Abstract
Rodent mast cells (MCs) are reported to play a pivotal role in both innate and adaptive immunity. However, there is so far no evidence that human MCs are involved in innate immunity. We found that a functional Toll-like receptor 4 (TLR4) was expressed on human MCs when it was up-regulated by interferon γ (IFN-γ). To systematically explore how human MCs modulate the immune system following TLR4-mediated activation and FcϵRI aggregation, we used high-density oligonucleotide probe arrays (GeneChip) to compare the lipopolysaccharide (LPS)-induced gene expression profile with the IgE/anti-IgE-mediated profile in MCs. Both a shared core response, and LPS- or anti-IgE-specific programs of gene expression were observed in MCs. Furthermore, MCs exhibited an antiviral response gene program in response to IFN-γ, and LPS sustained that expression. Compared with the LPS-stimulated gene expression profile of human peripheral blood mononuclear cells, LPS-stimulated MCs specifically induced a subset of genes that included a Th2 cytokine and chemokines that recruit Th2 cells and eosinophils. These results reveal that human MCs express tailored pathogen- and antigen-specific immune responses and that human MCs may play important roles in innate and adaptive immunity.(Blood. 2003;102:2547-2554)
Introduction
Human mast cells (MCs) express high-affinity IgE receptors (FcϵRI) on their surface, and they can be activated to secrete a variety of biologically active mediators by cross-linking of receptor-bound IgE.1 Among mediators, the production of Th2-type cytokines such as interleukin 5 (IL-5) and CC chemokines such as I-309 by human MCs is believed to be responsible for the initiation and maintenance of IgE-dependent allergic reactions.2,3
Recent studies in animal models indicate that mouse MCs may play a protective role in host defense against bacteria through production of tumor necrosis factor α (TNF-α) as a result of mainly Toll-like receptor 4 (TLR4)- or CD48-mediated activation.4-9 However, it has become clear that rodent MCs are somewhat different from their human counterparts in terms of their sensitivity to some cytokines and drugs and their receptor expression profiles such as for FcγR.3,10-13 It is also observed that the level of IgE-dependent TNF-α production by human MCs is quite low compared to that by rodent MCs.3,4,14 Therefore, it is now necessary to determine if human MCs express TLR4. If TLR4 is expressed on MCs, our question is how human MCs respond to pathogens and antigens they encounter via TLR4 and FcϵRI. We compared the expression of downstream target genes in human MCs induced via TLR4 and FcϵRI.
Human TLR4 was the first characterized mammalian Toll.15-17 It is expressed in a variety of cell types, most predominantly in the cells of the immune system, including macrophages and dendritic cells.18 TLR4 functions as the signal-transducing receptor for lipopolysaccharide (LPS).19 The TLR and IL-1R family members share several signaling components, including the adaptor MyD88, Toll-interacting protein, protein kinase IL-1R-associated kinase (IRAK), and TNF receptor-associated factor 6 (TRAF6).20-22
TRAF6 can activate nuclear factor κB (NF-κB), tumor growth factor (TGF)-β-kinase 6, c-Jun N-terminal kinase, and p38 mitogen-activated protein kinase kinase through mitogen-activated protein kinase 6.20 TLR4 signals via another adaptor in addition to MyD88-Toll/IL-1R domain-containing adaptor protein (TIRAP), which activates MyD88-independent signaling downstream of TLR4.23 The protein kinase PKR function is downstream of TIRAP. Importantly, recent reports described that TLR4-mediated nuclear translocation of interferon regulatory factor 3 (IRF3) occurs in MyD88-independent fashion and that it induces activation of a complex positive-feedback loop between type I IFNs and IRF family members, leading to induction of an antigrowth, antiviral response.22,24 IRF3 has been shown to mediate a TLR3/TLR4-specific antiviral gene program.24 Using DNA microarrays and serial analysis of gene expression, it has been reported that human macrophages and dendritic cells up-regulate many of the NF-κB pathway members as well as IFN-β-induced signal transducers and activators of transcription (STAT) 1α/β-dependent gene expression.25-27 However, the functional role of TLR4 on MCs is not known. We hypothesized that human MCs might play a different role in innate immunity from human peripheral blood mononuclear cells (PBMNCs) in response to LPS. To explore this hypothesis, we also compared the gene expression profile of LPS-stimulated human MCs with that of stimulated PBMNCs by using high-density oligonucleotide probe arrays.
Materials and methods
Cytokines and antibodies
Recombinant human (rh) IL-3, rhIL-6, and rh stem cell factor (SCF) were purchased from Intergen (Purchase, NY). The following monoclonal antibodies (mAbs) were purchased: mouse antihuman tryptase (clone G3, subclass IgG1; Chemicon, Temecula, CA; and clone AA1, subclass IgG1, Dako, Carpinteria, CA); mouse antihuman TLR4 (clone HTA125, subclass IgG2a; eBioscience, San Diego, CA); mouse antihuman kit (clone YB5, B8 subclass IgG1; BD PharMingen, San Diego, CA); mouse antihuman FcϵRI (clone CRA-1, subclass IgG2b; Kyokuto, Tokyo, Japan); and mouse antihuman TNF-α (clone MAb11, subclass IgG1; BD PharMingen).
Generation of adult peripheral blood-derived MCs
All human subjects in this study provided written informed consent; the study was approved by the ethical review boards of all hospitals. Lineage-negative mononuclear cells (MNCs) were selected from the PBMNCs and cultured in serum-free Iscove methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada) and Iscove modified Dulbecco medium (IMDM) containing SCF at 200 ng/mL, IL-6 at 50 ng/mL, and IL-3 at 1 ng/mL as previously described.28 On day 42 of culture, methylcellulose was dissolved in phosphate-buffered saline (PBS) and the cells were resuspended and cultured in IMDM containing SCF at 100 ng/mL and IL-6 at 50 ng/mL with 2% fetal calf serum (FCS).
Purification of human lung MCs
Macroscopically normal human lung resected during surgery was obtained and processed, after obtaining informed consent. Lung MCs were dispersed from chopped lung specimens by an enzymatic procedure and purified by magnetic bead affinity selection using the anti-kit mAb (clone YB5.B8) as described previously.3 Final purity of MCs was more than 99%.
Isolation of human PBMNCs and monocytes
Human PBMNCs were isolated by centrifugation on a Ficoll-Isopaque density gradient (Nycomed, Oslo, Norway). Human monocytes were selected from the PBMNCs using a magnetic separation column (Miltenyi Biotec, Auburn, CA) and a magnetic microbead-conjugated antibody against CD14 (Miltenyi Biotec) according to the manufacturer's instructions. The final purity of monocytes was more than 99%.
Activation of human MCs
Human MCs were precultured with or without 30 ng/mL IFN-γ (PeproTech EC, London, United Kingdom) for 24 hours, washed, and then challenged by incubation with 100 ng/mL LPS (derived from Salmonella typhimurium, Sigma-Aldrich, St Louis, MO) for the indicated time period. For aggregation of FcϵRI, MCs were sensitized with 1 μg/mL human myeloma IgE (CosmoBio, Tokyo, Japan) at 37°C for 24 hours. After washing, the cells were then challenged with either 1.5 μg/mL rabbit antihuman IgE antibody (Dako) or the culture medium alone at 37°C for the indicated time period. Under all conditions, the cells were suspended in complete IMDM containing SCF and IL-6.
Isolation of RNA and performance of RT-PCR
Total cellular RNA was isolated from MCs with RNeasy mini kits (Qiagen, Valencia, CA), according to the manufacturer's specifications. The yield of RNA per 106 MCs was 2.2 μg (median, with range, 0.6-3.2 μg; n = 6). An equal amount of RNA (100 ng) was used for reverse transcription-polymerase chain reaction (RT-PCR) analysis. PCR was performed in a thermocycler as follows: 94°C for 5 minutes, followed by 30 amplification cycles (94°C, 1 minute; 56°C, 2 minutes; 72°C, 3 minutes). The sequences of the primers for TLR4 were 5′ primer, 5′-TGGATACGTTTCCTTATAAG-3′ and 3′ primer, 5′-GAAATGGAGGCACCCCTTC-3′. Final extension was performed at 72°C for 10 minutes. Equal amounts of PCR-amplified products were visualized by ethidium bromide. In addition to TLR4-specific primers, mRNA for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected as a positive control. PCR without cDNA was performed to exclude contamination.
Real-time quantitative RT-PCR
Real-time quantitative RT-PCR was performed according to the manufacturer's instructions (Applied Biosystems, Weiterstadt, Germany). cDNA (10 ng) was amplified in 25 μL final volume, in the presence of 1.25 μL of the following “Assays-on-Demand” oligonucleotides (TLR4 and GAPDH; Applied Biosystems). Test gene primer and FAM-labeled probe sets were optimized for concentration, amplification efficiency, and faithful coamplification with one or more housekeeper gene primer and VIC-labeled probe sets—the latter including GAPDH. Real-time quantitative PCR was set up in 96-well plates using the above reagents and TaqMan master mix and as indicated by the optimization data, and it was run on 7700 ABI thermal cyclers (Applied Biosystems). Real-time data were acquired and analyzed using Sequence Detection System software (Applied Biosystems) with manual adjustment of the baseline and threshold parameters. Relative expression levels were determined using cycle threshold values and the Compared Ct method to adjust for coamplified housekeeper gene levels, 2-fold amplification/cycle rates, and the reference expression level of control samples.29
Flow cytometric analyses and confocal laser scanning microscopy
MCs were suspended in blocking medium consisting of triethanolamine-buffered saline (TBS), 0.1% bovine serum albumin (BSA), 1% nonfat milk, and 10 mg/mL human IgG (ICN Biomedicals, Aurora, OH). The cells were then incubated with fluorescein isothiocyanate (FITC)-conjugated anti-FcϵRI, biotin-conjugated anti-TLR4, Cy-Chrome-conjugated anti-kit, or the isotype control immunoglobulin in the blocking medium for 30 minutes at 4°C in the dark. When biotin-conjugated anti-TLR4 was used, cells were then incubated with streptavidin-allophycocyanin (APC; Becton Dickinson, San Jose, CA). When the cells were stained with antihuman tryptase, cells were then fixed and permeabilized with Fix and Perm cell permeabilization kits (Caltag Laboratories, Burlingame, CA). For intracellular TNF-α staining, cells were incubated with 1 μM monensin (Sigma-Aldrich) when LPS or anti-IgE was added. Cells were then fixed, permeabilized, and stained with APC-conjugated anti-TNF-α mAb. After washing, the cells were analyzed using FACScalibur (Becton Dickinson) and Cell Quest software (Becton Dickinson). The mean fluorescence intensities (MFIs) of MCs stained with specific antibody and those stained with control antibody were obtained. For confocal microscopic analysis, the stained cells were fixed with 4% paraformaldehyde, washed, and treated with SlowFade antifade kit with DAPI (4,6 diamidino-2-phenylindole; Molecular Probes, Eugene, OR). Confocal microscopy was done on an FV500 model confocal laser scanning microscope (Olympus, Tokyo, Japan).
GeneChip expression analysis
Human genome-wide gene expression was examined by using the Human Genome U133A probe array (GeneChip, Affymetrix, Santa Clara, CA), which contains the oligonucleotide probe set for approximately 23 000 full-length genes and expressed sequence tags (ESTs), according to the manufacturer's protocol (Affymetrix), and previous reports.28,30,31 Total RNA (0.1-10 μg) was extracted from approximately 105 to 107 cells. Double-stranded cDNA was synthesized, and the cDNA was subjected to in vitro transcription in the presence of biotinylated nucleoside triphosphates. The biotinylated cRNA was hybridized with a probe array for 16 hours at 45°C, and the hybridized biotinylated cRNA was stained with streptavidinphycoerythrin (PE) and then scanned with a Hewlett-Packard Gene Array Scanner (Palo Alto, CA). The fluorescence intensity of each probe was quantified using a computer program, GeneChip Analysis Suite 5.0 (Affymetrix). The expression level of a single mRNA was determined as the average fluorescence intensity among the intensities obtained by 11 paired (perfect-matched and single nucleotide-mismatched) probes consisting of 25-mer oligonucleotides. If the intensities of mismatched probes were very high, gene expression was judged to be absent even if a high average fluorescence was obtained with the GeneChip Analysis Suite 5.0 program. The level of gene expression was determined as the average difference (AD) using GeneChip software. The percentages of the specific AD level versus the mean AD level of 6 probe sets for housekeeping genes (β-actin and GAPDH) were then calculated.
Data analysis was further performed with Genespring software version 5 (Silicon Genetics, San Carlos, CA). To normalize the staining intensity variations among chips, the AD values for all genes on a given chip were divided by the median of all measurements on that chip. To eliminate changes within the range of background noise and to select the most differentially expressed genes, data were used only if raw data values were less than 100 AD and the gene expression was judged to be present by Affymetrix data analysis. Hierarchical clustering analysis with standard correlation was used to identify gene clusters. The separation ratio was set at 0.5. Normalization values below 0 were set to 0. Data were considered significant when (1) expression changed by at least 2-fold (activation program) and (2) increased gene expression included at least one “present absolute call” (Affymetrix algorithm). Normalized values were averaged for 2 donors and used for the data analysis. Based on those normalized values, the genes were classified as up-regulated or down-regulated. The expression levels of genes of the same cells analyzed twice showed a statistically significant correlation (r = 0.997). Under these criteria, the reproducibility of the differences that were seen between different cells under different conditions was confirmed.
ELISA for TNF-α, IL-5, CCL1, and CCL5
Human TNF-α, IL-5, CCL1 (also known as I-309), and CCL5 (RANTES) were measured with enzyme-linked immunosorbent assay (ELISA) kits purchased from R&D Systems (Minneapolis, MN). The sensitivities of the assays of human TNF-α, IL-5, CCL1, and CCL5 were 4.71, 1.0, 2.37, and 8 pg/mL, respectively. MCs (2 × 104) were used for each assay.
Statistical analysis
Differences between 2 paired groups were analyzed by the paired Student t test and considered significant at P < .05. Values are expressed as the mean ± SEM.
Results
Analysis of TLR4 expression by human MCs
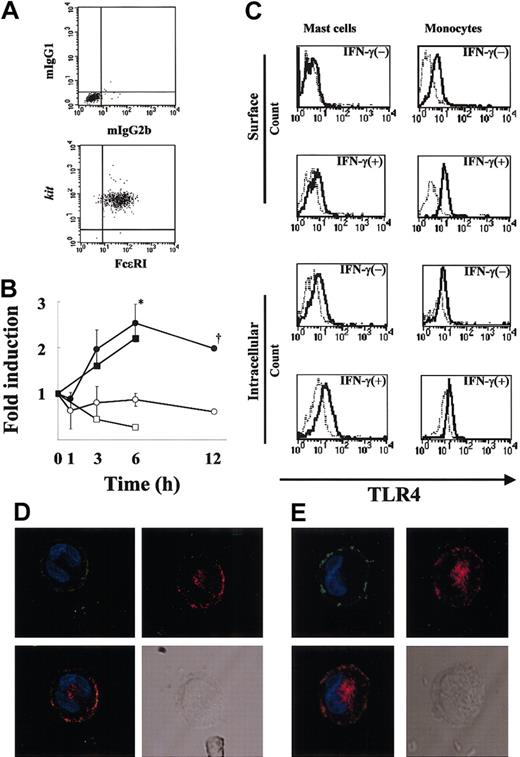
To demonstrate the purity of human MCs under the present culture conditions, flow cytometric analysis (Figure 1A) was first performed, and the purity of FcϵRI+kit+ cells was confirmed to be more than 99%. To examine whether MCs express TLR4, we performed RT-PCR using mRNA extracted from resting MCs and MCs treated with IFN-γ (6-hour treatment). The mRNA for TLR4 was detectable after 30 cycles of amplification for both the resting and treated MCs (data not shown). We next examined semiquantitatively the expression levels of TLR4 in MCs before and after IFN-γ treatment for up to 12 hours (Figure 1B). Figure 1B reveals an increase in TLR4 mRNA expression after 3 to 12 hours of IFN-γ stimulation compared with the control. TLR4 mRNA expression was significantly higher in MCs treated with IFN-γ compared with untreated MCs at 6 hours (P < .05), which was the maximal value and represented an increase of about 3-fold. TLR4 mRNA expression in human monocytes was also up-regulated by IFN-γ (Figure 1B). To verify that TLR4 is expressed on MCs and determine the effects of IFN-γ on this expression, we used FACS analysis using anti-TLR4 mAb. In agreement with the up-regulation of TLR4 mRNA by IFN-γ, the MFI ratio of TLR4 to control on the MC surface increased from 1.08 to 1.77 (Figure 1C). Analysis of intracellular staining of TLR4 also showed up-regulation of the expression by IFN-γ from 1.47 to 1.83 (MFI ratio of TLR4 to control in IFN-γ-untreated and -treated MCs, respectively; Figure 1C). We further compared the TLR4 expression levels on monocytes with those on MCs (Figure 1C). The MFI ratio of TLR4 to control on the monocyte surface increased from 2.09 to 3.50. The intensity of surface TLR4 expression on MCs was half of that on monocytes in the presence of IFN-γ. Thus, human MCs express TLR4, and the level of this expression is up-regulated by IFN-γ.
Expression of TLR4 by human MCs. (A) More than 99% of 12-week-old human MC populations stained for FcϵRI and kit (bottom panel) and the isotype controls (mIgG1 and mIgG2b; top panel). (B) Time-course of TLR4 mRNA expression by IFN-γ-treated human MCs (n = 3 donors). Total RNA (100 ng) was used for cDNA synthesis, and the fold change in the TLR4 mRNA level was evaluated by TaqMan analysis as described in “Materials and methods.” The results were expressed as the fold change in the TLR4 mRNA level of IFN-γ-treated cells (•) and untreated cells (○). *P < .05, for the comparison of MCs incubated with IFN-γ versus cells incubated without IFN-γ. Dagger indicates that the experimental number is 2. As a positive control, the fold change in the TLR4 mRNA level of IFN-γ-treated monocytes (▪) and untreated cells (□) is shown. (C) Surface and intracellular expression of TLR4 by human MCs in the presence or absence of IFN-γ. MCs and monocytes were cultured in medium with or without IFN-γ for 24 hours. TLR4 was detected in FcϵRI+ gated cells using biotin-conjugated anti-TLR4 mAb and streptavidin-PE and analyzed by flow cytometry. The isotype control is shown as a light line. As a positive control, monocytes were cultured similarly, and surface and intracellular TLR4 expressions were analyzed by FACS. (D-E) Localization of TLR4 in human cultured MCs (D) and human lung MCs (E). IFN-γ-treated MCs were first incubated with anti-FcϵRI (green) and anti-TLR4 (red), then stained with DAPI (blue). TLR4 is shown alone or merged with DAPI, whereas FcϵRI is shown merged with DAPI. A differential interference image is also shown. Original magnification, × 800.
Expression of TLR4 by human MCs. (A) More than 99% of 12-week-old human MC populations stained for FcϵRI and kit (bottom panel) and the isotype controls (mIgG1 and mIgG2b; top panel). (B) Time-course of TLR4 mRNA expression by IFN-γ-treated human MCs (n = 3 donors). Total RNA (100 ng) was used for cDNA synthesis, and the fold change in the TLR4 mRNA level was evaluated by TaqMan analysis as described in “Materials and methods.” The results were expressed as the fold change in the TLR4 mRNA level of IFN-γ-treated cells (•) and untreated cells (○). *P < .05, for the comparison of MCs incubated with IFN-γ versus cells incubated without IFN-γ. Dagger indicates that the experimental number is 2. As a positive control, the fold change in the TLR4 mRNA level of IFN-γ-treated monocytes (▪) and untreated cells (□) is shown. (C) Surface and intracellular expression of TLR4 by human MCs in the presence or absence of IFN-γ. MCs and monocytes were cultured in medium with or without IFN-γ for 24 hours. TLR4 was detected in FcϵRI+ gated cells using biotin-conjugated anti-TLR4 mAb and streptavidin-PE and analyzed by flow cytometry. The isotype control is shown as a light line. As a positive control, monocytes were cultured similarly, and surface and intracellular TLR4 expressions were analyzed by FACS. (D-E) Localization of TLR4 in human cultured MCs (D) and human lung MCs (E). IFN-γ-treated MCs were first incubated with anti-FcϵRI (green) and anti-TLR4 (red), then stained with DAPI (blue). TLR4 is shown alone or merged with DAPI, whereas FcϵRI is shown merged with DAPI. A differential interference image is also shown. Original magnification, × 800.
Localization of TLR4 in human cultured and lung MCs
To examine the localization of TLR4 in MCs, we performed confocal laser scanning microscopy of IFN-γ-treated MCs using antihuman TLR4 mAb and antihuman FcϵRI mAb. As can be seen in Figure 1D, TLR4 was expressed on the surface and around the nucleus of the MCs. We confirmed the expression of TLR4 in IFN-γ-treated human lung MCs using confocal laser scanning with anti-TLR4 mAb and found the expression pattern of TLR4 to be similar to that in human cultured MCs (Figure 1E). Confocal laser scanning also found a similar pattern of TLR4 expression in IFN-γ-treated monocytes (data not shown).
TNF-α production by human MCs on activation via TLR4
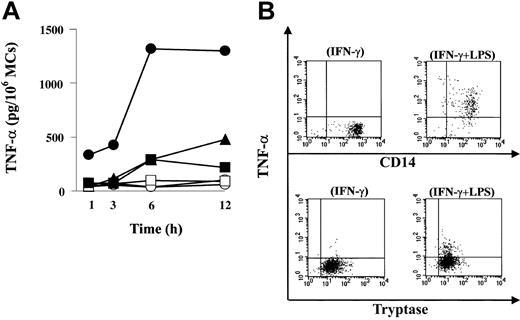
Next, we investigated whether LPS could activate MCs to undergo degranulation and to secrete TNF-α via TLR4. When evaluated by histamine release, LPS did not induce degranulation of resting or IFN-γ-treated MCs, whereas IgE receptor aggregation induced about 30% of total histamine release (data not shown). In contrast, the same concentration of LPS did induce TNF-α production (Figure 2A). Human MCs were precultured with or without IFN-γ, washed, and then challenged by incubation with LPS. Positive control cells were precultured with IgE but not with IFN-γ, washed, and then activated with anti-IgE. Production of TNF-α peaked at 12 hours after LPS stimulation of IFN-γ-untreated (477 pg/106 MCs) and IFN-γ-treated MCs (1301 pg/106 MCs), whereas anti-IgE activation induced TNF-α (216 pg/106 MCs), which peaked at 6 hours. IFN-γ enhanced LPS-induced TNF-α production, and the increase was approximately 3-fold. The amount of TNF-α induced by IgE-mediated activation was similar to that induced by LPS alone.
Functional expression of TLR4 on human MCs. (A) TNF-α release from MCs after LPS stimulation or FcϵRI aggregation. Human MCs were precultured with (•) or without (▴) IFN-γ and then exposed to LPS. Control cells were incubated with (○) or without (▵) IFN-γ for 24 hours, but LPS was omitted. Positive control cells were precultured with IgE and then activated with anti-IgE (▪). Control cells were incubated with IgE, but anti-IgE was omitted (□). Cell supernatants were harvested at various times up to 12 hours for ELISA of TNF-α. (B) The source of TNF-α produced by LPS was MCs. MCs were precultured with IFN-γ as described, and monensin was added to the MCs suspension simultaneously with LPS. Cells were incubated with LPS for 6 hours. Double-intracellular staining was performed with antitryptase and anti-TNF-α in human MCs (lower panels) following stimulation with IFN-γ alone (lower left) or IFN-γ LPS (lower right). The dot plots are representative of 7 separate analyses. As a positive control, human monocytes were precultured with IFN-γ and then activated with LPS for 6 hours. Double-intracellular staining with anti-CD14 and antitryptase in monocytes (upper panels) following IFN-γ (upper left) or LPS plus IFN-γ (upper right) was performed.
Functional expression of TLR4 on human MCs. (A) TNF-α release from MCs after LPS stimulation or FcϵRI aggregation. Human MCs were precultured with (•) or without (▴) IFN-γ and then exposed to LPS. Control cells were incubated with (○) or without (▵) IFN-γ for 24 hours, but LPS was omitted. Positive control cells were precultured with IgE and then activated with anti-IgE (▪). Control cells were incubated with IgE, but anti-IgE was omitted (□). Cell supernatants were harvested at various times up to 12 hours for ELISA of TNF-α. (B) The source of TNF-α produced by LPS was MCs. MCs were precultured with IFN-γ as described, and monensin was added to the MCs suspension simultaneously with LPS. Cells were incubated with LPS for 6 hours. Double-intracellular staining was performed with antitryptase and anti-TNF-α in human MCs (lower panels) following stimulation with IFN-γ alone (lower left) or IFN-γ LPS (lower right). The dot plots are representative of 7 separate analyses. As a positive control, human monocytes were precultured with IFN-γ and then activated with LPS for 6 hours. Double-intracellular staining with anti-CD14 and antitryptase in monocytes (upper panels) following IFN-γ (upper left) or LPS plus IFN-γ (upper right) was performed.
To confirm that the source of TNF-α induced by LPS was MCs, we performed double-intracellular staining of MCs with antitryptase and anti-TNF-α following stimulation with LPS. To enhance the immunoreactivity of LPS-activated MCs for TNF-α, MCs were precultured with IFN-γ as described, and monensin was added to the MC suspension simultaneously with LPS. Cells were incubated with LPS for 6 hours. As can be seen in Figure 2B, tryptase-positive MCs expressed TNF-α following activation with LPS. Thus, LPS activation of human MCs via TLR4 induces production of TNF-α.
LPS-induced human MC activation program
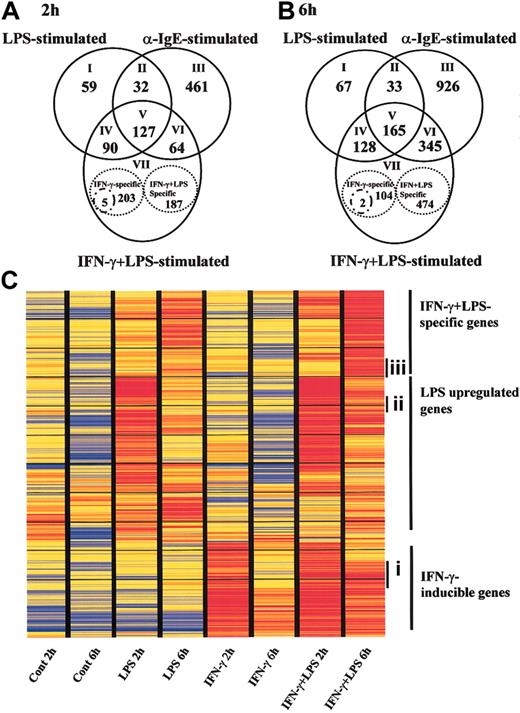
To clarify the specific gene expression profile in human MCs via TLR4, we evaluated approximately 23 000 genes by comparing the expression levels between LPS-stimulated MCs and anti-IgE-activated MCs. Because the transcriptomes for innate immune response and inflammation were reported to be up-regulated at early time points in dendritic cells and monocytes following TLR4-mediated activation,25,26 we chose 2 and 6 hours as the LPS stimulation periods in our experiments. MCs were preincubated either with or without IFN-γ and then activated with LPS for 2 and 6 hours. For comparison, IgE-sensitized MCs were activated with anti-IgE for 2 and 6 hours. Genes with expression levels that changed in response to stimulation were selected on the basis of repeated differences in the expression levels of the treated and untreated samples. Of the approximately 23 000 genes represented on GeneChip, 308, and 676, and 684 genes changed their expression significantly on 2-hour LPS stimulation of IFN-γ-untreated MCs, LPS stimulation of IFN-γ-treated MCs, and anti-IgE activation of MCs, respectively (Figure 3A).
TLR4- or FcϵRI-mediated gene expression profiles in human MCs. Human MCs were precultured with or without IFN-γ and then exposed to LPS for 2 hours (A) and 6 hours (B). Positive control cells were precultured with IgE and then activated with anti-IgE for 2 (A) and 6 hours (B). RNA was collected and used to conduct GeneChip analysis. Each experiment contained 1 or 2 independent donors, and each experiment was repeated using different sets of donors. The data are the average of 2 separate analyses. (A-B) Overlapping sets of LPS-, IFN-γ plus LPS-, and anti-IgE-regulated genes. Numbers in the overlapping region of the Venn diagram represent shared regulated genes. Numbers of LPS-, IFN-γ plus LPS-, or anti-IgE-specific genes are shown inside the stippled circles. Stimulus-specific genes were selected if the ratio of the relative expression level between stimuli was larger than 2.0, or if the data passed a stringent stimulus-specific filter based on the score. See the Supplemental Data Sets link at the top of the article on the Blood website. (C) Representation of mRNA expression levels in control MCs, LPS-stimulated MCs, IFN-γ-treated MCs, and LPS plus IFN-γ-stimulated MCs. One row of colored bars represents one gene, and each column represents one stimulus. Color bars capture the magnitude of the response for each gene, according to the scale shown. Genes are placed in groups corresponding to pair-wise overlaps shown in the accompanying Venn diagrams. From top to bottom: IFN-γ plus LPS-specific genes; LPS-specific genes; and IFN-γ-inducible genes. Ci indiates a set of genes that were up-regulated by pretreatment of IFN-γ following incubation with medium for 2 hours, but it waned during subsequent incubation with medium for 6 h. (Cii) a set of genes that were up-regulated by LPS for 2 hours, but decreased at 6 hours. (Ciii) a set of genes that were at the basal level at 2 hours under all conditions, but up-regulated by LPS at 6 hours in IFN-γ-pretreated cells.
TLR4- or FcϵRI-mediated gene expression profiles in human MCs. Human MCs were precultured with or without IFN-γ and then exposed to LPS for 2 hours (A) and 6 hours (B). Positive control cells were precultured with IgE and then activated with anti-IgE for 2 (A) and 6 hours (B). RNA was collected and used to conduct GeneChip analysis. Each experiment contained 1 or 2 independent donors, and each experiment was repeated using different sets of donors. The data are the average of 2 separate analyses. (A-B) Overlapping sets of LPS-, IFN-γ plus LPS-, and anti-IgE-regulated genes. Numbers in the overlapping region of the Venn diagram represent shared regulated genes. Numbers of LPS-, IFN-γ plus LPS-, or anti-IgE-specific genes are shown inside the stippled circles. Stimulus-specific genes were selected if the ratio of the relative expression level between stimuli was larger than 2.0, or if the data passed a stringent stimulus-specific filter based on the score. See the Supplemental Data Sets link at the top of the article on the Blood website. (C) Representation of mRNA expression levels in control MCs, LPS-stimulated MCs, IFN-γ-treated MCs, and LPS plus IFN-γ-stimulated MCs. One row of colored bars represents one gene, and each column represents one stimulus. Color bars capture the magnitude of the response for each gene, according to the scale shown. Genes are placed in groups corresponding to pair-wise overlaps shown in the accompanying Venn diagrams. From top to bottom: IFN-γ plus LPS-specific genes; LPS-specific genes; and IFN-γ-inducible genes. Ci indiates a set of genes that were up-regulated by pretreatment of IFN-γ following incubation with medium for 2 hours, but it waned during subsequent incubation with medium for 6 h. (Cii) a set of genes that were up-regulated by LPS for 2 hours, but decreased at 6 hours. (Ciii) a set of genes that were at the basal level at 2 hours under all conditions, but up-regulated by LPS at 6 hours in IFN-γ-pretreated cells.
The intersection of LPS-stimulated MCs in the presence or absence of IFN-γ- and anti-IgE-stimulated MCs revealed common sets of 127 and 165 regulated genes at 2 and 6 hours, respectively (Figure 3A-B; V, core set of genes). The common set of regulated genes at both time points contained such NF-κB pathway members as granulocyte-macrophage colony-stimulating factor (GM-CSF), CXCL1 (GRO1; 2 hours only), CXCL2 (GRO2), CXCL3 (GRO3), CXCL8 (IL-8), CCL3 (macrophage inflammatory protein 1α [MIP-1α]), CCL4 (MIP-1β), intercellular adhesion molecule 1 (ICAM-1), TNF-α (6 hours), IL-6 (2 hours), TNF-α-induced protein 3 (TNFAIP3), TANK (I-TRAF), NFκ-B1 (6 hours), NFκ-B2, and RelB (6 hours). It also included immune receptors (IL-15Rα (6 hours), TNFR super family 9 [TNFRSF9], TNFRSF4, prostaglandin ER4, and CD83), cytokines/chemokines (IL-5, pre-B cell colony-enhancing factor and CCL1), and cell stress genes (Sod2, Dusp5).
In addition to the core set of genes (Figure 3A-B; V), a total of 90 and 128 genes comprised a common set of genes that was expressed by both IFN-γ-treated and IFN-γ-untreated MCs in response to LPS at 2 and 6 hours, respectively (Figure 3A-B; IV). It included NF-κB pathway members (IL-1β, IL-6 [6 hours], TNF-α [6 hours], CXCL1 [6 hours], CCL5 [RANTES; 6 hours], vascular cell adhesion molecule 1 [VCAM-1], TNFAIP2), immune receptors (CCR7, TNFRSF5, and IL-15Rα; 2 hours), cytokines/chemokines (IL-10, IL-15 [6 hours], CCL8 [MCP2; 6 hours]), and an antiviral gene (adenosine deaminase [Ada] Epstein-Barr virus-induced gene-3 [EBI3]).
Proinflammatory cytokines such as TNF-α and IL-6 were up-regulated via both TLR4 and FcϵRI at 2 hours. LPS sustained the expression of these genes compared with the anti-IgE-induced signal. The mRNA for cytokines (IL-1β, IL-10, IL-15) and some chemokines (CCL5, CCL8) were significantly up-regulated in LPS-stimulated MCs compared with anti-IgE-stimulated MCs.
Next, we looked at the specific gene expression profile in LPS-induced MCs without IFN-γ treatment. Totals of 59 and 67 genes were specifically up-regulated in LPS-stimulated MCs at 2 and 6 hours, respectively (Figure 3A-B; I). The genes expressed following exposure to LPS included immune receptors (IL-2Rα [2 hours], paired immunoglobulin-like receptor β [6 hours]) and signal molecules (TRAF3). Totals of 395 and 580 regulated genes comprised the set of genes that was expressed by IFN-γ-treated MCs in response to LPS for 2 and 6 hours, respectively. The set of genes consisted of 3 groups: (1) 203 and 104 regulated genes were up-regulated in MCs by IFN-γ pretreatment alone following incubation of cells with medium for 2 and 6 hours, respectively (Figure 3A-B; VII). The expression levels of these genes were not affected by addition of LPS (expression changed by < 2-fold). At 2 hours, these genes included antiviral response genes such as Mx1 and Oas1, interferon-inducible genes such as interferon-induced protein 4 (IFI4), IFI with tetratricopeptide 4 (IFIT4) and IFIT1, and major histocompatibility complex (MHC) molecules. At 6 hours, the expression levels of some antiviral response genes and interferon-inducible genes had waned. (2) Only 5 and 2 regulated genes were up-regulated by IFN-γ and further enhanced by incubation of cells with LPS for 2 and 6 hours, respectively (expression changed by at least 2-fold) (Figure 3A-B; VII, small circles). The former included CXC10 and RIG-I. The latter were RGS7 and P5-1. (3) A total of 187 and 474 regulated genes were up-regulated in IFN-γ-treated MCs in response to LPS for 2 and 6 hours, respectively (Figure 3A-B; VII). The expression levels of these genes were not affected by IFN-γ alone at 2 hours. They included IL-12, CCL5, and CXC11. At 6 hours, the set of genes consisted of antiviral response genes such as Mx1 and Oas1, and IFN-inducible genes such as IFIT1 and IFI4. The expression levels of antiviral response genes and interferon-inducible genes reverted to the basal levels by 6 hours after removal of IFN-γ. These data indicate that a set of genes including CXC11, CCL5, and IL-12 is specifically up-regulated in IFN-γ pretreated MCs stimulated by LPS, that human MCs up-regulate antiviral response genes such as Mx1 and Oas1 in response to IFN-γ alone, and that LPS sustains the expression levels of the genes (Figure 4).
Induction of antiviral genes (Mx-1, Oas-1, IFIT1, Ada, and EBI3) by MCs. Bar graphs display expression of antiviral genes selected from Figure 3A (2 hours; □) and Figure 3B (6 hours; ▪), in human MCs stimulated by LPS in the presence or absence of IFN-γ. It should be noted that EBI3 and Ada were up-regulated by LPS alone, whereas Mx1, Oas-1, and IFIT 1 were up-regulated by IFN-γ pretreatment and the expression levels were sustained by addition of LPS at 6 hours. As a positive control, PBMNCs were stimulated by LPS for 6 hours (▪). Expression levels are shown as normalized values (“Materials and methods”). ND indicates not determined.
Induction of antiviral genes (Mx-1, Oas-1, IFIT1, Ada, and EBI3) by MCs. Bar graphs display expression of antiviral genes selected from Figure 3A (2 hours; □) and Figure 3B (6 hours; ▪), in human MCs stimulated by LPS in the presence or absence of IFN-γ. It should be noted that EBI3 and Ada were up-regulated by LPS alone, whereas Mx1, Oas-1, and IFIT 1 were up-regulated by IFN-γ pretreatment and the expression levels were sustained by addition of LPS at 6 hours. As a positive control, PBMNCs were stimulated by LPS for 6 hours (▪). Expression levels are shown as normalized values (“Materials and methods”). ND indicates not determined.
The analysis of FcϵRI-mediated specific genes (Figure 3A-B; III) showed that MCs induced growth factors (IL-3, M-CSF, amphiregulin [6 hours], epiregulin, inhibin β A [6 hours]), chemokines (CXCL5, CCL7, CCL11 [6 hours]), cell adhesion molecules (thrombospondin 1, a disintegrin and metalloproteinase domain 9 [6 hours], catenin [cadherin-associated protein] α1 [6 hours]), and apoptosis-related molecules (CASP8 and FADD-like apoptosis regulator [6 hours], TNFSF14, Fas ligand [6 hours]) in response to IgE-dependent stimulation. The intersection of LPS- and anti-IgE-stimulated MCs revealed only 32 and 33 regulated common genes, including CXCL5 (2 hours) and IL-2Rα (6 hours); Figure 3 A-B; II). Thus, human MCs are capable of exhibiting different gene expression profiles for IgE-dependent stimulation compared with LPS-mediated stimulation.
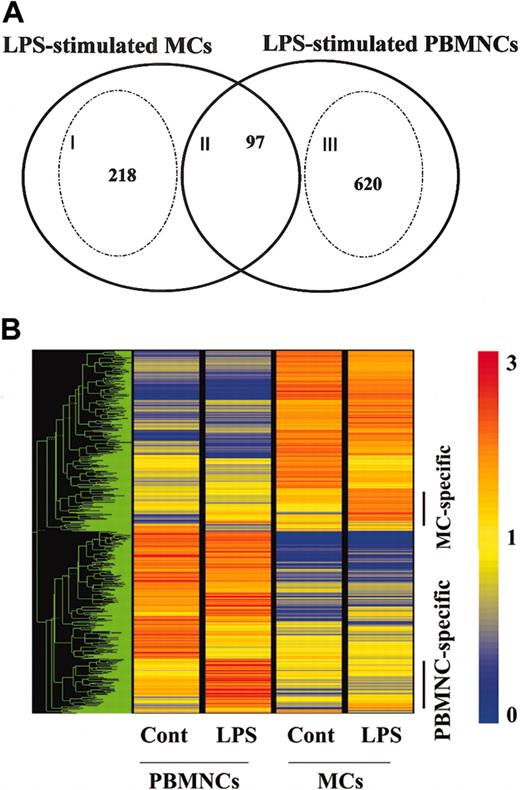
To examine the specific gene expression profile of human MCs via TLR4, we next compared the gene profiles induced between MCs and PBMNCs in response to LPS (Figure 5). The intersection of LPS-stimulated PBMNCs and MCs revealed a core set of 97 genes (Figure 5A; II). As expected, it included NF-κB pathway members (IL-1β, IL-6, TNF-α, GM-CSF, CXCL1, CXCL2, CXCL3, CXCL8, CCL3, CCL4, ICAM-1, TNFAIP3, TNFAIP2, NFκ-B1, NFκ-B2), immune receptors (IL-2Rα, IL-15Rα, TNFRSF9, CD83), cytokines (IL-10, pre-B-cell colony-enhancing factor), and signal molecules (STAT4, STAT inhibitor-2).
Comparison of LPS-mediated gene profiles between MCs and PBMNCs. Human MCs and PBMNCs were exposed to LPS for 6 hours. Each experiment contained one or 2 independent donors, and each component was repeated using different sets of donors. The data are representative of 2 separate analyses. (A) Overlapping sets of LPS-regulated genes expressed in human MCs and PBMNCs. Numbers in the overlapping region of the Venn diagram represent shared regulated genes. Numbers of MC- or PBMNC-specific genes are shown inside the stippled circles. Cell-specific genes were selected if the ratio of the relative expression level between stimuli was larger than 2.0, or if the data passed a stringent stimulus-specific filter based on the score. See the Blood website for supplemental data. (B) Representation of mRNA expression levels in control MCs, LPS-stimulated MCs, control PBMNCs, and LPS-stimulated PBMNCs. One row of colored bars represents one gene, and each column represents one stimulus. Color bars capture the magnitude of the response for each gene, according to the scale shown. Genes are placed in groups corresponding to pair-wise overlaps shown in the accompanying Venn diagrams. From top to bottom: LPS-stimulated MC-specific genes; and LPS-stimulated PBMNC-specific genes. A dendrogram shows overall similarity of the expression profiles of the representative samples.
Comparison of LPS-mediated gene profiles between MCs and PBMNCs. Human MCs and PBMNCs were exposed to LPS for 6 hours. Each experiment contained one or 2 independent donors, and each component was repeated using different sets of donors. The data are representative of 2 separate analyses. (A) Overlapping sets of LPS-regulated genes expressed in human MCs and PBMNCs. Numbers in the overlapping region of the Venn diagram represent shared regulated genes. Numbers of MC- or PBMNC-specific genes are shown inside the stippled circles. Cell-specific genes were selected if the ratio of the relative expression level between stimuli was larger than 2.0, or if the data passed a stringent stimulus-specific filter based on the score. See the Blood website for supplemental data. (B) Representation of mRNA expression levels in control MCs, LPS-stimulated MCs, control PBMNCs, and LPS-stimulated PBMNCs. One row of colored bars represents one gene, and each column represents one stimulus. Color bars capture the magnitude of the response for each gene, according to the scale shown. Genes are placed in groups corresponding to pair-wise overlaps shown in the accompanying Venn diagrams. From top to bottom: LPS-stimulated MC-specific genes; and LPS-stimulated PBMNC-specific genes. A dendrogram shows overall similarity of the expression profiles of the representative samples.
Analysis of the individual responses to LPS in PBMNCs and MCs showed that a unique number of genes were regulated (Figure 5B). The specific gene expression profile of PBMNCs via TLR4 revealed a total of 620 genes (Figure 5A; III), which included chemokines (CCL2, CCL7, CCL18, CCL20, Mig-2), cytokines (IFN-γ, IL-1Ra, IL-12 p40, IL-15, IL-24), immune receptors (IL-12Rβ2, IL-21R, TLR1, TLR3), immune transcription genes (STAT2, STAT3, IRF2, ISGF3, Mx1), and tissue remodeling genes (mmp19, mmp14, mmp10, mmp8). Many IFN-inducible genes, such as IFIT1, IFIT2, IFITRF2, IFITM1, IFITM2, IFITM3, and IFN-stimulated gene 20, were up-regulated only in this group. This profile of LPS-induced genes in PBMNCs is in agreement with previous reports.26
In MCs, LPS up-regulates mRNA for enzymes (protease serine 22), cytokines/chemokines (IL-5, CCL1, CCL5, CCL8), receptors (PGER3, TLR1, FcγRIIa, activated leukocyte cell adhesion molecule), and STAT5a (Figure 5A; I). These results suggest that human MCs might recruit eosinophils and Th2 cells to inflammatory sites through the release of IL-5, CCL5, and CCL1 in response to LPS.
Confirmation of specific cytokine profiles using ELISA
We demonstrated that LPS up-regulated IL-5 and CCL1 mRNA in human MCs, but not human PBMNCs. IgE-dependent stimulation also up-regulated IL-5 and CCL1 mRNA. LPS selectively up-regulated mRNA for CCL5 in human MCs with or without IFN-γ pretreatment. Thus, we measured IL-5, CCL1, and CCL5 production by LPS-stimulated MCs, anti-IgE-stimulated MCs, and LPS-activated human monocytes. As can be seen in Figure 6, IL-5 and CCL1 were released from LPS- and anti-IgE-stimulated MCs, but these cytokines were below their levels of detection in LPS-stimulated monocytes. CCL5 was detectable in supernatants from LPS plus IFN-γ-stimulated MCs and LPS-stimulated monocytes. We thus confirmed the mRNA profiles of cytokine expression under the individual stimulation conditions.
IL-5, CCL1, and CCL5 production by human MCs and human monocytes. Human MCs were precultured with or without IFN-γ and then exposed to LPS for 16 hours. Control cells were incubated similarly, except that LPS was omitted. Positive control cells were precultured with IgE and then activated with anti-IgE for 16 hours. Human monocytes (Mono) were exposed to LPS for 16 hours. The cell supernatants were used for cytokine ELISAs.
IL-5, CCL1, and CCL5 production by human MCs and human monocytes. Human MCs were precultured with or without IFN-γ and then exposed to LPS for 16 hours. Control cells were incubated similarly, except that LPS was omitted. Positive control cells were precultured with IgE and then activated with anti-IgE for 16 hours. Human monocytes (Mono) were exposed to LPS for 16 hours. The cell supernatants were used for cytokine ELISAs.
Discussion
In this paper, we demonstrate that human MCs express TLR4 and that this receptor expression and its function are up-regulated by IFN-γ (Figures 1, 2). We further demonstrate that LPS induces TNF-α production by MCs via TLR4 (Figure 2). We compared the LPS-induced gene expression profile and FcϵRI-mediated profile in MCs by using high-density oligonucleotide probe arrays to find molecules similarly and differently regulated by LPS- and anti-IgE-mediated stimulation. Both a shared core response and a stimulation-specific program of gene expression were observed following TLR4- and FcϵRI-mediated MC activation (Figure 3). MCs exhibited an antiviral response gene program in response to IFN-γ and LPS sustained that expression (Figures 3C-4). Furthermore, we identified an LPS-induced MC-specific gene expression profile by comparison with the LPS-stimulated gene profile of PBMNCs (Figure 5). We confirmed the mRNA profiles of cytokine expression under individual stimulation conditions by measuring stimulus-specific cytokines/chemokines (Figure 6). This demonstration of functional TLR4 on human MCs may help explain in vivo observations that document the participation of MCs in host defense mechanisms against bacteria and viruses.
Although we believe this to be the first definitive report that human MCs express TLR4, there have been extensive studies on TLR4 on murine MCs.4,5,32,33 Murine bone marrow-derived mast cells (BMMCs) and the murine MC line MC/9 express TLR4 mRNA. After activation with Escherichia coli-derived LPS, murine MCs produce TNF-α, IL-1β, IL-6, and IL-13, but not IL-4 or IL-5.33 A study using an MC-dependent model of acute sepsis found that higher mortality was shown by TLR4-mutated BMMC-reconstituted W/Wv mice and that TLR4 deficiency of BMMCs in mice results in significantly higher mortality because of defective neutrophil recruitment and production of proinflammatory cytokines in the peritoneal cavity.4 These findings in murine models suggest that murine MCs play important roles in the expression of innate immunity through TLR4.
We first investigated the purity of the human cultured MCs that we used in our experiments by flow cytometry (Figure 1A) and immunocytochemistry (data not shown), and we confirmed that the purity of tryptase-positive cells was 99.9%, and the purity of kit+ FcϵRI+ cells was more than 99%. We also performed FACS analysis of the cultured MCs using anti-CD14 and anti-CD19 mAbs and found no CD14+ or CD19+ cells in the population (data not shown). Using those cultured MCs, we demonstrated the expression of both mRNA for TLR4 (Figure 1B) and cell surface TLR4 protein (Figure 1C-D). Comparison of TLR4 expression levels on monocytes with those on MCs found them to be comparable. Further, human lung MCs also expressed TLR4 on their cell surface (Figure 1E). As analyzed by semiquantitative RT-PCR, IFN-γ treatment of MCs significantly up-regulated TLR4 mRNA expression (Figure 1B). After up-regulation of mRNA for TLR4 by IFN-γ, the resulting cell surface expression of the protein was evident by 24 hours (Figure 1C). This was also confirmed in monocytes.
The activation of human MCs via TLR4 by LPS did not induce histamine release, which was not affected by IFN-γ pretreatment. This is in agreement with the findings of previous reports for murine BMMCs.4,5 However, triggering of MCs via TLR4 led to induction of TNF-α production (Figure 2A). IFN-γ enhanced LPS-induced TNF-α production, and the increase was about 3-fold. Thus, IFN-γ up-regulated both TLR4 expression and TLR4-mediated TNF-α production. Next, we confirmed that the source of TNF-α produced by LPS was MCs by using double-intracellular staining with antitryptase and anti-TNF-α (Figure 2B). We recently reported that FcγRI is expressed on human MCs and is up-regulated by IFN-γ and that some proinflammatory cytokines such as TNF-α are significantly up-regulated via aggregation of FcγRI compared with FcϵRI.3,34 The level of TNF-α produced by human MCs via TLR4 in the presence of IFN-γ was comparable to the level produced by LPS-stimulated monocytes (1.3 ng/106 MCs versus 19 ng/106 monocytes, data not shown). These findings suggest that human MCs, after recognizing enterobacteria through TLR4 on their surface in the presence of IFN-γ may play important roles in host defense not only in acquired immunity through FcγRI but also in innate immunity by releasing a large amount of TNF-α, which recruits neutrophils.
Studying FcϵRI- and TLR4-mediated reactions by using a parallel, comparative analysis of gene expression permitted the identification of both shared and distinct gene expression responses in MCs. Of the approximately 23 000 genes represented on the GeneChip, a total of about 680 genes changed their expression significantly on exposure to an antigen (FcϵRI-mediated activation) or a bacterial component (LPS, TLR4-mediated activation). Such large-scale changes in gene expression suggest that human MCs are able to undergo a marked transformation in their cellular phenotype. The MC activation program shared by FcϵRI- and TLR4- mediated reactions includes mainly NF-κB pathway members, immune receptors, and cell stress genes, indicating that MCs are major effector cells in inflammation and can communicate with other cells.
Analysis of the individual responses to the activation conditions showed that a unique number of genes were regulated by each activation condition. LPS was able to modulate an exclusive subset of genes (Figure 3A-B; IV). This includes cytokines (IL-1β, IL-10, and IL-15) and chemokines (CCL5, CCL8). The proinflammatory component of the activation program may constitute a genetic “alarm signal” that marshals antibacterial defenses. This was confirmed by a comparison of the LPS-induced gene profiles expressed by MCs and PBMNCs (Figure 5). The intersection of LPS-stimulated PBMNCs and MCs revealed a core set of genes (Figure 5) that included NF-κB pathway members and immune receptors. Thus, MCs might be able to undergo a marked transformation in their cellular phenotype, like monocytes in response to LPS. It has been reported that IRF3 and NF-κB are key transcription factors due to their stimulation of TLR3/TLR4, and that IRF3 directs the specific induction of a set of primary and secondary genes involved in host defense in murine B cells. In human MCs, due to the lack of induction of type I IFNs mRNA in response to LPS, which we also confirmed by RT-PCR (data not shown), a group of secondary response genes that are part of the autocrine paracrine loop activated by the primary response gene product, type I IFNs, was absent.
However, analysis of the kinetics of the responses with or without IFN-γ pretreatment (Figure 3C) revealed that expression of genes such as Mx1, Oas1, and IRF7 was up-regulated by IFN-γ pretreatment following incubation with medium for 2 hours, after which it decreased and became almost undetectable at 6 hours. We confirmed by real-time RT-PCR that IFN-γ alone up-regulated the expression of Mx1. The Mx1 mRNA expression was maximal at 8 hours of IFN-γ stimulation, at which time the increase was 4.8-fold (data not shown). Addition of LPS to the MCs kept the IFN-γ-induced gene levels high even at 6 hours (Figure 3Ci). We further realized that expression of another set of genes such as IRF-1 and IL-1α was up-regulated by LPS alone at 2 hours but had waned by 6 hours. However, after IFN-γ pretreatment, the up-regulation of the LPS-induced gene expression level was sustained for another 4 hours (Figure 3Cii). On the other hand, expression of yet another set of genes, including annexin A2, was minimal at 2 hours under all conditions, but it was up-regulated by LPS at 6 hours in MCs pretreated with IFN-γ (Figure 3Ciii).
Finally, we identified a subset of genes that is specifically induced in LPS-stimulated MCs but not in LPS-activated PBMNCs. Further gene expression analyses established that several response genes were dependent on NF-κB in both activated MCs and activated PBMNCs. Also, up-regulation of a Th2 cytokine (IL-5) and the CC chemokines, such as CCL5 and CCL1, which recruit Th2 cells and eosinophils, was found to confer MC specificity. We further confirmed that the up-regulation of IL-5 and the CC chemokines was not observed in LPS-stimulated PBMNCs with IFN-γ pretreatment (data not shown). These results suggest that direct activation of human MCs via TLR4 contributes to the innate immunity and allergic responses.
We confirmed the mRNA profiles of cytokine expression obtained with GeneChip by measuring stimulus-specific cytokines. However, discrepancies between the GeneChip and ELISA data were sometimes observed. This was due mainly to 2 reasons: (1) GeneChip data are considered significant when expression is changed at least 2-fold (“Materials and methods”). For example, up-regulation of IL-6 mRNA by IgE/anti-IgE at 6 hours was 1.37-fold. Thus, this up-regulation is not considered significant although the production of IL-6 was enhanced in MCs stimulated with IgE/anti-IgE. (2) The sensitivity of ELISA in our experimental conditions may be another reason. For example, LPS alone up-regulated CCL5 mRNA, but CCL5 was below the detection level of the assay.
In this report, we demonstrated for the first time functional TLR4 expression on human MCs. We identified a subset of genes that is specifically induced by stimulation through TLR4 but not by aggregation of FcϵRI. Furthermore, in response to IFN-γ, MCs underwent induction of a variety of antiviral response genes and LPS prolonged that expression. Finally, by comparison with LPS-stimulated PBMNCs, we identified a subset of genes that is specifically induced in LPS-stimulated MCs. This includes a Th2 cytokine and chemokines that induce or enhance allergic inflammation. Taken together, human MCs exhibit tailored pathogen- and antigen-specific immune responses, suggesting that human MCs may play important roles in innate and adaptive immunity.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2002-12-3929.
Supported in part by grants from RIKEN Yokohama Institute, a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (project no. 14570402; Y.O.), Hokuriku Seiyaku Co, Ltd-Japanese Allergy Foundation (Y.O.), AstraZeneca Asthma Research Award 2002 (Y.O.), and the Organization for Pharmaceutical Safety and Research and the Ministry of Health, Labour and Welfare (Millennium Genome Project, MPJ-5).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Mr T. Honma, Mr A. Kato, and Ms N. Hashimoto of National Research Institute for Child Health and Development for their skillful technical assistance and helpful suggestions. We thank Prof K. Ohta and Assoc Prof N. Yamashita and the surgical staff of Teikyo University School of Medicine for supplying human lung specimens and for their capable assistance. We also thank Prof K. Okuda of Tokyo Dental College for his critical review and helpful suggestions.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal