Abstract
Acute promyelocytic leukemia (APL) is characterized by the PML-RARA fusion gene. To identify genetic changes that cooperate with PML-RARA, we performed spectral karyotyping analysis of myeloid leukemias from transgenic PML-RARA mice and from mice coexpressing PML-RARA and BCL2, IL3, activated IL3R, or activated FLT3. A cooperating mutation that enhanced survival (BCL2) was not sufficient to complete transformation and was associated with multiple numeric abnormalities, whereas cooperating mutations that deregulated growth and enhanced survival were associated with normal karyotypes (IL3) or simple karyotypic changes (IL3R, FLT3). Recurring abnormalities included trisomy 15 (49%), trisomy 8 (46%), and -X/-Y (54%). The most common secondary abnormality in human APL is +8 or partial trisomy of 8q24, syntenic to mouse 15. These murine leukemias have a defined spectrum of changes that recapitulates, in part, the cytogenetic abnormalities found in human APL. Our results demonstrate that different cooperating events may generate leukemia via different pathways.
Introduction
Acute promyelocytic leukemia (APL) is a distinct clinicopathologic entity characterized by the t(15;17)(q22;q11.2-12) and the resultant PML-RARA fusion gene.1,2 Expression of PML-RARA in transgenic mice initiates myeloid leukemias with features of human APL.3 We have reported a transgenic mouse model for APL, in which the human MRP8 promoter drives expression of PML-RARA in myeloid cells.4 The long latency before disease onset (median, 8.5 months), together with the incomplete penetrance (64% at 1 year), suggest that leukemogenesis initiated by PML-RARA requires additional genetic changes. This hypothesis is consistent with other transgenic mouse models of acute myeloid leukemia (AML), in which cooperating mutations are required for leukemogenesis by fusion genes (eg, RUNX1/ETO, CBFB/MYH11, and MLL-ELL).5
An important aspect of leukemia biology is the elucidation of the spectrum of chromosomal abnormalities and molecular mutations that cooperate in the pathways leading to leukemogenesis.6 To identify genetic changes that cooperate with PML-RARA in vivo, we performed spectral karyotyping analysis of leukemias arising in PML-RARA mice and mice expressing PML-RARA plus cooperating mutations. Our results suggest that different cooperating events may generate acute leukemia via different pathways and that the particular combination leads to a specific pattern of additional mutations that complete leukemic transformation.
Study design
Generation of leukemias
Mice (FVB/N background) were bred and maintained at the University of California at San Francisco in accordance with institutional guidelines. PML-RARA/BCL2 mice were reconstituted with MRP8–PML-RARA/MRP8-BCL2 doubly transgenic bone marrow or with MRP8–PML-RARA bone marrow that had been transduced with a retroviral vector expressing BCL2.4,7 PML-RARA/FLT3W51, PML-RARA/IL3, and PML-RARA/IL3R mice were reconstituted with MRP8–PML-RARA bone marrow that had been transduced with retroviral vectors expressing an activated version of FMS-related tyrosine kinase 3 (FLT3; murine stem cell virus [MSCV]-FLT3W51-internal ribosomal entry site [IRES]-green fluorescent protein [GFP]),8,9 interleukin 3 (IL3; pMPZen-IL-3),10,11 or an activated IL3R (βcV449E),11,12 respectively.
Spectral karyotyping (SKY) analysis
Cytogenetic analysis was performed on fresh or cryopreserved spleen or bone marrow cells obtained at the time of development of leukemia. The initiation of short-term (24-72 hours) cultures, metaphase cell preparation, and spectral karyotyping were performed as described previously.13 A minimum of 10 metaphase cells were analyzed per leukemia.
Results and discussion
Cytogenetic analysis of murine leukemias initiated by PML-RARA
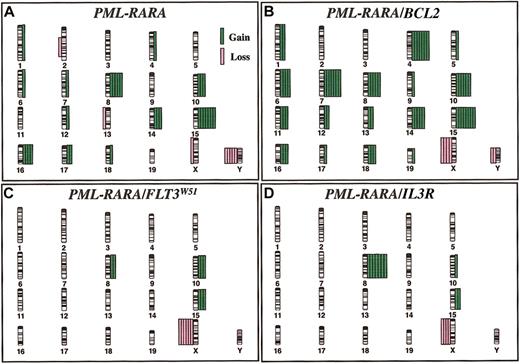
The results of SKY analysis and features of the murine leukemias are given in Table 1. The distribution of chromosome abnormalities in the genetic cohorts is illustrated in Figure 1; the complete karyotypes are provided on the laboratory's website (http://LeBeauLab.uchicago.edu, Table D), and in previous reports.9,11,13 Each of the additional mutations cooperated with PML-RARA, as evidenced by the increase in leukemia incidence from 64% to between 70% and 100% and the decrease in latency period from 257 days to between 14 and 127 days. The incidence of clonal chromosomal abnormalities was high, ranging from 80% to 100% of cases, in all but the cohort expressing PML-RARA and IL3, in which all cases had a normal karyotype. The most common abnormality was loss of a sex chromosome, an X in females or a Y chromosome in males, noted in 22 (54%) of 41 cases. A gain of chromosome 15 was observed in 20 (49%) cases, and trisomy 8 was observed in 19 (46%) cases.
Characteristics of murine leukemias initiated by PML-RARA or PML-RARA and cooperating mutations
. | Cooperating mutations . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter . | None . | BCL2 . | FLT3 . | IL3R . | IL3 . | ||||
| Leukemia, % | 64 | 100 | 70 | 100 | 100 | ||||
| Median latency, d | 257 | 127 | 105 | 49 | 14-21 | ||||
| Disease | AML | AML | AML | AML | AML* | ||||
| Leukocytosis | No | No | Yes | Yes | Yes | ||||
| Anemia | Yes | Yes | Yes | Yes | Yes | ||||
| Thrombocytopenia | Yes | Yes | Yes | Yes | Yes | ||||
| Disseminated disease | Yes | Yes | Yes | Yes | Yes | ||||
| Transplantable | Yes | Yes | Yes | Yes | No | ||||
| ATRA response | Yes | Yes | Yes | Yes | Yes | ||||
| SKY analysis | |||||||||
| No. cases | 11 | 8 | 7 | 9 | 6 | ||||
| Abnormal (%) | 10 (91) | 8 (100) | 7 (100) | 7 (78) | 1 (17)† | ||||
| Karyotype complexity | Intermediate | Complex | Simple | Simple | Normal | ||||
| Numeric abnormality (%) | 10 (91) | 8 (100) | 7 (100) | 7 (78) | 0 | ||||
| Structural Abnormality (%) | 2 (18) | 1 (13) | 0 | 0 | 1 (17)† | ||||
| Recurring abnormalities (%) | |||||||||
| +4 | 1 (9) | 7 (88) | 0 | 0 | 0 | ||||
| +6 | 1 (9) | 4 (50) | 0 | 0 | 0 | ||||
| +7 | 1 (9) | 7 (88) | 0 | 0 | 0 | ||||
| +8 | 5 (45) | 5 (63) | 2 (29) | 7 (78) | 0 | ||||
| +10 | 3 (27) | 6 (75) | 3 (43) | 1 (11) | 0 | ||||
| +14 | 3 (27) | 5 (63) | 0 | 0 | 0 | ||||
| +15 | 7 (64) | 8 (100) | 3 (43) | 2 (22) | 0 | ||||
| +16 | 4 (36) | 3 (38) | 0 | 0 | 0 | ||||
| -X/-Y | 6 (55) | 6 (75) | 6 (86) | 4 (44) | 0 | ||||
. | Cooperating mutations . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter . | None . | BCL2 . | FLT3 . | IL3R . | IL3 . | ||||
| Leukemia, % | 64 | 100 | 70 | 100 | 100 | ||||
| Median latency, d | 257 | 127 | 105 | 49 | 14-21 | ||||
| Disease | AML | AML | AML | AML | AML* | ||||
| Leukocytosis | No | No | Yes | Yes | Yes | ||||
| Anemia | Yes | Yes | Yes | Yes | Yes | ||||
| Thrombocytopenia | Yes | Yes | Yes | Yes | Yes | ||||
| Disseminated disease | Yes | Yes | Yes | Yes | Yes | ||||
| Transplantable | Yes | Yes | Yes | Yes | No | ||||
| ATRA response | Yes | Yes | Yes | Yes | Yes | ||||
| SKY analysis | |||||||||
| No. cases | 11 | 8 | 7 | 9 | 6 | ||||
| Abnormal (%) | 10 (91) | 8 (100) | 7 (100) | 7 (78) | 1 (17)† | ||||
| Karyotype complexity | Intermediate | Complex | Simple | Simple | Normal | ||||
| Numeric abnormality (%) | 10 (91) | 8 (100) | 7 (100) | 7 (78) | 0 | ||||
| Structural Abnormality (%) | 2 (18) | 1 (13) | 0 | 0 | 1 (17)† | ||||
| Recurring abnormalities (%) | |||||||||
| +4 | 1 (9) | 7 (88) | 0 | 0 | 0 | ||||
| +6 | 1 (9) | 4 (50) | 0 | 0 | 0 | ||||
| +7 | 1 (9) | 7 (88) | 0 | 0 | 0 | ||||
| +8 | 5 (45) | 5 (63) | 2 (29) | 7 (78) | 0 | ||||
| +10 | 3 (27) | 6 (75) | 3 (43) | 1 (11) | 0 | ||||
| +14 | 3 (27) | 5 (63) | 0 | 0 | 0 | ||||
| +15 | 7 (64) | 8 (100) | 3 (43) | 2 (22) | 0 | ||||
| +16 | 4 (36) | 3 (38) | 0 | 0 | 0 | ||||
| -X/-Y | 6 (55) | 6 (75) | 6 (86) | 4 (44) | 0 | ||||
ATRA indicates all-trans-retinoic acid
Diseased animals show pathologic features of AML
Robertsonian translocation (centric fusion) noted in 2 cells has no known phenotypic consequences
The distribution of chromosomal abnormalities in murine leukemias. The distribution of chromosomal abnormalities in (A) PML-RARA, (B) PML-RARA/BCL2, (C) PML-RARA/FLT3W51, and (D) PML-RARA/IL3R (βcV449E) mice reveals a defined spectrum of numeric abnormalities. Chromosome gain is depicted by green bars on the right of each chromosome, and chromosome loss is depicted by pink bars on the left. Each bar represents a single case.
The distribution of chromosomal abnormalities in murine leukemias. The distribution of chromosomal abnormalities in (A) PML-RARA, (B) PML-RARA/BCL2, (C) PML-RARA/FLT3W51, and (D) PML-RARA/IL3R (βcV449E) mice reveals a defined spectrum of numeric abnormalities. Chromosome gain is depicted by green bars on the right of each chromosome, and chromosome loss is depicted by pink bars on the left. Each bar represents a single case.
Of the 11 cases analyzed in PML-RARA transgenic mice, clonal abnormalities were identified in 10 (91%). The most common abnormality was a gain of chromosome 15 (7 cases, 64%; Figure 1A). Loss of a sex chromosome was noted in 6 cases (55%), and trisomy 8 was observed in 5 (45%) cases.
BCL2 is expressed in most APLs.14 In the PML-RARA/BCL2 cohort, all leukemias had a complex karyotype and were characterized by multiple abnormal clones with multiple numeric abnormalities in each clone (Table 1; Figure 1B). All but 2 cases had at least 45 chromosomes in the abnormal clone(s), compared with 1 of 11 leukemias arising in PML-RARA mice and none in the other genetic cohorts. All 8 cases were characterized by trisomy 15 (100%). Of interest is that 6 of 8 cases had the identical pattern of +4, +7, +10, and +15.
The FLT3 receptor tyrosine kinase enhances the proliferation and survival of hematopoietic progenitors in response to the FLT3 ligand. Activating FLT3 mutations have been identified in approximately 35% of APLs, most of which result from an internal tandem duplication (ITD) of the juxtamembrane domain.15 All of the leukemias arising in the PML-RARA/FLT3W51 mice had abnormal karyotypes; however, the karyotypes were very simple with 6 (86%) of 7 showing loss of an X chromosome, together with either +10 or +15 (each in 3 of 7 cases, 43%; Figure 1C).9
Activating mutations of the common β chain of the IL3, IL5, and granulocyte-macrophage colony-stimulating factor (GM-CSF) receptors confer cytokine independence.10,12 In the PML-RARA/IL3R (βcV449E) leukemias, 7 (78%) of 9 cases were abnormal with simple karyotypes. Trisomy 8 (7 [78%] of 9 cases) was most common, followed by loss of an X chromosome (4 [44%] of 9 cases), and trisomy 15 (2 [22%] of 9 cases; Figure 1D).11
We have reported that complete trisomy 8 or partial trisomy of bands 8q23-24 (12.4%) are the most common secondary chromosomal abnormalities in human APL.13 Trisomy 15 is the most frequent abnormality in murine T-cell lymphomas (40%-90%), as well as in B/myeloid leukemias arising in irradiated Eμ-BCL2 transgenic mice.16,17 The smallest region gained is bands 15D2-3; this region is syntenic to human 8q23-24 and contains the Myc and Pvt1 genes.17 At present, the identity of the relevant gene(s) on human 8q is unknown; however, AMLs characterized by trisomy 8 overexpress genes on chromosome 8, suggesting that gene dosage effects mediate leukemogenesis.18 Perhaps the best candidate gene is MYC, which is overexpressed in a number of human tumors.
Genetic pathways to APL
We have demonstrated that the particular combination of cooperating events leads to an obligate pattern of additional mutations that complete transformation. The initiating event, the PML-RARA fusion protein, impairs differentiation; however, leukemogenesis requires additional mutations. The addition of a cooperating mutation that enhances survival, such as BCL2 expression, is not sufficient to complete transformation and is associated with instability and the acquisition of multiple numeric abnormalities. In contrast, cooperating mutations that deregulate growth, such as activated cytokine signaling pathways (FLT3W51, IL3R, and IL3), are associated with the rapid development of leukemia and simple karyotypic changes or normal karyotypes. There is redundancy in the pathways activated by FLT3 and IL3/IL3R. Both induce MYC expression via RAS/MAPK (rat sarcoma viral oncogene/mitogen-activated protein kinase) activation, BCLXL expression via STAT5 (signal transducer and activator of transcription 5) and PI3K/Akt (phosphatidylinositol 3–kinase/v-akt murine thymoma viral oncogene homolog), and BCL2 phosphorylation via ERK1/2 (extracellular signal-related kinase 1/2) or PLCγ (phospholipase C–γ) promoting its potent antiapoptotic function.19 Moreover, FLT3-ITD mutants induce a transcriptional program that resembles activation of the IL3R, rather than that of wild-type FLT3.20 Overexpression of MYC induces apoptosis, and tumor cell viability is dependent upon cooperating overexpression of BCL2 family members.21 We speculate that the increase in MYC levels by activation of the IL3 receptor (IL3R) or FLT3 might decrease selective pressure for gain of a third MYC allele via acquisition of an additional chromosome 15. The observation of trisomy 15 in 100% of PML-RARA/BCL2 leukemias, but in only 43% of PML-RARA/FLT3W51, 22% of PML-RARA/IL3R, and 0% of PML-RARA/IL3 mice, is consistent with this hypothesis. Identifying the genes that drive chromosome gains or losses in human and murine myeloid leukemias may offer insights into the pathogenesis or progression of leukemias and may provide new therapeutic targets.
Prepublished online as Blood First Edition Paper, April 10, 2003; DOI 10.1182/blood-2003-01-0155.
Supported by National Institutes of Health (NIH) grants U01 CA84221 (M.M.L., S.C.K.), K08 CA75986 (S.C.K.), and CA 95274 (S.C.K.). S.C.K. is the 32nd Edward Mallinckrodt Junior Scholar and is a recipient of a Burroughs Wellcome Fund Career Award.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Kevin Shannon, James Downing, Nigel Killeen, David Largaespada, Nancy Jenkins, and Neal Copeland for helpful discussions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal