embryonic day-14.5 (E14.5) embryos were obtained from pregnant female mice that had been killed by cervical dislocation, in accordance with the recommendations of the Panel on Euthanasia of the American Veterinary Association. Yolk sacs were removed, and the embryos were examined with an SZH-10 research stereomacroscope (DF Plan, 1.5 × objective) (Olympus, Tokyo, Japan) and digitally imaged with a DX1 SLR camera (Nikon, Tokyo, Japan).
RNA analysis
Cells were obtained from the blood and bone marrow of adult mice and the fetal livers of E12.5 to E14.5 embryos. Total RNA was extracted by the method of Chomczynski and Sacchi47 (RNazol B; Tel-Test, Friendswood, TX). cDNA was made from 0.2 μg RNA with an oligo-dT primer. The cDNA was diluted 1:40 with water, and 1 μL was used in the polymerase chain reaction (PCR) mix. The primers used in the PCR reaction were 5′-gatgtaagccacggctctgc-3′ and 5′-aaggtcaccagcaggcagtg-3′. The region of the murine α-globin mRNA amplified by these primers was 175 to 362, inclusive (GenBank accession number NM_008218). The PCR reaction was set up according to the manufacturer's instructions (Roche Molecular Systems, Branchburg, NJ). The final magnesium concentration was 2 mM. The reaction was spiked with 32 P dCTP. PCR cycle conditions were 1 cycle at 95°C for 3 minutes; 20 cycles at 95°C for 30 seconds, 58°C for 1 minute, and 72°C for 1 minute; and 1 cycle at 72°C for 10 minutes. The PCR product was purified over a G-25 spin column, digested with MspI, and resolved on a 10% nondenaturing polyacrylamide gel. Expression was quantitated with the PhosphorImager and ImageQuant software (Amersham Biosciences, Uppsala, Sweden). Data were analyzed in Excel (Microsoft, Redmond, WA) using the t test for 2 samples assuming unequal variances.
HS-26 hypersensitive site mapping
Heterozygous ΔNFE2 mice (without the Cre transgene) were set up in a timed mating. Fetal liver cells from E14.5 embryos were washed once in PBS and resuspended in 1 mL ice-cold RSB (10 mM NaCl, 3 mM MgCl2, 10 mM Tris [pH 7.5]). After 5-minute incubation in the hypotonic buffer on ice, the cells were pipetted to disrupt the plasma membrane. Nuclei were pelleted and resuspended in 1.5 mL RSB, and 0.2 mL was aliquoted into 6 tubes on ice. DNase I (DPRF; Worthington, Lakewood, NJ) was added to the nuclei, the nuclei were incubated for 10 minutes at 37°C, and genomic DNA was extracted (Puregene; Gentra, Minneapolis, MN). Genomic DNA was digested with HindIII, Southern blotted, and probed with a labeled BglI 94272-HindIII 94694 fragment.
Results
Role of AP1/NFE2 binding sites in α-globin regulation
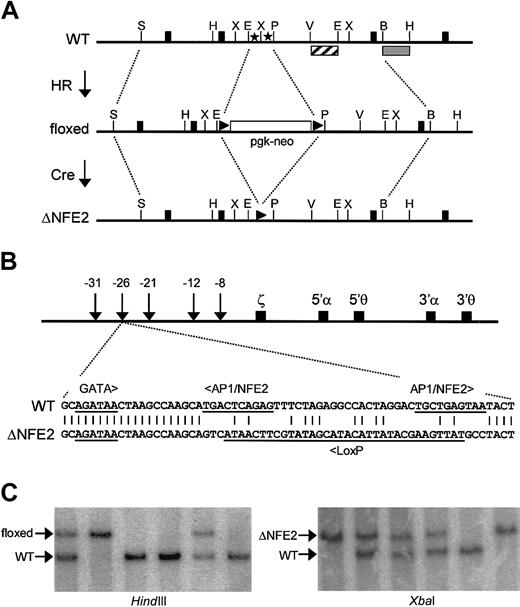
We used a 2-step strategy to inactivate the AP1/NFE2 binding sites in HS-26 (Figure 1A). In the first step, we replaced the AP1/NFE2 binding sites with a floxed neo gene by homologous recombination in ES cells. The frequency of homologous recombination was 19%. Five chimeras, from 4 independent ES clones, provided germline transmission of the mutation. This strain of mice, with an inserted neomycin resistance gene, is called floxed. In the second step, we bred floxed mice with CMV-Cre transgenic mice (Cre-deletor strain).44 All the progeny that coinherited the floxed neo gene and the CMV-Cre transgene showed excision of the floxed neo gene. Cre-mediated recombination of the floxed neo gene resulted in replacement of the tandem AP1/NFE2 binding sites with a single LoxP site (Figure 1B). Thus, the AP1/NFE2 sites are replaced with 40 nucleotides of heterologous sequence with no known binding sites for mammalian transcription factors. All other transcription factor binding sites associated with HS-26, and the exact spacing between them, are preserved. This mutation, and the strain of mice that have this mutation, are called ΔNFE2. To date, all the experimentally induced mutations of the β-globin LCR and α-globin PRE in mice have been large (larger than 1 kb) deletions.5-9,18 This is the first example of a transcription factor binding site mutation, and the availability of these mice permits us to take a genetic approach to identify the proteins that activate expression through these sites.
Site-directed mutagenesis of AP1/NFE2 binding sites in HS-26. (A) AP1/NFE2 binding sites (stars) are replaced with a neo gene, which is flanked by LoxP sites (triangles), through homologous recombination (HR). This is the floxed mutation. The selectable marker is removed by Cre-mediated recombination (Cre). This is the ΔNFE2 mutation. Solid boxes represent exons of the gene C16orf35. The restriction enzyme sites are StuI (S), HindIII (H), EcoRI (E), XbaI (X), Pst I (P), EcoRV (V), and Bgl I (B). (B) Murine α-globin locus with the sequence of the ΔNFE2 mutation. (C) Southern blot analysis of tail DNA from F2 mice. With HindIII, an external probe (▦) hybridizes to a 4.9-kb wild-type band (WT) or a 6.7-kb band (floxed). With XbaI, an internal probe (▨) hybridizes to a 1.8-kb band (WT) or a 2.4-kb band (ΔNFE2). The probes are shown in panel A.
Site-directed mutagenesis of AP1/NFE2 binding sites in HS-26. (A) AP1/NFE2 binding sites (stars) are replaced with a neo gene, which is flanked by LoxP sites (triangles), through homologous recombination (HR). This is the floxed mutation. The selectable marker is removed by Cre-mediated recombination (Cre). This is the ΔNFE2 mutation. Solid boxes represent exons of the gene C16orf35. The restriction enzyme sites are StuI (S), HindIII (H), EcoRI (E), XbaI (X), Pst I (P), EcoRV (V), and Bgl I (B). (B) Murine α-globin locus with the sequence of the ΔNFE2 mutation. (C) Southern blot analysis of tail DNA from F2 mice. With HindIII, an external probe (▦) hybridizes to a 4.9-kb wild-type band (WT) or a 6.7-kb band (floxed). With XbaI, an internal probe (▨) hybridizes to a 1.8-kb band (WT) or a 2.4-kb band (ΔNFE2). The probes are shown in panel A.
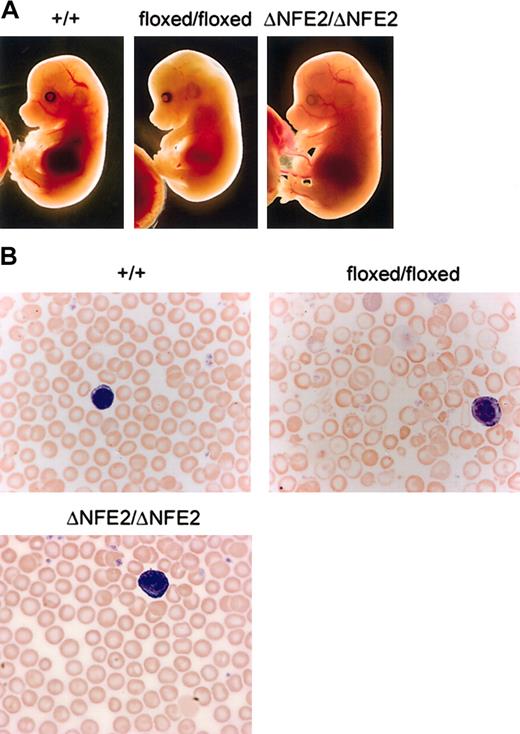
Mice that were homozygous for the floxed mutation were viable, but present at one third the expected Mendelian frequency at weaning (Figure 1C; Table 1). E14.5 homozygous floxed embryos were pale, edematous, and anemic (Figure 2A and data not shown). In contrast, ΔNFE2 homozygotes were present at the expected frequency, and the embryos appeared normal. Adult homozygous floxed mice were moderately anemic and the erythrocytes showed hypochromia and marked variation in size and shape (Figure 2B; Table 2). Overall, the presentation was consistent with α-thalassemia intermedia. In contrast, adult homozygous ΔNFE2 mice had normal erythrocyte indices and occasional target cells. Because the ΔNFE2 mice were derived from the floxed mice, it is unlikely that the subtle phenotype in the ΔNFE2 mice was caused by improper gene targeting. In addition, we confirmed the presence of the ΔNFE2 mutation by Southern blotting and sequencing (Figure 1 and data not shown).
Genotypes of HS-26 mice at weaning
+/+ . | +/floxed . | floxed/floxed . | +/+ . | +/ΔNFE2 . | ΔNFE2/ΔNFE2 . |
|---|---|---|---|---|---|
| n = 53 | n = 111 | n = 15 | n = 10 | n = 18 | n = 11 |
| 0.30 | 0.62 | 0.08 | 0.26 | 0.46 | 0.28 |
+/+ . | +/floxed . | floxed/floxed . | +/+ . | +/ΔNFE2 . | ΔNFE2/ΔNFE2 . |
|---|---|---|---|---|---|
| n = 53 | n = 111 | n = 15 | n = 10 | n = 18 | n = 11 |
| 0.30 | 0.62 | 0.08 | 0.26 | 0.46 | 0.28 |
Floxed mutation causes α-thalassemia. (A) Dissected E14.5 embryos. HS-26 genotypes are indicated at the top. (B) Blood smears from adult mice, stained with Wright-Giemsa (original magnification, × 100). Note the target cells, hypochromic cells, and prominent fragments in the blood from the floxed/floxed mice. A white blood cell (purple) is shown in each panel for size comparison.
Floxed mutation causes α-thalassemia. (A) Dissected E14.5 embryos. HS-26 genotypes are indicated at the top. (B) Blood smears from adult mice, stained with Wright-Giemsa (original magnification, × 100). Note the target cells, hypochromic cells, and prominent fragments in the blood from the floxed/floxed mice. A white blood cell (purple) is shown in each panel for size comparison.
Erythrocyte indices of adult HS-26 mice
HS-26 genotype . | RBC, × 106/μL . | MCV, fL . | Hb, g/dL . | MCH, pg/cell . | Reticulocytes, % . |
|---|---|---|---|---|---|
| +/+, n = 10 | 8.5 ± 0.4 | 54 ± 2 | 15.2 ± 0.7 | 17.8 ± 0.9 | 2.9 ± 0.4 |
| floxed/floxed, n = 9 | 9.8 ± 0.7 | 44 ± 4 | 10.9 ± 1.1 | 11.1 ± 1.1 | 32.4 ± 10.7 |
| ΔNFE2/ΔNFE2, n = 10 | 8.5 ± 0.4 | 54 ± 3 | 14.8 ± 0.6 | 17.4 ± 0.7 | 3.7 ± 1.4 |
HS-26 genotype . | RBC, × 106/μL . | MCV, fL . | Hb, g/dL . | MCH, pg/cell . | Reticulocytes, % . |
|---|---|---|---|---|---|
| +/+, n = 10 | 8.5 ± 0.4 | 54 ± 2 | 15.2 ± 0.7 | 17.8 ± 0.9 | 2.9 ± 0.4 |
| floxed/floxed, n = 9 | 9.8 ± 0.7 | 44 ± 4 | 10.9 ± 1.1 | 11.1 ± 1.1 | 32.4 ± 10.7 |
| ΔNFE2/ΔNFE2, n = 10 | 8.5 ± 0.4 | 54 ± 3 | 14.8 ± 0.6 | 17.4 ± 0.7 | 3.7 ± 1.4 |
Values presented as ± SD.
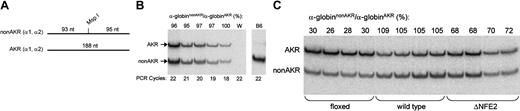
Next, we determined the effect of the floxed and ΔNFE2 mutations on α-globin expression. To accomplish this, we used an allele-specific RNA PCR assay. We identified a polymorphism of the α-globin transcript, which mutated a restriction enzyme site (MspI). This polymorphism is present in both adult α-globin genes of the AKR strain and is not present in most other inbred (non-AKR) strains.48 After digestion with MspI, the PCR product from the non-AKR α-globin allele (α-globinnonAKR) is cleaved into 2 fragments of 93 and 95 nucleotides (Figure 3A). These fragments migrate as a single band. The PCR product from the AKR α-globin allele (α-globinAKR) does not cut with MspI and gives rise to a single band of 188 nucleotides. Experiments are performed on the F1 progeny of non-AKR and AKR mice. With regard to HS-26, the test allele is linked to the α-globinnonAKR gene and is wild type, floxed, or ΔNFE2. The control allele is linked to the α-globinAKR gene and is wild type. The activity of the test allele is expressed as the ratio of α-globinnonAKR to α-globinAKR expression. We found that this ratio was constant over a limited range of PCR cycles and signal intensities (Figure 3B). Consequently, small differences in the efficiency of PCR amplification should not affect the validity of the results. This approach has been previously used in studies of the β-globin locus.5,6
Allele-specific RNA PCR. (A) The RNA PCR product from both adult α-globin genes of most non-AKR strains is bisected by an MspI restriction site. (B) Allele-specific RNA PCR on blood from HS26+(nonAKR)/+(AKR) non-AKR × AKR F1 mice. Controls are water (W) and C57BL6 (B6). (C) Allele-specific RNA PCR on bone marrow from HS26floxed(nonAKR)/+(AKR), HS26+(nonAKR)/+(AKR), and HS26ΔNFE2(nonAKR)/+(AKR) non-AKR × AKR F1 mice.
Allele-specific RNA PCR. (A) The RNA PCR product from both adult α-globin genes of most non-AKR strains is bisected by an MspI restriction site. (B) Allele-specific RNA PCR on blood from HS26+(nonAKR)/+(AKR) non-AKR × AKR F1 mice. Controls are water (W) and C57BL6 (B6). (C) Allele-specific RNA PCR on bone marrow from HS26floxed(nonAKR)/+(AKR), HS26+(nonAKR)/+(AKR), and HS26ΔNFE2(nonAKR)/+(AKR) non-AKR × AKR F1 mice.
Relative to wild type, the ΔNFE2 mutation caused a 25% or a 35% reduction in α-globinnonAKR/α-globinAKR expression in the fetal liver cells of E14.5 embryos or the bone marrow of adult mice, respectively (Figure 3C; Table 3). The Cre transgene, which was present in a few embryos, had no effect. There was a smaller decrease in expression in the blood of adult mice, possibly reflecting selection against the most severely affected cells. These percentages represent an average; therefore, we cannot say whether there is a 25% to 35% reduction in expression in all cells or a more severe defect in a fraction of the cells assayed. The floxed mutation had a more severe effect on expression, consistent with previous reports on the effect of a selectable marker gene in the β-globin LCR.5,49
Role of AP1/NFE2 binding sites in α-globin expression
. | α-globinnonAKR/α-globinAKRexpression, % . | . | . | ||
|---|---|---|---|---|---|
| HS-26 test allele . | Fetal liver, E 14.5 . | Adult bone marrow . | Adult blood . | ||
| Wild type | 93 ± 4 | 106 ± 2 | 95 ± 6 | ||
| n = 19* | n = 4* | n = 13* | |||
| Floxed | 22 ± 2 | 28 ± 2 | 37 ± 4 | ||
| n = 6 | n = 4 | n = 7 | |||
| ΔNFE2 | 70 ± 4 | 69 ± 2 | 81 ± 4 | ||
| n = 10* | n = 4* | n = 8* | |||
. | α-globinnonAKR/α-globinAKRexpression, % . | . | . | ||
|---|---|---|---|---|---|
| HS-26 test allele . | Fetal liver, E 14.5 . | Adult bone marrow . | Adult blood . | ||
| Wild type | 93 ± 4 | 106 ± 2 | 95 ± 6 | ||
| n = 19* | n = 4* | n = 13* | |||
| Floxed | 22 ± 2 | 28 ± 2 | 37 ± 4 | ||
| n = 6 | n = 4 | n = 7 | |||
| ΔNFE2 | 70 ± 4 | 69 ± 2 | 81 ± 4 | ||
| n = 10* | n = 4* | n = 8* | |||
Values presented as ± SD.
t test; P < .001.
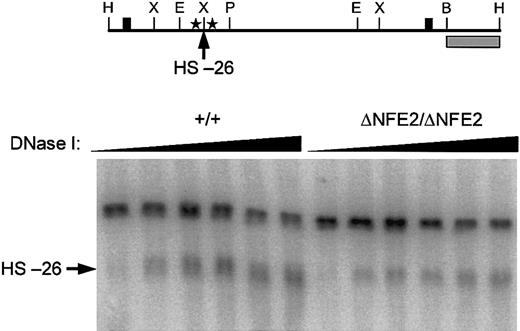
Because we specifically mutated the AP1/NFE2 binding sites in the HS-26 enhancer, we were able to assess the contribution of these sites to hypersensitive site formation. In fetal liver cells from E14.5 wild-type embryos, HS-26 formed a broad band that appeared to be composed of several subbands (Figure 4). In fetal liver cells from homozygous ΔNFE2 embryos HS-26 still formed, though in some experiments the intensity of the hypersensitive site, and possibly the number of subbands, was diminished (Figure 4 and data not shown).
HS-26 is formed in the absence of AP1/NFE2 binding sites. (Top) Diagram of 4.9-kb wild-type HindIII DNA fragment. Cleavage at HS-26 yields a 3.6-kb band in Southern blots. The probe (▦) and other symbols are the same as in Figure 1. (Bottom) DNase I hypersensitivity in E14.5 fetal liver cells. Genotypes are indicated at the top.
HS-26 is formed in the absence of AP1/NFE2 binding sites. (Top) Diagram of 4.9-kb wild-type HindIII DNA fragment. Cleavage at HS-26 yields a 3.6-kb band in Southern blots. The probe (▦) and other symbols are the same as in Figure 1. (Bottom) DNase I hypersensitivity in E14.5 fetal liver cells. Genotypes are indicated at the top.
Role of p45Nfe2 in long-range α-globin gene activation
Various BZIP proteins are present in erythroid cells, which potentially could activate globin gene expression through AP1/NFE2 binding sites. These include p45Nfe2, Nfe2-related factors, and members of the AP-1 or CREB/ATF transcription factor families. Of these, Nfe2 is the most abundant AP1/NFE2 binding activity in fetal liver cells.50 To test the hypothesis that p45Nfe2 activates globin gene expression through these sites, we used a genetic approach in mice (Figure 5). We bred p45Nfe2 knock-out mice with ΔNFE2 mice and AKR mice, then intercrossed the progeny to obtain embryos with the following genotypes: the ΔNFE2 allele of HS-26 linked to α-globinnonAKR on one chromosome, wild-type HS-26 linked to α-globinAKR on the other, and 1 of 3 possible p45Nfe2 genotypes (+/+, +/-, -/-). If p45Nfe2 is required for globin gene activation through these sites, then the absence of p45Nfe2 should cause activity of the wild-type HS-26 allele to decrease to the level of the ΔNFE2 mutant, and α-globinnonAKR/α-globinAKR expression should normalize. Instead, we observed no change in this ratio (Table 4). We conclude that p45Nfe2 is dispensable for α-globin activation through the AP1/NFE2 sites.
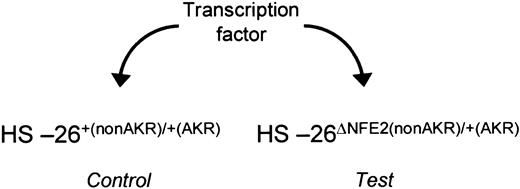
Assay for activators. In fetal liver cells from HS-26ΔNFE2(nonAKR)/+(AKR) embryos, the activity of the wild-type allele of HS-26 can be directly compared with the mutated allele, under different trans-acting conditions. This assay can be used to determine the ability of a transcription factor to function through the intact AP1/NFE2 binding sites.
Assay for activators. In fetal liver cells from HS-26ΔNFE2(nonAKR)/+(AKR) embryos, the activity of the wild-type allele of HS-26 can be directly compared with the mutated allele, under different trans-acting conditions. This assay can be used to determine the ability of a transcription factor to function through the intact AP1/NFE2 binding sites.
Role of p45Nfe2 in α-globin expression
. | α-globinnonAKR/α-globinAKRexpression, % . | . | . | ||
|---|---|---|---|---|---|
| HS-26 test allele . | p45Nfe2+/+ . | p45Nfe2+/− . | p45Nfe2−/− . | ||
| Wild type | 100 ± 2 | 101 ± 4 | 97 ± 3 | ||
| n = 5 | n = 17 | n = 8 | |||
| ΔNFE2 | 72 ± 2 | 69 ± 5 | 71 ± 2 | ||
| n = 8* | n = 19 | n = 6* | |||
. | α-globinnonAKR/α-globinAKRexpression, % . | . | . | ||
|---|---|---|---|---|---|
| HS-26 test allele . | p45Nfe2+/+ . | p45Nfe2+/− . | p45Nfe2−/− . | ||
| Wild type | 100 ± 2 | 101 ± 4 | 97 ± 3 | ||
| n = 5 | n = 17 | n = 8 | |||
| ΔNFE2 | 72 ± 2 | 69 ± 5 | 71 ± 2 | ||
| n = 8* | n = 19 | n = 6* | |||
Values presented as ± SD. Expression occurred in E12.5 to E13.5 fetal liver cells.
t test; P = .58.
Discussion
Recently, it has been shown that targeted disruption of HS-26 causes a 50% decrease in α-globin expression in mice.18 Based on the 25% reduction in α-globin expression seen with the ΔNFE2 mutant, we estimate that the tandem AP1/NFE2 binding sites account for approximately 50% of the activity of the HS-26 enhancer. To date, all experimentally induced mutations of the β-globin LCR and the α-globin PRE have been large (larger than 1 kb) deletions.5-9,18 The 25% reduction in α-globin expression, caused by the ΔNFE2 mutation, is not sufficient to cause α-thalassemia. Nonetheless, our results demonstrate that a small transcription factor binding site motif can function over large distances to activate globin gene expression. The persistence of the HS-26 hypersensitive site in the ΔNFE2 mutant strain suggests that other proteins remain bound to the enhancer in the absence of the AP1/NFE2 sites. There are 3 GATA binding sites and 1 CACCC binding site in the ΔNFE2 mutant of HS-26. Proteins bound to these or other sites may account for the residual activity of the ΔNFE2 allele of HS-26.
p45Nfe2 has been implicated in the function of the β-globin LCR and globin gene activation. In MEL cells, α- and β-globin gene expression depend on p45Nfe2.51,52 The activation domains of p45Nfe2 have been mapped.53 An amino-terminal activation domain has been shown to interact with TAFII130, CREB-binding protein, and several ubiquitin ligases.54-57 Interactions between both subunits of Nfe2 and the murine β-globin locus have been studied by chromatin immunoprecipitation assay. The tandem AP1/NFE2 binding sites of 5′HS2 are occupied by p45Nfe2 in MEL cells and fetal liver cells.58 In MEL cells, histone acetylation and recruitment of RNA polymerase II to the β-globin promoter is p45Nfe2 dependent.59 MEL differentiation is accompanied by a 10-fold increase in MafK and p45Nfe2 binding to 5′HS2.60 This correlates with a shift in the nuclear location of MafK from centromeric heterochromatin to euchromatin.61 p45Nfe2 has been identified bound to the β-globin promoter.60 This binding, which is presumably indirect, is independent of the LCR.62 Together, these experiments demonstrate that Nfe2 binds to the LCR, and possibly to globin promoters, in erythroid cells. Direct evidence is lacking, however, for globin gene activation by LCR-bound p45Nfe2.
Despite the sensitivity of the allele-specific RNA PCR assay, we failed to identify a role for p45Nfe2 in activating α-globin expression through the AP1/NFE2 sites in HS-26. One explanation for this result may be that another protein, or proteins, is fully redundant with p45Nfe2. A second may be that another protein is uniquely required and p45Nfe2 is not involved. In either case, our experiments provide genetic evidence that a protein other than p45Nfe2 can activate globin gene expression through these sites. The AP1/NFE2 sites in HS-26 match the NFE2 consensus sequence at 10 of 11 nucleotides. As part of a heterodimer with a small Maf protein, members of the cap'n collar family (p45Nfe2, Nrf1, Nrf2, and Nrf3) require the full NFE2 site to bind DNA.32 Conservation of this binding site, albeit imperfect, implies that one or more proteins from this family may be involved in globin gene activation. We have shown that Nrf2, like p45Nfe2, is not required for α-globin activation through the AP1/NFE2 sites in HS-26 (Martin et al63 ; and P.A.N., unpublished results, January 2003). Potentially, no single protein is essential, but several members of this family or other members of the BZIP transcription factor family can function through these sites. In this regard, chromatin immunoprecipitation of these sites may provide useful information.
The mechanism whereby remote cis-acting sequences activate high-level α- and β-globin expression is an unsolved problem in the field of gene regulation. Considerable evidence supports the view that the active upstream sequences are concentrated into functionally redundant enhancers. For HS-26, we have shown that a pair of AP1/NFE2 binding sites is required for full enhancer activity and α-globin gene transcription. This effect does not depend on p45Nfe2. Future studies will be directed toward identifying the protein or proteins that activate globin gene expression through these sites.
Prepublished online as Blood First Edition Paper, August 14, 2003; DOI 10.1182/blood-2003-02-0574.
Supported by National Institutes of Health grant DK53469 and by the American, Lebanese, and Syrian Associated Charities.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Beatrice Allnutt and Lawryn Kasper for technical assistance. We thank Janet Partridge and Paul Brindle for reviewing the manuscript and Chris Lowrey for help with the hypersensitivity assay. We thank the Transgenic Core of St Jude Children's Research Hospital for the production of genetically modified mice. We thank Stuart Orkin for the gift of p45Nfe2-deficient mice and Steve Fiering for the Diphtheria A toxin gene cassette. We also thank Doug Higgs for sharing sequence information on the α-globin locus in advance of publication.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal