Abstract
Hemophilia A is a life-threatening, hemorrhagic, X-linked recessive disorder resulting in deficient factor VIII (FVIII) activity. After the infusion of therapeutic FVIII, 25% of patients develop anti-FVIII antibodies that inhibit FVIII procoagulant activity, thus precluding further administration of FVIII. Here we report a novel approach aimed at neutralizing the activity of FVIII inhibitors by peptide epitope surrogates. To illustrate our concept, we chose the human anti-FVIII monoclonal antibody, Bo2C11, as a representative of anti-FVIII antibodies and a phage-displayed peptide library approach to obtain surrogate peptides. We selected a series of constrained dodecapeptides with the core sequence W-NR, which specifically interacts with the combining site of Bo2C11. The peptides mimic the epitope recognized by Bo2C11 and are able to inhibit specifically and in a dose-dependent manner the binding of Bo2C11 to FVIII. Peptide 107, in particular, neutralized the activity of Bo2C11 in vitro and restored normal hemostasis in hemophilic mice. Thus, the use of peptide decoys may be a promising new approach for the neutralization of pathologic antibodies.
Introduction
Factor VIII (FVIII) is a cofactor in the generation of thrombin by the intrinsic pathway tenase complex. It is synthesized as a 330-kDa precursor protein with an A1-a1-A2-a2-B-a3-C1-C2 domain structure, which subsequently undergoes proteolytic processing.1 FVIII circulates as a series of heterodimers, consisting of a heavy chain (domains A1-a1-A2-a2) and variable lengths of the B domain, linked by a metal bridge to a light chain (domains a3-A3-C1-C2). Dysfunction or deficiency in FVIII results in a bleeding disorder called hemophilia A. The therapeutic administration of exogenous FVIII to patients with hemophilia results in the emergence of antibodies (Abs) that inhibit FVIII procoagulant activity in up to 25% of patients.2
Inhibitor antibodies are preferentially directed against epitopes in the A2, A3, and C2 domains of FVIII.3 Two major distinct determinants have been identified in the C2 domain that encompass residues Val2248 to Ser2313 and residues Glu2181 to Val2243 and that are brought together by a disulfide bridge between cysteines Cys2174 and Cys2236.4 Most anti-C2 domain Abs inhibit FVIII procoagulant activity by interfering with the physiologic interaction of C2 with von Willebrand factor (VWF) and phospholipids (PLs). Bo2C11, a human immunoglobulin G4κ (IgG4κ) monoclonal antibody (mAb) derived from an inhibitor-positive patient with hemophilia A, is representative of this class of anti-C2 inhibitors.5 Bo2C11 binds to a conformational epitope composed of side chains of various discontinuous regions of C2.6
Although the mechanisms by which Abs inhibit FVIII activity are now well elucidated, the presence of FVIII inhibitors remains a major threat for patients. Available therapeutic approaches to bypass FVIII inhibitors are prohibitively expensive, and their efficacy is often limited to patients with recent or low-titer inhibitors. An alternative approach to neutralize FVIII inhibitors may reside in the use of surrogate epitopes that mimic major FVIII epitopes and disrupt the interaction between FVIII and anti-FVIII antibodies, thus restoring the biologic activity of FVIII. Because most antibodies recognize discontinuous epitopes,7 we have chosen to isolate peptides that functionally mimic discontinuous epitopes using libraries of random peptides displayed by filamentous phages. Such libraries have proven to be powerful tools for the selection of peptides that mimic linear, conformational, and even nonproteinaceous epitopes.8 Selected epitopes do not necessarily resemble natural ligands but rather mimic their binding properties and are called mimotopes.9 To establish the feasibility of this approach, we have investigated, both in vitro and in vivo, the ability of such peptides to disrupt the interaction between the C2 domain and Bo2C11.
Study design
Isolation of Bo2C11-binding peptides from random peptide libraries
Two M13 phage libraries were obtained that express either linear 15-mer random peptides (X15) or constrained 12-mer random peptides (XCX8CX) at the surface of the pVIII protein.10 Two additional phage libraries, expressing either linear random nonapeptides (X9) or constrained random dodecapeptides (CX10C) at the surface of the pIII protein, were obtained.11 Generation and characterization of Bo2C11 has been described elsewhere.5
Pannings were performed as described.12 Briefly, Petri dishes were coated with Bo2C11 in 100 mM NaHCO3, pH 8.6, at 1 μg/mL for the first 2 pannings and 0.1 μg/mL for the last one, overnight at 4°C with gentle shaking. For the first panning, 5 × 1012 transducting units (TUs) of each library in Tris-buffered saline (TBS)–0.05% Tween-20 were incubated with the immobilized mAb. Unbound phages were washed with TBS-0.5% Tween-20. Bound phages were eluted (0.1 N HCl–5 M glycine–1 mg/mL bovine serum albumin [BSA], pH 2.2) and were used to infect Escherichia coli K91 cells. After 3 rounds of enrichment, individual phage clones were isolated and further analyzed. Peptides expressed on the pVIII or pIII proteins were sequenced using the primer 5′-TCG GCA AGC TCT TTT AGG-3′ or 5′-GTT TTG TCG TCT TCC AGA CG-3′ for annealing.
Screening ELISA of specific phages
Enzyme-linked immunosorbent assay (ELISA) plates were coated with 0.1 μg/mL Bo2C11 or human IgG4κ in 100 mM NaHCO3, pH 8.6, overnight at 4°C. Plates were washed with phosphate-buffered saline (PBS)–0.1% Tween-20 blocked with PBS-0.1% Tween-20–2% nonfat dried milk (wt/vol) (blocking buffer) for 1 hour at 37°C. A mixture of 50 μL phage dilution and 50 μL blocking buffer was incubated for 90 minutes at 37°C. Bound phages were revealed using a peroxidase-conjugated anti-M13 IgG.
Synthesis of soluble peptides
Soluble peptides were synthesized on an Abimed AMS422 synthesizer (Abimed GmbH, Langenfeld, Germany) by Fmoc chemistry.13 Peptides were then deprotected and released from the resin by trifluoroacetic acid treatment in the presence of appropriate scavengers and lyophilized. The purity of peptides was assessed by reverse-phase high-performance liquid chromatography (HPLC) and mass spectrometry, and it generally reached 95%. When necessary, the peptides were further purified by HPLC.
Competitive ELISA assay with synthetic peptides
Bo2C11 (3.5 nM) was preincubated overnight at 4°C with synthetic peptides (2 nM-200 μM). ELISA plates were coated with FVIII (1 μg/mL; Kogenate Bayer, Berkeley, CA) in PBS overnight at 4°C. Plates were washed and blocked for 1 hour at 37°C. The mixture of Bo2C11 and peptides was added to the plates for a 2-hour incubation at 37°C. The binding of Bo2C11 was revealed using a peroxidase-conjugated antihuman IgG. Reference absorbance was obtained with the incubation of Bo2C11 alone.
Neutralization of the inhibitory activity of Bo2C11 in the Bethesda assay
We used the Bethesda test, modified as described by Nijmegen.14 Briefly, Bo2C11 at 3.5 nM, a concentration yielding less than 5% residual FVIII activity, was incubated with peptides diluted in veronal buffer (0.5-100 μM each peptide) overnight at 4°C. Samples (35 μL) were then incubated at equal volumes with normal plasma buffered with 0.1 M imidazole, pH 7.4, for 2 hours at 37°C. Coagulation assays were then carried out. Results were expressed as the percentage of residual FVIII activity. Each dilution was tested in triplicate.
In vivo neutralization of FVIII inhibition
Peptide-mediated neutralization of FVIII inhibition was evaluated in vivo using FVIII knockout (KO) mice as a model of hemophilia A.15 Briefly, exon 16 disrupted FVIII KO males aged 2 months were used in groups of 3 individuals. Recombinant human FVIII (0.5 IU in 100 μL physiologic saline) was injected into the tail vein. Preliminary experiments demonstrated that 0.25 μg Bo2C11 was sufficient to inhibit 100% of the activity of infused FVIII. Single tail injections of 0.25 μg Bo2C11 (16.7 nM) alone or preincubated with 1 mg peptide (0.65 mM)—followed 30, 60, and 120 minutes later by an injection of 0.5 IU FVIII—were injected in 4, 2, and 2 mice, respectively. Blood samples were obtained by cardiac puncture 15 minutes after FVIII injection, and FVIII concentration in plasma was measured in a chromogenic assay (Dade Behring).
Results and discussion
Selection and characterization of Bo2C11-binding peptides
Four peptide libraries were used to identify FVIII mimotopes. Two libraries of random pentadecapeptides (X15) or dodecapeptides (XCX8CX) fused to the N-terminal of the major coat protein (pVIII) and 2 libraries of nonapeptides (X9) and dodecapeptides (CX10C) fused to the N-terminal minor coat protein (pIII), respectively, were sorted against immobilized Bo2C11. Two of the 4 libraries, X15 and X9, displayed linear peptides, whereas the others, XCX8CX and CX10C, displayed peptides constrained by 2 fixed flanking cysteine residues. After 3 cycles of panning and phage amplification, 600 individually isolated bacterial colonies were randomly picked from the third round, and the corresponding phages were assayed for binding to Bo2C11 in ELISA. Analysis revealed that 88 of the 600 clones exhibited a strong binding capacity to Bo2C11—that is, at least 4 times higher than background. Their specificity for Bo2C11 was confirmed by testing them against other anti-C2 domain mAbs and against control Abs of irrelevant specificity. No significant cross-reactivity was observed (data not shown). Peptide sequences of the positive clones are shown in Table 1. Twenty-seven distinct sequences were obtained, 23 of which shared the consensus motif WXNR, with X being any amino acid. This motif was found on pIII- and pVIII-displayed peptides. All selected peptides had a higher-than-expected frequency of positive residues and were derived from the 2 constrained dodecapeptide libraries: 6 peptides originated from the CX10C library, and the remaining was derived from the XCX8CX library, suggesting that a constrained conformation is required for optimal peptide recognition by Bo2C11. The primary structure of the C2 domain did not reveal any homology with the motif WXNR, suggesting that the selected peptides mimic the discontinuous epitope recognized by Bo2C11 on the C2 domain.5 The different sequences were synthesized on a cellulose membrane using the Spot technique.16 All membrane-bound peptides, except peptides KCEPDDPWPQCI and ACKRNHRWGACV, were reactive with Bo2C11 (data not shown), demonstrating that the phage framework is not involved in the binding of the peptides to Bo2C11. The most reactive peptides (103, 104, 105, 106, 107, 108, 109; Table 1) were synthesized in soluble form for further characterization. In addition, alanine scanning was performed in which each residue of the lead sequences was systematically replaced by an alanine. This allowed us to identify Trp5, Asn7, and Arg8 as the most critical residues for Bo2C11 binding (data not shown). Interestingly, these 3 residues are part of the consensus motif WXNR.
Sequences of peptides isolated after screening the phage libraries
Prevalence . | Sequence . | Peptide no. . |
|---|---|---|
| 1 | C M K W S N R S S R W C | 106 |
| 1 | Q C S K W V N R S R C A | — |
| 1 | C S K W H N R S K R H C | 105 |
| 1 | C S K W A N R L V S I C | — |
| 3 | N C G K W T N R R T C L | — |
| 1 | Q C S R W S N R T S C T | — |
| 2 | K C S R W T N R H L C D | — |
| 2 | K C T R W T N R H L C S | — |
| 2 | K C T R W T N R A H C P | — |
| 13 | E C T R W S N R S R C F | — |
| 3 | C G R W F N R S D L H C | — |
| 4 | Q C G R W S N R S Y C S | — |
| 8 | K C G R W S N R S S C T | 103 |
| 2 | T C H R W G N R T S C Q | — |
| 1 | Q C H R W A N R I S C S | — |
| 13 | Q C H T W S N R R S C L | 108 |
| 4 | A C T T W S N R S K C P | — |
| 1 | R C T Q W T N R A Y C P | — |
| 1 | A C T Q W S N R H M C G | — |
| 2 | S C H A W S N R R T C R | 107 |
| 8 | R C H A W S N R K S C V | 109 |
| 2 | A C H E W S N R S T C T | — |
| 1 | K C G P W S N R S S C T | — |
| 1 | T C H P F S N R S T C T | — |
| 7 | K C E P D D P W P Q C I | — |
| 2 | A C K R N H R W G A C V | — |
| 1 | E C G S H A W G R R C K | 104 |
Prevalence . | Sequence . | Peptide no. . |
|---|---|---|
| 1 | C M K W S N R S S R W C | 106 |
| 1 | Q C S K W V N R S R C A | — |
| 1 | C S K W H N R S K R H C | 105 |
| 1 | C S K W A N R L V S I C | — |
| 3 | N C G K W T N R R T C L | — |
| 1 | Q C S R W S N R T S C T | — |
| 2 | K C S R W T N R H L C D | — |
| 2 | K C T R W T N R H L C S | — |
| 2 | K C T R W T N R A H C P | — |
| 13 | E C T R W S N R S R C F | — |
| 3 | C G R W F N R S D L H C | — |
| 4 | Q C G R W S N R S Y C S | — |
| 8 | K C G R W S N R S S C T | 103 |
| 2 | T C H R W G N R T S C Q | — |
| 1 | Q C H R W A N R I S C S | — |
| 13 | Q C H T W S N R R S C L | 108 |
| 4 | A C T T W S N R S K C P | — |
| 1 | R C T Q W T N R A Y C P | — |
| 1 | A C T Q W S N R H M C G | — |
| 2 | S C H A W S N R R T C R | 107 |
| 8 | R C H A W S N R K S C V | 109 |
| 2 | A C H E W S N R S T C T | — |
| 1 | K C G P W S N R S S C T | — |
| 1 | T C H P F S N R S T C T | — |
| 7 | K C E P D D P W P Q C I | — |
| 2 | A C K R N H R W G A C V | — |
| 1 | E C G S H A W G R R C K | 104 |
Prevalence indicates number of identical sequences found
Cysteine residues involved in disulfide bridges appear in italics; consensus motifs appear in boldface
— indicates that the corresponding peptide was not synthesized
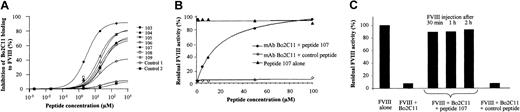
Experiments were performed to evaluate the capacity of peptides 103 to 109 to compete for the FVIII binding site of Bo2C11. The 7 selected peptides inhibited the binding of Bo2C11 to immobilized FVIII in a dose-dependent manner (Figure 1A). Peptide 107 showed a peptide concentration giving 50% inhibition of binding (IC50) of 3 μM. This inhibition was found to be specific because an irrelevant peptide showed no inhibitory effect. The IC50 of other peptides ranged from 15 to 35 μM. The rest of the study was therefore focused on peptide 107. The epitope recognized by Bo2C11 overlaps with the binding sites of PL and VWF on FVIII.5,6 Interestingly, peptide 107 did not inhibit the binding of FVIII either to PL or to VWF at concentrations up to 200 μM (data not shown).
Capacity of peptides selected by phage display to neutralize Bo2C11 to FVIII. (A) Inhibition of the binding of Bo2C11 to immobilized FVIII by increasing concentrations of specific peptides (peptides 103-109) or irrelevant ones (controls 1 and 2). (B) Bo2C11 preincubated overnight at 4°C with increasing concentrations of peptide 107 or an irrelevant peptide was incubated in the presence of FVIII for 2 hours at 37°C. Residual FVIII activity was then measured in a 1-stage clotting assay. Results are expressed as percentage residual FVIII activity, which represents the percentage of neutralization of Bo2C11 by peptide 107. The concentration of Bo2C11 (3.5 nM) was chosen as providing close to complete (98%) inhibition of the FVIII residual activity (data not shown). (C) Bo2C11 alone (16.7 nM) or after preincubation with peptide 107 or a control peptide was injected into the tail vein of FVIII KO mice. Recombinant human FVIII (0.5 IU) was injected 30, 60, and 120 minutes later. Residual FVIII activity in plasma was measured in a chromogenic assay 15 minutes later. One hundred percent activity corresponded to 0.3 IU/mL.
Capacity of peptides selected by phage display to neutralize Bo2C11 to FVIII. (A) Inhibition of the binding of Bo2C11 to immobilized FVIII by increasing concentrations of specific peptides (peptides 103-109) or irrelevant ones (controls 1 and 2). (B) Bo2C11 preincubated overnight at 4°C with increasing concentrations of peptide 107 or an irrelevant peptide was incubated in the presence of FVIII for 2 hours at 37°C. Residual FVIII activity was then measured in a 1-stage clotting assay. Results are expressed as percentage residual FVIII activity, which represents the percentage of neutralization of Bo2C11 by peptide 107. The concentration of Bo2C11 (3.5 nM) was chosen as providing close to complete (98%) inhibition of the FVIII residual activity (data not shown). (C) Bo2C11 alone (16.7 nM) or after preincubation with peptide 107 or a control peptide was injected into the tail vein of FVIII KO mice. Recombinant human FVIII (0.5 IU) was injected 30, 60, and 120 minutes later. Residual FVIII activity in plasma was measured in a chromogenic assay 15 minutes later. One hundred percent activity corresponded to 0.3 IU/mL.
Capacity of peptide 107 to neutralize Bo2C11 in functional and in vivo assays
Using the Bethesda assay, we evaluated the ability of peptide 107 to restore FVIII activity in the presence of Bo2C11. At a concentration of 3.5 nM Bo2C11, which resulted in 98% inhibition of FVIII activity, peptide 107 neutralized in a dose-dependent manner the inhibitory activity of Bo2C11, with an IC50 of 19 μM (Figure 1B), whereas a control peptide exerted no neutralizing effect. Peptide 107 incubated with FVIII alone did not alter the activity of FVIII. Other peptides (103, 104, 105, 106, 108, 109) also efficiently neutralized the inhibitory activity of Bo2C11, with an IC50 ranging from 30 to 50 μM (data not shown).
We then investigated the neutralizing capacity of peptide 107 toward Bo2C11 in vivo. Intravenous administration of 0.5 IU recombinant human FVIII to FVIII KO mice restored FVIII activity in plasma of approximately 0.3 IU/mL. Injection of 0.25 μg (16.7 nM) Bo2C11 30 minutes before FVIII administration was sufficient to inhibit 100% of FVIII activity. The inhibitory activity of Bo2C11 was completely abrogated when Bo2C11 was preincubated with 650 μM peptide 107 (Figure 1C). A similar effect was observed when the Bo2C11–peptide 107 mixture was administered to the mice 1 hour and 2 hours before FVIII injection (Figure 1C). This was not observed on the preincubation of Bo2C11 with a control-irrelevant peptide (700 μM). Administration of peptide 107 alone had no effect on FVIII half-life (data not shown). Recent evidence demonstrates that low-density lipoprotein receptor-related protein (LRP) plays an important role in FVIII clearance and catabolism.17 Binding of FVIII to LRP involves the C2 and A2 domains of FVIII. Peptide 107 (up to 50 μM) did not inhibit the binding of sodium iodide125I–FVIII to immobilized LRP and did not interfere with the uptake and degradation of FVIII by LRP-expressing mouse embryonic fibroblast cells (data not shown). Altogether, the data indicate that the neutralizing capacity of peptide 107 toward Bo2C11-mediated FVIII inhibition, which was initially suggested by in vitro experiments, was also effective under in vivo conditions in a murine model of hemophilia A.
In this study, we focused on selecting and characterizing peptides that mimic FVIII epitopes and that interfere with the recognition of FVIII by anti-FVIII Abs. The human inhibitory anti-FVIII mAb Bo2C11 was used as a model antibody to screen various phage-displayed peptide libraries. The selected peptides were all disulfide-constrained dodecapeptides and share the consensus motif WXNR. Both their constrained conformation and the conserved nature of the consensus residues appeared important, both for direct binding to Bo2C11 and for the structural conformation of the peptide. Among the peptides studied, peptide 107 was the most potent inhibitor with an IC50 of 3 μM. Peptide 107 mimicked the epitope of Bo2C11: It neutralized the inhibitory activity of Bo2C11 both in functional in vitro assays and in a murine model of hemophilia A. Interestingly, peptide 107 retained its neutralizing properties for at least 2 hours in mice plasma, suggesting either that peptide 107 circulates essentially in a bound form and is not readily susceptible to protease degradation or that its half-life is increased because of the capping of its N- and C-termini by acetylation and amidation, respectively. Altogether, the stability and neutralizing efficiency of peptide 107 are particularly encouraging in view of a possible therapeutic application.
This study provides proof of the concept that challenging the interaction of human FVIII inhibitors with FVIII by using peptide decoys may restore FVIII procoagulant activity. The interaction of FVIII with VWF protects it from proteolytic degradation in the circulation, and the binding of activated FVIII is compulsory for the formation of the tenase complex.18 Although peptide 107 completely neutralized the inhibitory activity of Bo2C11, it did not interfere with the binding of the C2 domain of FVIII to VWF and PLs, which is relevant to its use in a clinical setup. Indeed, no competitive effect was observed at peptide 107 concentrations up to 200 μM, whereas the use of peptides 2302-2317 and 2308-2322, tested under similar conditions, inhibited FVIII binding to PLs with IC50 values of approximately 16 μM and 20 μM, respectively.19 Thus, the ability of the phage-display technique to select peptides able to mimic FVIII but without showing sequence homology is a great advantage for our approach given that the retained peptides target specifically the interaction between FVIII and inhibitors without hampering FVIII functions.
Because of their small size, peptides are poor immunogens and are inefficient in triggering strong T-cell responses. It is thus presumable that they might be ignored by the immune system on intravenous administration. Usage of peptides in therapy is generally challenged by their short in vivo half-life consequent to their susceptibility to degradation by proteases. Introduction of modifications to peptides, such as the replacement of L amino acids by D amino acids20 or by unnatural amino acids, or modification of peptide bonds21 may be an approach to obtain more stable analogs.
The immune response to FVIII is polyclonal22 and differs among patients. Peptide 107 was recognized by IgG in the plasma of 2 of 12 inhibitor-positive patients (S.V., unpublished data, 2002), suggesting that FVIII inhibitors similar to Bo2C11 are produced by other patients with hemophilia A. Indeed, anti-C2 Abs from different patients are mostly encoded by the DP-5 V gene,23 which is the case for Bo2C11.5 Peptide 107 might thus be successfully used in several patients. Three major clusters of functional epitopes have been delineated that span the A2, A3, and C2 domains of FVIII.3 Monoclonal antibody 413, whose epitope is restricted to region 484-508, is representative of anti-A2 inhibitors.24 A similar study with mAb 413 should provide us with an additional neutralizing mimic that could be part of a cocktail of peptide mimics necessary to efficiently neutralize polyclonal FVIII inhibitors. Alternatively, more complete mixtures of mimics may be derived according to the method described by Folgori et al,25 in which libraries of random peptides displayed on phages are subjected to sequential positive and negative screenings using IgG from the plasma of patients and healthy donors, respectively.
We believe that our approach offers a promising solution toward the design of small synthetic molecules suitable for restoring FVIII function in inhibitor-positive patients with hemophilia A.
Prepublished online as Blood First Edition Paper, May 1, 2003; DOI 10.1182/blood-2002-06-1886.
Supported by a grant from Wyeth Genetics Institute. S.V. is a recipient of a fellowship from the Fondation pour la Recherche Médicale.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr S. L. Salhi for the editorial revision of the manuscript. We also thank Ms C. Artaud for help with the Bethesda experiments, Mr C. Nguyen for help with the peptide synthesis, and Dr Thierry Lambert of the Hôpital Bicêtre (France) and Dr Jacqueline Reynaud of the Département d'Hématologie des Hôpitaux de Saint-Etienne (France) for providing us with plasma samples.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal