Abstract
We synthesized BβArg14His fibrinogen with histidine substituted for arginine at the Bβ thrombin-cleavage site. This substitution led to a 300-fold decrease in the rate of thrombin-catalyzed fibrinopeptide B (FpB, Bβ 1-14) release, whereas the rate of FpA release was normal with either thrombin or the FpA-specific enzyme, batroxobin. Both thrombin- and batroxobincatalyzed polymerization of BβArg14His fibrinogen were significantly impaired, with a longer lag time, slower rate of lateral aggregation, and decreased final turbidity. Moreover, desA monomer polymerization was similarly impaired, demonstrating that the histidine substitution itself, and not the lack of FpB cleavage, caused the abnormal polymerization of BβArg14His fibrin. Scanning electron microscopy showed BβArg14His fibrin fibers were thinner than normal (BβArg14His, approximately 70 nm; normal, approximately 100 nm; P < .0001), as expected from the decreased final turbidity. We conclude that the N-terminus of the Bβ chain is involved in the lateral aggregation of normal desAprotofibrils and that the Arg→His substitution disrupts these interactions in BβArg14His fibrinogen. (Blood. 2003;102:2466-2471)
Introduction
Fibrinogen is a 340-kDa plasma protein consisting of 2 sets of 3 nonidentical polypeptide chains, Aα, Bβ, and γ, that combine to form a symmetrical, trinodular molecule. The central E nodule contains the N-termini of all 6 chains, whereas both distal D nodules contain independently folded βC and γC domains that consist of the Bβ and γ C-termini, respectively. These 3 nodules are connected by helical coiled-coil regions to form a symmetrical, linear D-E-D arrangement. The Aα chains extend from the E nodule through the distal D nodules and then fold back toward the center of the molecule, where their C-termini form compact globular structures termed αC domains, which interact with each other and with the E nodule.1 To convert soluble fibrinogen to insoluble fibrin, thrombin cleaves fibrinopeptide A (FpA, Aα1-16) to expose the “A” polymerization site. This “A” site noncovalently interacts with the “a” polymerization site in the γC domain of another fibrinogen molecule. This A:a interaction results in the spontaneous formation of half-staggered, double-stranded protofibrils.2 As these protofibrils grow in length, thrombin cleaves fibrinopeptide B (FpB, Bβ1-14), which exposes the “B” site3-5 and dissociates the αC domains from the central E nodule.1,6 The release of FpB results in an enhanced rate of lateral aggregation of protofibrils3,7 to form thick fibers. Lateral aggregation is supported by multiple interactions, including: a specific, noncovalent interaction between the “B” site and the “b” polymerization site in the βC domain of another molecule,8,9 intermolecular interactions between αC domains of different fibrin molecules,1,6 and interactions between 2 βC domains of different protofibrils, specifically residues β330 to β375.10 The end result of thrombin-catalyzed polymerization is the formation of a complex branching network of insoluble fibers.
Fibrin clot formation also follows the release of only FpA, as catalyzed by snake venom enzymes such as batroxobin (Batroxobin moojeni; CenterChem, Stamford, CT), reptilase, and ancrod. The resultant desA fibrin monomers are capable of lateral aggregation despite the lack of FpB release and B:b interactions, although desA fibers are generally thinner than desAB fibers.11,12 Although the mechanism of desA lateral aggregation is not fully understood, published data point to several reasonable participants. High-resolution structures of the DD fragment co-crystallized with the peptide GPRP, which mimics the A:a interaction, suggest that the regions γ350-360 and γ370-380 participate in γD lateral association contacts between protofibrils.10 Other studies suggest that the N-termini of the Bβ chains participate in desA lateral aggregation. Work with desBβ1-42 fibrinogen, which is missing the N-terminal 42 residues of the Bβ chain, shows impaired desA polymerization.13,14 Reptilase-catalyzed polymerization of several dysfibrinogens, including BβArg14Cys and BβGly15Cys,15-17 also shows impaired desA polymerization. Taken together, these results suggest that the N-termini of the Bβ chains13-17 and residues in the γC domains of growing protofibrils10 participate in desA polymerization.
To further examine the role of FpB release, we synthesized and characterized recombinant BβArg14His fibrinogen, with an Arg→His substitution at the thrombin cleavage site. This substitution results in drastically delayed FpB release. Polymerization studies with this variant suggest that the N-terminal part of the Bβ chain, and specifically fibrinopeptide B, contributes to the lateral association of desA protofibrils.
Materials and methods
Construction of plasmid
Plasmids containing the cDNAs of all 3 fibrinogen chains—pMLP-Aα, pMLP-Bβ, and pMLP-γ—have been previously described.18 A plasmid encoding the substitution of His for Arg at position 14 in the Bβ chain (called pMLP-BβArg14His) was constructed using the Transformer Site Directed Mutagenesis Kit (ClonTech, Palo Alto, CA) based on the method of Deng and Nickloff.19 Two sets of polymerase chain reaction (PCR) primers were prepared (University of North Carolina Oligonucleotide Facility): selection, 5′ TCTAGGGCCCAGGCTTGTTTCC; mutagenic, 5′ GGTTTCTTCAGTGC CCA*TGGTCATCGACCC. Primer 1 eliminated a HindIII restriction site and was used for primary selection; primer 2 altered the codon for residue 14 from Arg to His by introducing a point mutation, denoted by the asterisk, and incorporated a unique NcoI restriction site, as denoted by the underlined sequence. To construct the pMLP-BβArg14His plasmid, selection and mutagenic primers were annealed to denatured pMLP-Bβ and extended with T4 DNA polymerase. The ends were joined with T4 DNA ligase. Selective digestion of the mutant plasmid was achieved with HindIII, and MutS Escherichia coli were transformed. Plasmid DNA was isolated using the Qiagen MiniPrep Kit (Chatsworth, CA). Positive clones were detected by NcoI digestion, and the Bβ cDNA was sequenced using an automated sequencer (Applied Biosystems, Foster City, CA).
Recombinant fibrinogen expression and purification
For synthesis of BβArg14His fibrinogen, the new plasmid, pMLPBβArg14His, was transfected into Chinese hamster ovary (CHO) cells expressing the Aα and γ chains, as described.20 Clones were selected for neomycin and histidinol resistance and were screened for fibrinogen secretion using an enzyme-linked immunosorbent assay (ELISA). The clone secreting the highest fibrinogen concentration was grown in roller bottles in serum-free medium, and the media were harvested and stored at -70°C with protease inhibitors.18 The same procedure was used to express normal recombinant fibrinogen. These proteins were purified as described.21 Purified protein was dialyzed against 20 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid), pH 7.4, 150 mM NaCl, 1 mM CaCl2 for one change and then extensively against 20 mM HEPES, pH 7.4, 150 mM NaCl and was stored at -70°C. The fibrinogen concentration was determined by absorbance at 280 nm using the extinction coefficient of ϵ= 1.506 for a 1 mg/mL fibrinogen solution.22 Purity of the proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel according to the method of Laemmli.23
Thrombin-catalyzed fibrinopeptide release
Thrombin (human α-thrombin; Enzyme Research Labs, South Bend, IN) digestion of fibrinogen was performed as previously described.20 Normal and BβArg14His fibrinogens were diluted to 0.1 mg/mL (0.3 μM) fibrinogen concentration. Initial reactions were performed with 0.1 mg/mL fibrinogen and 0.005 U/mL thrombin, and no FpB release was detected from BβArg14His after 6 hours. At a thrombin concentration that was 50 times higher (0.25 U/mL), 20% of FpB was released after 2 hours. To monitor FpA and FpB release from both normal BβArg14His fibrinogens on the same time scale, thrombin was added to the fibrinogen solutions to give a final thrombin concentration of 0.1 U/mL, and the experiment was repeated in duplicate. Incubations were as described previously, in the microscale assay.24 All fibrinopeptide release assays were performed at ambient temperatures.
Batroxobin-catalyzed fibrinopeptide release
Normal and BβArg14His fibrinogens were diluted to 0.1 mg/mL (0.3 μM). To initiate the reaction, batroxobin was added to a final concentration of 0.005 U/mL; the solutions were mixed by inversion and divided into 4 aliquots of 440 μL and 5 aliquots of 260 μL. All manipulations after the addition of batroxobin were completed within 45 seconds. An infinity time point was prepared with the addition of 1 μL of 100 U/mL batroxobin to a 260 μL aliquot of 0.3 μM fibrinogen and incubation at ambient temperature for 2 hours. Aliquots were incubated and prepared for high-performance liquid chromatography (HPLC) as described for the thrombin-catalyzed reactions. Batroxobin activity was determined relative to thrombin activity by equalizing the amounts of fibrinopeptide A released after a 20-minute incubation.
Analysis of fibrinopeptide release
Fibrinopeptide release was monitored by reverse-phase HPLC as described.20 Briefly, the stored samples were thawed at 37°C and loaded onto an ISCO System HPLC (Lincoln, NE) with a C18 column (Vydac, Hesperia, CA) equilibrated with buffer A (25 mM NaH2PO4/Na2HPO4, pH 6.0). Fibrinopeptides were eluted with a linear gradient from 0% buffer B to 40% buffer B (25 mM NaH2PO4/Na2HPO4, pH 6.0, with 50% acetonitrile) and monitored by the absorbance at 206 nm. The differences in molar absorptivities for FpA and FpB were not part of our analysis.25 No shift in the retention time of FpB(Arg14His) was detected compared with normal FpB. Fibrinopeptide peak area was determined using ISCO software (version 2.4; ChemResearch). FpA data for both fibrinogens were fitted with a simple first-order reaction; FpB data were fitted to a standard equation describing 2 first-order processes, as described,25 and the curves plotted using DeltaGraph (DeltaPoint, Monterey, CA). Rate constants for the release of FpA (k1) and FpB (k2) and the specificity constant kcat/KM were determined as described.25
Thrombin- and batroxobin-catalyzed polymerization
Polymerization of normal recombinant and BβArg14His fibrinogen was monitored at 350 nm in a SpectraMax-340PC 96-well microtiter plate reader at ambient temperatures (Molecular Devices, Sunnyvale, CA). Two separate experiments were performed in triplicate for each polymerization condition, as previously described.24 Briefly, 90 μL fibrinogen in 20 mM HEPES, pH 7.4, 148 mM NaCl, and 1 mM CaCl2 was added to each well. To initiate polymerization, 10 μL enzyme (thrombin or batroxobin) was added, and turbidity was monitored every 10 seconds for 1 hour. Values were normalized to a 1-cm path length by the PathCheck sensor within the instrument. Final concentrations were 0.2 mg/mL fibrinogen and 0.1 U/mL thrombin or batroxobin.
Analysis of polymerization results
Lag time, rate of lateral aggregation (rateagg), and final turbidity were determined for each polymerization reaction. Lag time was the time when the slope of the steepest part of the polymerization curve (rateagg) crossed the x-axis,26 and final turbidity represented the absorbance at 350 nm after 150 minutes. Statistical values comparing normal recombinant and BβArg14His fibrinogen were determined by unpaired t tests using StatView (Berkeley, CA). A significant difference was evident when P values were less than .05.
DesA fibrin monomer preparation
Two separate preparations of fibrin monomers were prepared as described.21,27 In brief, fibrinogen was clotted by batroxobin. The clot was wrapped around a glass rod, washed in 0.15 M NaCl, and dissolved in 0.125% acetic acid on ice. The dissolved fibrin monomer was repolymerized by dilution (10-fold) in 40 mM HEPES, pH 7.4, 0.2 M NaCl buffer. The resultant clot was wrapped around the glass rod, washed in 0.15 M NaCl, and dissolved in 0.125% acetic acid on ice; repolymerization was repeated twice. The resultant fibrin monomer was kept in 0.125% acetic acid at 4°C and used within 1 month of preparation. SDS-PAGE was run to ensure that conversion to desA monomers was complete and that degradation had not occurred.
DesA fibrin monomer polymerization
DesA fibrin monomer polymerization was performed in a 50-μL microcuvette (StarnaCells, Atascadero, CA), and the increase in turbidity at 350 nm was monitored on a BioSpec-1601 UV-spectrophotometer (Shimadzu, Tokyo, Japan) for 30 minutes. Polymerization was initiated by adding 10 μL fibrin monomer solution to 90 μL buffer (40 mM HEPES, pH 7.4, 148 mM NaCl, 1 mM CaCl2) and mixing with the pipette tip. Final concentration of fibrin monomer was 0.08 mg/mL. Polymerizations of both fibrin monomer preparations were performed in triplicate.
Scanning electron microscopy
Clots were polymerized as described, with final concentrations of 0.4 mg/mL fibrinogen and 0.4 U/mL thrombin. For each condition, scanning electron microscopy (SEM) was performed on 2 clots each with at least 2 separate microscopy preparations. Samples were prepared as originally described28 with the previously defined modifications.24 Briefly, the fibrinogens were allowed to polymerize at ambient temperature for 4 hours in a moist environment. The clots were then rinsed 3 times with cacodylate buffer, fixed in 2% glutaraldehyde overnight, rinsed 3 times with cacodylate buffer, and then stained with 2% osmium tetroxide for 30 minutes. The clots were rinsed with distilled water, dehydrated with a series of ethanol dehydrations up to 100% and then critical point-dried for 1 hour. Samples were then sputter-coated with approximately 20 nm gold-palladium and viewed on a Cambridge StereoScan S200 (LEO Electron Microscopy, Thornwood, NY). All images were taken at ×16 200 with a 17-mm working distance and 20-kV accelerating voltage. Fiber diameters were measured for at least 20 fields in each clot using ScionImage (Scion, Frederick, MD). Statistical analysis comparing fiber diameters was determined by unpaired t test using StatView (Berkeley, CA). A difference was considered significant at P values less than .05.
Results
Protein synthesis and characterization
We synthesized BβArg14His fibrinogen with histidine substituted for arginine at position 14 in the Bβ chain, where thrombin cleaves fibrinogen to release fibrinopeptide B. Nonreducing SDS-PAGE showed that normal and BβArg14His fibrinogen contained the expected bands representing high-molecular-weight (HMW) and low-molecular-weight (LMW) fibrinogen (data not shown). SDS-PAGE run under reducing conditions showed that both fibrinogens contained the expected bands representing the Aα, Bβ, and γ chains and that the Arg→His mutation did not affect the migration of the Bβ chain in BβArg14His fibrinogen relative to normal (Figure 1B, inset). No extra bands were detectable by SDS-PAGE. As shown previously, normal recombinant fibrinogen functions similarly to purified plasma fibrinogen,21 such that our results can be extrapolated to include plasma conditions.
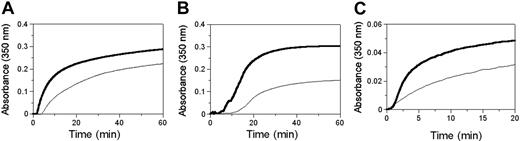
SDS-PAGE and fibrinopeptide release curves of normal and BβArg14His fibrinogens. (A) Thrombin-catalyzed release of FpA (squares) and FpB (circles) from normal [▪, •] and BβArg14His (□, •) fibrinogens. (B) Batroxobin-catalyzed release of FpA from normal (▪) and BβArg14His (•) fibrinogens. Conditions: 0.1 mg/mL fibrinogen, 0.1 U/mL thrombin or 0.005 U/mL Batroxobin in 20 mM HEPES, pH 7.4, 150 mM NaCl. (inset) 10% reducing SDS-PAGE gel of normal (lane 1) and BβArg14His (lane 2) fibrinogens, with molecular weight markers indicated on left.
SDS-PAGE and fibrinopeptide release curves of normal and BβArg14His fibrinogens. (A) Thrombin-catalyzed release of FpA (squares) and FpB (circles) from normal [▪, •] and BβArg14His (□, •) fibrinogens. (B) Batroxobin-catalyzed release of FpA from normal (▪) and BβArg14His (•) fibrinogens. Conditions: 0.1 mg/mL fibrinogen, 0.1 U/mL thrombin or 0.005 U/mL Batroxobin in 20 mM HEPES, pH 7.4, 150 mM NaCl. (inset) 10% reducing SDS-PAGE gel of normal (lane 1) and BβArg14His (lane 2) fibrinogens, with molecular weight markers indicated on left.
Thrombin-catalyzed fibrinopeptide release
We examined the rate of fibrinopeptide release from normal and BβArg14His fibrinogens by measuring the peak areas of FpA and FpB and plotting the data as percentage fibrinopeptide release. The substitution of histidine for arginine did not alter the retention time of FpB(Arg14His). All FpA release data were fittoa first-order rate equation, whereas FpB release data were fit to a standard equation describing 2 consecutive first-order processes.25 To gain information on the release of FpA and FpB from normal and BβArg14His fibrinogen in the same experiment, we performed duplicate fibrinopeptide release assays at an intermediate thrombin concentration of 0.1 U/mL (0.09 nM). Under these conditions, FpA was released from both fibrinogens at a similar rate (Figure 1A), and the average specificity constants for the release of FpA from normal and BβArg14His were not significantly different (P = .92). Average specificity constants revealed that FpB(Arg14His) was released from BβArg14His fibrinogen at a rate that was 300 times slower than from normal fibrinogen (P = .008).
Batroxobin-catalyzed fibrinopeptide release
We monitored the batroxobin-catalyzed release of FpA from normal and BβArg14His fibrinogens by measuring the peak areas of FpA on the chromatogram and plotting the data as percentage fibrinopeptide release. Standard first-order equations were used to fit the data. As shown in Figure 1B, the release of FpA from normal fibrinogen was similar to that from BβArg14His fibrinogen, with similar specificity constants (Tables 1 and 2; P = .57). No FpB release was detected for either fibrinogen over the 120-minute period. Of note, the data for FpA release with batroxobin were qualitatively different from FpA released with thrombin (compare Figure 1B with Figure 1A). Visual inspection suggests that the batroxobin data fit a biphasic curve rather than a standard first-order equation.
First-order rate constants for normal and BβArg14His fibrinogen with thrombin and batroxobin
. | Thrombin* . | . | Batroxobin† . | |
|---|---|---|---|---|
| Fibrinogen . | k1 ± SD . | k2 ± SD . | k1 ± SD . | |
| Normal | 0.85 ± 0.67 | 0.19 ± 0.072 | 0.071 ± 0.017 | |
| BβArg14His | 0.83 ± 0.51 | 0.00053 ± 0.000081 | 0.092 ± 0.041 | |
. | Thrombin* . | . | Batroxobin† . | |
|---|---|---|---|---|
| Fibrinogen . | k1 ± SD . | k2 ± SD . | k1 ± SD . | |
| Normal | 0.85 ± 0.67 | 0.19 ± 0.072 | 0.071 ± 0.017 | |
| BβArg14His | 0.83 ± 0.51 | 0.00053 ± 0.000081 | 0.092 ± 0.041 | |
Values are the averages from 2 experiments at 0.1 mg/mL fibrinogen and 0.1 U/mL thrombin and are expressed as min−1.
Values are the averages from 2 experiments with 0.005 U/mL Batroxobin and are expressed as min−1.
Specificity constants for normal and BβArg14His fibrinogen with thrombin and batroxobin
. | Thrombin* . | . | Batroxobin† . | |
|---|---|---|---|---|
. | kcat/Km, FpA . | kcat/Km, FpB . | kcat/Km, FpA . | |
| Normal | 14.3 ± 9.2 | 3.1 ± 1.1 | 1.12 ± 0.26 | |
| BβArg14His | 13.5 ± 7.5 | 0.011 ± 0.002 | 1.45 ± 0.64 | |
. | Thrombin* . | . | Batroxobin† . | |
|---|---|---|---|---|
. | kcat/Km, FpA . | kcat/Km, FpB . | kcat/Km, FpA . | |
| Normal | 14.3 ± 9.2 | 3.1 ± 1.1 | 1.12 ± 0.26 | |
| BβArg14His | 13.5 ± 7.5 | 0.011 ± 0.002 | 1.45 ± 0.64 | |
Values are the averages from 4 experiments-all with 0.1 mg/mL fibrinogen and 1 with 0.005 U/mL thrombin, 1 with 0.25 U/mL thrombin, and 2 with 0.1 U/mL thrombin-and are expressed as 106 M−1 sec−1.
Values are the averages from 2 experiments with 0.005 U/mL batroxobin and are expressed as 106 M−1 sec−1.
Enzyme-catalyzed fibrin polymerization
We monitored the enzyme-catalyzed polymerization of normal and BβArg14His fibrinogens to determine the effect of FpB (Arg14His) on thrombin and batroxobin-catalyzed polymerizations. Representative curves of thrombin-catalyzed polymerization (Figure 2A) show that polymerization of BβArg14His fibrinogen is impaired compared with normal fibrinogen, with a longer lag time, lower rateagg, and lower final turbidity (Table 3). Representative curves of batroxobin-catalyzed polymerization (Figure 2B) show that BβArg14His polymerization was again impaired compared with normal, with a significantly increased lag time, lower rateagg, and noticeable decrease in final turbidity compared with normal fibrinogen polymerized with either thrombin or batroxobin.
Polymerization. Polymerization of normal (thick lines) and BβArg14His (thin lines) fibrinogens by (A) thrombin, (B) batroxobin, and (C) desA fibrin monomers. Fibrinogen (0.2 mg/mL) was polymerized with thrombin or batroxobin (0.1 U/mL) in 20 mM HEPES, pH 7.4, 148 mM NaCl, 1 mM CaCl2. DesA fibrin monomers in 0.125% acetic acid were diluted 1:10 to a final concentration of 0.076 mg/mL in 100 μL of 40 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM CaCl2.
Polymerization. Polymerization of normal (thick lines) and BβArg14His (thin lines) fibrinogens by (A) thrombin, (B) batroxobin, and (C) desA fibrin monomers. Fibrinogen (0.2 mg/mL) was polymerized with thrombin or batroxobin (0.1 U/mL) in 20 mM HEPES, pH 7.4, 148 mM NaCl, 1 mM CaCl2. DesA fibrin monomers in 0.125% acetic acid were diluted 1:10 to a final concentration of 0.076 mg/mL in 100 μL of 40 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM CaCl2.
Parameters of thrombin- and batroxobin-catalyzed polymerization of control and BβArg14His fibrinogens
. | Control . | BβArg14His . | P . |
|---|---|---|---|
| Thrombin | |||
| Lag time, sec | 90 ± 23 | 230 ± 28 | < .0001 |
| Rateagg, × 10−4 U/sec | 7.0 ± 0.2 | 2.3 ± 0.2 | .0004 |
| Final OD | 0.36 ± 0.08 | 0.28 ± 0.05 | .07 |
| Batroxobin | |||
| Lag time, sec | 320 ± 46 | 710 ± 88 | < .0001 |
| Rateagg, × 10−4U/sec | 2.8 ± 0.2 | 1.5 ± 0.1 | < .0001 |
| Final OD | 0.30 ± 0.03 | 0.16 ± 0.01 | < .0001 |
| desA fibrin monomers | |||
| Lag time, sec | 45 ± 20 | None observed | NA |
| Rateagg, × 10−4U/sec | 154 ± 44 | 39 ± 11 | .002 |
| Final OD | 0.051 ± 0.01 | 0.041 ± 0.01 | .22 |
. | Control . | BβArg14His . | P . |
|---|---|---|---|
| Thrombin | |||
| Lag time, sec | 90 ± 23 | 230 ± 28 | < .0001 |
| Rateagg, × 10−4 U/sec | 7.0 ± 0.2 | 2.3 ± 0.2 | .0004 |
| Final OD | 0.36 ± 0.08 | 0.28 ± 0.05 | .07 |
| Batroxobin | |||
| Lag time, sec | 320 ± 46 | 710 ± 88 | < .0001 |
| Rateagg, × 10−4U/sec | 2.8 ± 0.2 | 1.5 ± 0.1 | < .0001 |
| Final OD | 0.30 ± 0.03 | 0.16 ± 0.01 | < .0001 |
| desA fibrin monomers | |||
| Lag time, sec | 45 ± 20 | None observed | NA |
| Rateagg, × 10−4U/sec | 154 ± 44 | 39 ± 11 | .002 |
| Final OD | 0.051 ± 0.01 | 0.041 ± 0.01 | .22 |
OD indicates optical density at 350 nm; NA, not applicable.
desA fibrin monomer polymerization
To eliminate the effect of the enzymatic catalysis step on polymerization and to determine whether the substitution in FpB alone altered polymerization, we prepared desA monomers from both fibrinogens. As described in “Materials and methods,” these monomers were completely desA and undegraded. Fibrin monomer polymerization curves showed a slow, steady increase in turbidity with BβArg14His desA fibrin (Figure 2C), whereas normal desA fibrin monomer polymerization proceeded with a lag time followed by an increase in turbidity. Overall, polymerization of BβArg14His desA fibrin monomers proceeded without a noticeable lag time and with a lower rateagg and lower final turbidity than normal (Table 3).
Scanning electron microscopy
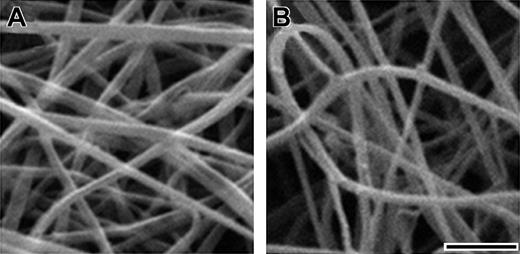
To confirm that the lower final turbidity was indicative of thinner fibers and to quantitate the differences in fiber thickness, we examined thrombin-catalyzed fibrin clots using scanning electron microscopy. As shown in Figure 3, fibers in BβArg14His clots were thinner than fibers in normal clots. We examined 2 clots of each fibrinogen and measured fiber diameters as described in “Materials and methods.” We found the average fiber diameter for BβArg14His fibrinogen was 70 ± 8 nm, significantly (P < .0001) thinner than the average fiber diameter for normal fibrinogen (102 ± 9 nm). Based on the previous data that show final turbidity is directly proportional to fiber thickness,29 we infer from the data in Figure 2B-C that desA BβArg14His fibrin fibers are thinner than normal desA fibers.
SEM of fibrinogens. Scanning electron micrographs of normal (A) and BβArg14His (B) fibrinogens polymerized with thrombin. Clots used for scanning electron microscopy were formed with 0.4 mg/mL fibrinogen and 0.4 U/mL thrombin in 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM CaCl2. SEM was performed as detailed in “Materials and methods.” Scale bar equals 1 μm.
SEM of fibrinogens. Scanning electron micrographs of normal (A) and BβArg14His (B) fibrinogens polymerized with thrombin. Clots used for scanning electron microscopy were formed with 0.4 mg/mL fibrinogen and 0.4 U/mL thrombin in 20 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM CaCl2. SEM was performed as detailed in “Materials and methods.” Scale bar equals 1 μm.
Discussion
Previous work had shown that the substitution of histidine for arginine at the P1 residue of the FpA cleavage site in fibrinogen Petoskey, AαR16H fibrinogen, reduced thrombin specificity by 160-fold.30 Here we report that the analogous substitution at the P1 residue of the FpB cleavage site reduced thrombin specificity by 300-fold. Thus, thrombin is able to cleave a His/Gly bond on either the N-terminus of the Aα chain or the Bβ chain, though at a dramatically reduced rate compared with the natural substrate, Arg/Gly. We had expected this result and synthesized BβArg14His fibrinogen to characterize the impact of delayed FpB release on thrombin-catalyzed polymerization. With this variant we were able to examine thrombin-catalyzed polymerization with essentially no FpB released. We found that polymerization of BβArg14His fibrinogen was impaired relative to normal fibrinogen.
Based on the prevailing idea that FpB release initiates lateral aggregation through the B:b interaction3,9 and an accompanying conformational change in the βC domain,10,31 we expected that the loss of the “B” site would result in a clot with thinner fibers. Seemingly, our assumption was correct. We also expected that batroxobin-catalyzed polymerization of normal and BβArg14His fibrinogens would be similar. Surprisingly, batroxobin-catalyzed polymerization of BβArg14His fibrin was even more impaired compared with normal desA fibrin than thrombin-catalyzed polymerization of BβArg14His fibrin was compared with normal desAB fibrin. This result suggested that the Arg→His substitution within FpB, and not the loss of the B:b interaction, directly impaired polymerization of BβArg14His fibrinogen. This conclusion was further supported by the impaired polymerization of desA BβArg14His fibrin.
To determine which specific step in polymerization was altered by the BβArg14His substitution, we compared our results with the kinetic model of polymerization set forth by Weisel.32 The model considered a 3-step mechanism—fibrinopeptide A release, protofi-bril formation, and lateral aggregation—and showed that 5 kinetic constants would generate curves comparable to those found under a variety of experimental conditions. These constants described the rates of (1) FpA cleavage, (2) the association of fibrin monomers initiating protofibril formation, (3) the addition of monomers to protofibrils where protofibrils lengthen, (4) the aggregation of 2 protofibrils to initiate fiber formation, and (5) the addition of protofibrils to a growing fiber. Our results demonstrated that FpA release was normal, eliminating the need to consider this step. The best fit of this model to our data—longer lag time, lower Vmax, and lower final turbidity—was observed when the rate of protofibril addition to a growing fiber was decreased. Thus, within the context of this model, we conclude that the BβArg14His substitution decreased the rate of fiber growth, or lateral aggregation, for both thrombin-catalyzed and batroxobin-catalyzed polymerization. This conclusion is consistent with our electron microscope images of clots, which are similar to those previously observed for batroxobincatalyzed polymerization.33
There are 2 reasonable explanations for our finding that a substitution in fibrinopeptide B impaired the lateral aggregation of desA protofibrils. First, it is possible that the N-terminus of the Bβ chain, specifically FpB, is normally involved in the desA polymerization mechanism and contributes to the mechanism by which desA monomers laterally aggregate. This possibility suggests that the N-terminus of the Bβ chain participates in lateral aggregation, irrespective of whether fibrinopeptide B is present or absent, and that by incorporating our mutation, we have perturbed the normal role of FpB in this process. Second, it is possible that the N-terminus of the Bβ chain does not normally play an active role in polymerization; rather, by imposing a mutation, we have impaired the proper interactions needed for desA lateral aggregation.
We favor the direct interpretation that fibrinopeptide B contributes to desA lateral aggregation, based on our findings and those of others that have previously reported on the participation of the N-terminus of the Bβ chain in desA polymerization.13,14,34 Studies with desBβ1-42 fibrinogen, formed by the specific cleavage of fibrinogen by protease III from Crotalus atrox venom, showed that the desA polymerization of this variant was 180-fold slower than normal desAB fibrin and that this impairment was not caused by a gross conformational change in the molecule imposed by the deletion.13 Further studies on the reptilase-catalyzed polymerization of this desBβ1-42 fibrin have shown that both protofibril formation and lateral aggregation of this fibrinogen were impaired compared with normal desA polymerization.14 In a different study, photo-oxidation of Bβ16His was reported to impair batroxobincatalyzed polymerization.34 Additionally, dysfibrinogens with BβArg14Cys mutations have shown delayed reptilase times,15,16 though this delay may be attributed to albumin binding to the free sulfhydryl group available in these variants.15 To further complicate matters, dysfibrinogens with a BβGly15Cys mutation have been shown to have both normal reptilase times, in the case of fibrinogen Ise,35 and prolonged reptilase times, as shown for fibrinogen Fukuoka II.17 The genetic heterozygous nature of these variants may account for the different results seen in different plasma milieus. Our recombinant BβArg14His fibrinogen is fundamentally different from these natural variants, based on its homogeneity, and provides a clearer picture of the participation of the N-terminus of the Bβ chain in desA polymerization. Combining our data with previously reported results, we suggest that the N-terminus of the Bβ chain is involved in desA polymerization and contributes to the lateral aggregation of desA protofibrils. Specifically, our data support a role for fibrinopeptide B in the lateral aggregation of desA monomers.
In contrast, previous data do not support the alternative explanation that the N-terminus of the Bβ chain indirectly impairs lateral aggregation by blocking desA lateral aggregation sites or by hindering the normal exposure of such sites. Based on crystal structure data,10 it has been proposed that the contacts between γC domains contribute to lateral associations upon A:a interactions. These contacts are localized to the region γ340-380, which is distant from the E nodule. Thus, the BβArg14His substitution in the E nodule is unlikely to alter the proposed contacts between γC domains other than by impairing A:a interactions. It has been proposed that intermolecular interactions between αC domains contribute to lateral aggregation.6 These studies showed, however, that the αC domains likely do not contribute to batroxobincatalyzed polymerization, because dissociation of these domains from the E nodule occurs after FpB release.6 Thus, we favor the conclusion that desA lateral aggregation was decreased because the BβArg14His substitution directly disrupts the normal participation of fibrinopeptide B in desA lateral aggregation.
In conclusion, we report the synthesis of recombinant BβArg14His fibrinogen, which has a drastically reduced rate of thrombin-catalyzed FpB release and impaired thrombin- and batroxobin-catalyzed polymerization. We demonstrated that impaired polymerization was attributed solely to the Arg→His substitution in fibrinopeptide B, as shown by desA monomer polymerization, and not simply to the delay in FpB release. We conclude that this implies a role for the N-termini of the Bβ chains, specifically fibrinopeptide B, in desA polymerization. Additionally, we present the idea that this variant fibrinogen, which has virtually no FpB release in the presence of thrombin, could serve as a model fibrinogen in cellular or molecular studies to examine the role of FpB in coagulation.
Prepublished online as Blood First Edition Paper, June 12, 2003; DOI 10.1182/blood-2003-01-0204.
Supported by National Institutes of Health grants R01 HL 31048 (S.T.L.) and HL30954 (J.W.W).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Li Fang Ping and Kasim McLain for their excellent technical assistance in protein purification and production.
Jennifer L. Moen previously published as Jennifer L. Mullin.

![Figure 1. SDS-PAGE and fibrinopeptide release curves of normal and BβArg14His fibrinogens. (A) Thrombin-catalyzed release of FpA (squares) and FpB (circles) from normal [▪, •] and BβArg14His (□, •) fibrinogens. (B) Batroxobin-catalyzed release of FpA from normal (▪) and BβArg14His (•) fibrinogens. Conditions: 0.1 mg/mL fibrinogen, 0.1 U/mL thrombin or 0.005 U/mL Batroxobin in 20 mM HEPES, pH 7.4, 150 mM NaCl. (inset) 10% reducing SDS-PAGE gel of normal (lane 1) and BβArg14His (lane 2) fibrinogens, with molecular weight markers indicated on left.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/7/10.1182_blood-2003-01-0204/6/m_h81935034001.jpeg?Expires=1766062428&Signature=Shic-epUZhgt3j6m3C1JxhW7p1cJxaYVMw1353XuuExLwR-JqNPBQxS5wgkrNU6HuRozuKFvPD5TXOq9dUs8KtPbPIWU6SaNEPHKE0C-3GO-tnRy1IjreLKWDMITmvlGmHcGsksGH7ZMjax8ByLnbxXU-hWSeP0ounzOHBbUzrtDFULEIqeThevumBpHa2MEPNqrRvQRUQ5CAZr2-C123C4r2p67zoLe8ipveMurMEpD46nY55iJPh6JVHpBxEVBXDTQuYezSq-YrRXk0z13U2tmeTCDFMVYfHw0paq3~F2u2-fhoR6xpH1CSTGk6q4gdZ~Q6xypct01KPgIylr1eQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal