Abstract
Imatinib mesylate is effective in the treatment of hematologic malignancies that are characterized by either abl- or PDGFRβ- activating mutations. The drug is also active in a subset of patients with eosinophilic disorders and systemic mast cell disease (SMCD). Recently, a novel tyrosine kinase that is generated from fusion of the Fip1-like 1 (FIP1L1) and PDGFRα (PDGFRA) genes has been identified as a therapeutic target for imatinib mesylate in hypereosinophilic syndrome (HES). We used fluorescence in situ hybridization (FISH) to detect deletion of the CHIC2 locus at 4q12 as a surrogate for the FIP1L1-PDGFRA fusion. CHIC2 deletion was observed in bone marrow cells for 3 of 5 patients with SMCD associated with eosinophilia. Deletion of this locus and expression of the FIP1L1–platelet-derived growth factor receptor α (PDGFRA) fusion was also documented in enriched eosinophils, neutrophils, or mononuclear cells by both FISH and reverse transcriptase–polymerase chain reaction (RT-PCR) for one patient. While all 3 patients with the FIP1L1-PDGFRA rearrangement achieved a sustained complete response with imatinib mesylate therapy, the other two, both carrying the c-kit Asp816 to Val (Asp816Val) mutation, did not. These observations suggest that the FIP1L1-PDGFRA rearrangement occurs in an early hematopoietic progenitor and suggests that the molecular pathogenesis for a subset of SMCD patients is similar to that of HES. Screening for the FIP1L1-PDGFRA rearrangement and Asp816Val mutation will advance rational therapy decisions in SMCD.
Introduction
Imatinib mesylate (Gleevec), a small-molecule tyrosine kinase inhibitor from the 2-phenylaminopyrimidine class of compounds, has shown activity in the treatment of malignancies that are associated with the constitutive activation of a specific subgroup of tyrosine kinases. Imatinib mesylate–sensitive malignancies include chronic myeloid leukemia (CML)1,2 and gastrointestinal stromal tumors (GISTs)3,4 that are associated with the activation of bcr-abl and c-kit tyrosine kinases, respectively. More recently, imatinib mesylate has proven effective in the treatment of hypereosinophilic syndrome (HES)5-10 and chronic myeloproliferative disorders that are associated with rearrangement of the PDGFRβ gene.11 Both types of disorders are characterized by prominent peripheral blood and bone marrow eosinophilia, with symptoms and signs of organ infiltration by eosinophils. In a recent study, a novel tyrosine kinase, generated from fusion of the Fip1-like 1 (FIP1L1) gene to the PDGFRA gene, was identified in 9 of 16 patients (56%) with HES.10 This fusion results from an approximately 800-kb interstitial chromosomal deletion that includes the cysteine-rich hydrophobic domain 2 (CHIC2) locus. FIP1L1–platelet-derived growth factor receptor α (PDGFRA) is a constitutively activated tyrosine kinase that transforms hematopoietic cells and is a therapeutic target for imatinib mesylate in a subset of HES patients.
Mast cell disease (MCD) is a clinically heterogenous disorder wherein accumulation of mast cells (MCs) may be limited to the skin (cutaneous mastocytosis) or involve one or more extracutaneous organs (systemic MCD [SMCD]).12 Because SMCD is considered a c-kit–driven malignancy13 and is often associated with eosinophilia (SMCD-eos),14-16 we recently tested the therapeutic activity of imatinib mesylate in 12 adults with this disease.17 In the current study we demonstrate that FIP1L1-PDGFRA is the therapeutic target of imatinib mesylate in the specific subset of patients with SMCD and associated eosinophilia (SMCD-eos). Furthermore, we provide evidence using both fluorescence in situ hybridization (FISH) and reverse transcriptase–polymerase chain reaction (RT-PCR) that the FIP1L1-PDGFRA rearrangement occurs in an early hematopoietic progenitor in SMCD.
Patients and methods
Twelve adult SMCD patients, including 5 with associated eosinophilia, were prospectively treated with imatinib mesylate. The accrual of patient data and collection of biologic specimens were approved by Mayo Clinic's institutional review board, and written informed consent was obtained from all patients. Those patients who presented consecutively were symptomatic, had bone marrow biopsy–proven MCD, and were deemed to require cytoreductive therapy. All patients met the World Health Organization (WHO) criteria for SMCD, including the presence of multifocal dense infiltrates of morphologically atypical mast cells in the bone marrow (Figure 1).18 No additional inclusion or exclusion criteria were used.
Histologic response with imatinib mesylate therapy (case 3). Hematoxylin-eosin (H&E)–stained (A,D) and tryptase-immunostained (B-C,E) bone marrow biopsy tissue showing the markedly hypercellular marrow with characteristic aggregates of atypical mast cells before treatment with imatinib mesylate (A-C). Panels D-E show the dramatic reduction in marrow mast cell burden and overall cellularity after imatinib mesylate therapy (at 4 weeks). Only scattered tryptase-positive mast cells are observed following therapy. Original magnification, × 80.
Histologic response with imatinib mesylate therapy (case 3). Hematoxylin-eosin (H&E)–stained (A,D) and tryptase-immunostained (B-C,E) bone marrow biopsy tissue showing the markedly hypercellular marrow with characteristic aggregates of atypical mast cells before treatment with imatinib mesylate (A-C). Panels D-E show the dramatic reduction in marrow mast cell burden and overall cellularity after imatinib mesylate therapy (at 4 weeks). Only scattered tryptase-positive mast cells are observed following therapy. Original magnification, × 80.
Genomic DNA isolated from peripheral blood mononuclear cells (PBMCs) and bone marrow mononuclear cells (BMMCs) from all patients was screened for known activating c-kit mutations (codons Asp816 and Val560) by direct sequencing of exons 11-12 and exon 17.19 Presence of t(5;12)(q33;p13) (ETV6-PDGFRβ) was screened for by conventional cytogenetic analysis of bone marrow cells. In addition, all coding exons of PDGFRβ (exons 2-23) and c-kit (exons 1-21) were amplified from PBMCs and/or BMMCs (cases 1, 2, 4, and 5) and purified eosinophils (cases 1 and 4) for mutational analysis by denaturing high-performance liquid chromatography (WAVE; Transgenomics, Crewe, United Kingdom). Eosinophils were purified as previously described.20 Briefly, a 30-mL sample of peripheral blood was subjected to double–density gradient centrifugation (Histopaque-1077 layered over Histopaque-1119; Sigma Diagnostics, St Louis, MO) in order to isolate the granulocyte layer. The cells were then further fractionated using magnetic-activated cell separation (MACS; Miltenyi Biotech, Auburn, CA) by the incubation of an antibody/magnetic bead complex specific for neutrophils (CD16). Suspensions were processed per the manufacturer's protocol and both the positive and negative fractions were kept: CD16+ for purified neutrophils and CD16– for eosinophils. The cell purity of various fractions was analyzed using the FACScalibur flow cytometer (Becton Dickinson [BD], Franklin Lakes, NJ; data not shown). Samples were incubated with 5 μL of the CD45 fluorescein isothiocyanate (FITC)–conjugated monoclonal antibody (BD 347463) or isotype control (BD 349526), washed, and resuspended in 1% paraformaldehyde. Ten thousand events were collected using FITC/side-scatter. Smears were made from separate aliquots of CD16+ and CD16– cell populations and sample purity (≥ 97%) was confirmed by a manual differential count of Wright-Giemsa–stained slides. For case 3, first-strand cDNA was synthesized from RNA isolated from eosinophils or neutrophils using Superscript (Invitrogen, Carlsbad, CA) and random primers. Fusion of FIP1L1 to PDGFRA was analyzed by nested PCR using primers FIP1L1-F1 (5′-ACCTGGTGCTGATCTTTCTGAT-3′) and PDGFRA-R1 (5′TGAGAGCTTGTTTTTCACTGGA-3′) during the first PCR and primers FIP1L1-F2 (5′-AAAGAGGATACGAATGGGACTTG-3′) and PDGFRA-R2 (5′-GGGACCGGCTTAATCCATAG-3′) for the second PCR.
To detect the CHIC2 deletion, direct-labeled FISH probes were designed from 2 bacterial artificial chromosome (BAC) clones (RP11-98G22, accession number AQ317591 and PR11-89B16, accession number AC105384) covering the area of interest on 4q12. Glycerol stocks of the BAC clones were received from ResGen Invitrogen. Clones were plated and propagated immediately upon arrival. Isolation and purification of DNA were performed using the QIAGEN Plasmid Maxi Kit according to the manufacturer's instructions (Qiagen, Valencia, CA). Primers were designed using unique gene sequences (http://www.ncbi.nlm.nih.gov/) for CHIC2, FIP1L1, PDGFRA, and sequence tag site (STS) markers for flanking clones. PCR and gel electrophoresis were done to verify the presence of the gene or region of interest for each clone. Nick translation was performed using the Vysis Nick Translation Kit (Vysis, Downers Grove, IL) to produce the fluorescence-labeled DNA probes for FISH. Probes were labeled with Spectrum Orange–dUTP (deoxyuridine triphosphate; Vysis), precipitated, and applied to normal peripheral blood samples. Signal validation was verified by observing both interphase and metaphase cells.
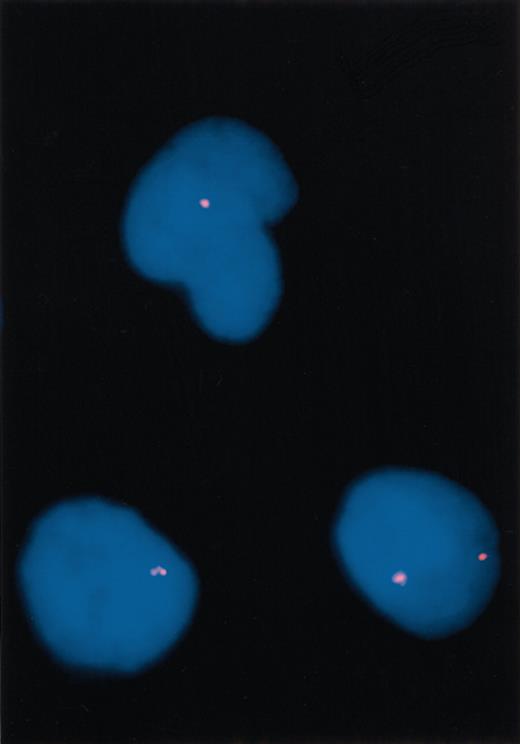
Whole bone marrow specimens were processed for FISH and conventional cytogenetics using standard procedures.21 For each patient, bone marrow cell pellets were stored at –70°C in methanol–glacial acetic acid (2:1) fixative. Each sample had a change of fixative and FISH was performed by standard procedures.22 Two individuals independently scored 100 nuclei (200 total) in a blinded fashion for 10 healthy patients and the 5 study patients. Representative signal patterns for normal and abnormal interphase nuclei are demonstrated in Figure 2.
Fluorescence in situ hybridization (FISH) results. Three interphase cells from case 2, with 1 cell (bottom right) showing 2 CHIC2 signals (normal pattern) and 2 cells (top and bottom left) showing deletion of 1 CHIC2 signal (abnormal). Original magnification, × 1000.
Fluorescence in situ hybridization (FISH) results. Three interphase cells from case 2, with 1 cell (bottom right) showing 2 CHIC2 signals (normal pattern) and 2 cells (top and bottom left) showing deletion of 1 CHIC2 signal (abnormal). Original magnification, × 1000.
Results
All patients described here met the WHO criteria for SMCD.18 Of the 5 SMCD-eos patients, 4 had aggressive SMCD, 1 had indolent SMCD, and all had peripheral blood eosinophilia (median, 10.2; range, 3.5 to 16.1 × 109/L) and bone marrow eosinophilia at diagnosis (Table 1). Known causes of reactive eosinophilia and presence of both the bcr-abl translocation (by FISH) and t(5;12)(q33; p13) (by karyotype) were excluded in each instance. In addition, none of the 4 patients tested (cases 1, 2, 4, and 5) carried additional activating mutations in coding exons of c-kit or PDGFRβ genes. Patients were treated with imatinib mesylate at a dose of 100 to 400 mg per day and the primary end point was mast cell cytoreduction as monitored by serial bone marrow biopsies.
Patient characteristics: SMCD with associated eosinophilia
Cases . | 1 . | 2 . | 3 . | 4 . | 5 . |
|---|---|---|---|---|---|
| Age, y | 46 | 31 | 72 | 78 | 85 |
| Organomegaly | |||||
| Before treatment | S | H,S | S | H,S | S |
| After treatment | CR | CR | CR | No change | No change |
| Diagnosis date, BM | 05/31/01 | 06/12/01 | 2/19/03 | 06/09/98 | 10/04/02 |
| Prior treatment | None | HU, IFN-α | HU | IFN-α | HU |
| Follow-up, mos | 10+ | 19+ | 1+ | 9+ | 5+ |
| WHO classification | ISM | ASM | ASM | ASM | ASM |
| Imatinib mesylate dose: induction/maintenance, mg/d | 100, 100 | 400, 100 (biw) | 100, 100 | 100-400, NA | 100-400, NA |
| Hgb concentration, g/dL | |||||
| Before treatment | 12.1 | 9.2 | 13.9 | 10.1 | 10 |
| After treatment | 15.1 | 15.3 | 13.3 | 10.0 | 11.4 |
| WBC count, × 109/L | |||||
| Before treatment | 14.5 | 69.8 | 15.2 | 19.7 | 18 |
| After treatment | 4.9 | 6.1 | 3.1 | 21.7 | 17.5 |
| AEC count, × 109/L | |||||
| Before treatment | 10.7 | 16.1 | 10.2 | 3.5 | 6.1 |
| After treatment | 0.2 | 0.3 | 0.4 | 2.7 | 3.2 |
| Platelet count, × 109/L | |||||
| Before treatment | 118 | 31 | 213 | 210 | 166 |
| After treatment | 169 | 179 | 166 | 252 | 213 |
| Serum tryptase, less than 11.5 ng/mL | |||||
| Before treatment | 137 | ND | 56 | 904 | 331 |
| After treatment | 25.7 | ND | 19.8 | 1370 | 342 |
| c-kit Asp816Val mutation | A | A | A | P | P |
| FIP1L1-PDGFRA | P | P | P | A | A |
| CHIC2 deletion, % of nuclei positive | |||||
| Before treatment | 61.0 | 58.0 | 76.0 | 2.5 | 0.5 |
| After treatment | 0.0 | 1.5 | 0.5 | ND | ND |
| ETV6-PDGFRB | A | A | A | A | A |
| Cytogenetics | 46,XY | 46,XY | 46,XY | 46,XX | 46,XX |
| Response | Complete response | Complete response | Complete response | NR | NR |
| BM overall cellularity/mast cell % | |||||
| Before treatment | 90/30 | 95/15 | 90/20 | 90/70 | > 95/30 |
| After treatment | 35/CR | 45/CR | 60/CR | 90/70 | ND/ND |
Cases . | 1 . | 2 . | 3 . | 4 . | 5 . |
|---|---|---|---|---|---|
| Age, y | 46 | 31 | 72 | 78 | 85 |
| Organomegaly | |||||
| Before treatment | S | H,S | S | H,S | S |
| After treatment | CR | CR | CR | No change | No change |
| Diagnosis date, BM | 05/31/01 | 06/12/01 | 2/19/03 | 06/09/98 | 10/04/02 |
| Prior treatment | None | HU, IFN-α | HU | IFN-α | HU |
| Follow-up, mos | 10+ | 19+ | 1+ | 9+ | 5+ |
| WHO classification | ISM | ASM | ASM | ASM | ASM |
| Imatinib mesylate dose: induction/maintenance, mg/d | 100, 100 | 400, 100 (biw) | 100, 100 | 100-400, NA | 100-400, NA |
| Hgb concentration, g/dL | |||||
| Before treatment | 12.1 | 9.2 | 13.9 | 10.1 | 10 |
| After treatment | 15.1 | 15.3 | 13.3 | 10.0 | 11.4 |
| WBC count, × 109/L | |||||
| Before treatment | 14.5 | 69.8 | 15.2 | 19.7 | 18 |
| After treatment | 4.9 | 6.1 | 3.1 | 21.7 | 17.5 |
| AEC count, × 109/L | |||||
| Before treatment | 10.7 | 16.1 | 10.2 | 3.5 | 6.1 |
| After treatment | 0.2 | 0.3 | 0.4 | 2.7 | 3.2 |
| Platelet count, × 109/L | |||||
| Before treatment | 118 | 31 | 213 | 210 | 166 |
| After treatment | 169 | 179 | 166 | 252 | 213 |
| Serum tryptase, less than 11.5 ng/mL | |||||
| Before treatment | 137 | ND | 56 | 904 | 331 |
| After treatment | 25.7 | ND | 19.8 | 1370 | 342 |
| c-kit Asp816Val mutation | A | A | A | P | P |
| FIP1L1-PDGFRA | P | P | P | A | A |
| CHIC2 deletion, % of nuclei positive | |||||
| Before treatment | 61.0 | 58.0 | 76.0 | 2.5 | 0.5 |
| After treatment | 0.0 | 1.5 | 0.5 | ND | ND |
| ETV6-PDGFRB | A | A | A | A | A |
| Cytogenetics | 46,XY | 46,XY | 46,XY | 46,XX | 46,XX |
| Response | Complete response | Complete response | Complete response | NR | NR |
| BM overall cellularity/mast cell % | |||||
| Before treatment | 90/30 | 95/15 | 90/20 | 90/70 | > 95/30 |
| After treatment | 35/CR | 45/CR | 60/CR | 90/70 | ND/ND |
S indicates splenomegaly; CR, complete remission; H, hepatomegaly; BM, bone marrow; HU, hydroxyurea; IFN-α, interferon-alpha; WHO, World Health Organization; ISM, indolent systemic mastocytosis; ASM, aggressive systemic mastocytosis; biw, twice per week; NA, not applicable; Hgb, hemoglobin; WBC, white blood cell; AEC, absolute eosinophil; ND, not done; A, absent; P, present; and NR, no response.
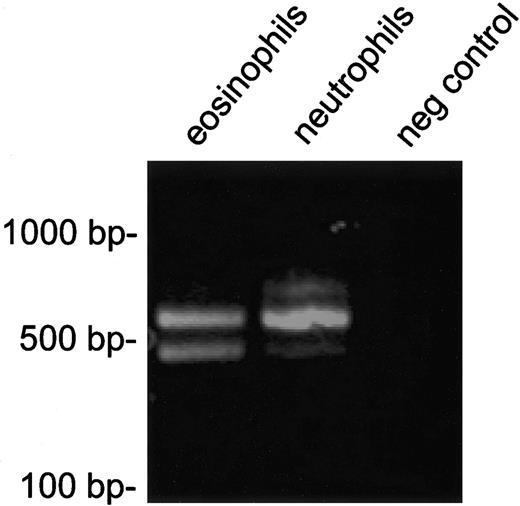
We developed a one-color interphase FISH strategy to identify the FIP1L1-PDGFRA rearrangement. Loss of a probe at the CHIC2 locus, which lies between the FIP1L1 and PDGFRA genes, has been previously demonstrated for 1 patient who carried the FIP1L1-PDGFRA rearrangement.10 We assayed for the loss of 1 of the 2 CHIC2 alleles using interphase FISH as a surrogate for the FIP1L1-PDGFRA fusion. Fixed bone marrow interphase cells (cases 1-5) and enriched eosinophils, neutrophils, and mononuclear cells from peripheral blood (case 3) were used for the FISH studies. The purity of separated cell fractions has been previously demonstrated to be at least 97%.20 As controls, samples from 10 bone marrow transplant donors were studied. Analysis of data from these donors revealed that 200 nuclei can be analyzed to confidently detect an abnormal percentage as low as 5% (normal cutoff established at 4.5%). Three patients (60%) carried the FIP1L1-PDGFRA rearrangement, as demonstrated by CHIC2 deletion by FISH (cases 1, 2, and 3 had 61.0%, 58.0%, and 72.0% percent abnormal nuclei, respectively), all of whom achieved a complete clinical and histologic remission with imatinib mesylate therapy (Table 1; Figure 2). In each instance, the peripheral eosinophil count normalized in less than a week and a follow-up bone marrow analysis revealed complete resolution of both mast cell and eosinophil infiltration (Figure 1). Posttreatment analysis of bone marrow cells for the 3 patients who had loss of the CHIC2 allele by FISH at baseline demonstrated a normal signal pattern (Table 1). The other 2 patients (cases 4 and 5) carried the c-kit Asp816Val mutation, but not the FIP1L1-PDGFRA rearrangement, and both were refractory to imatinib mesylate (Table 1). None of the remaining 7 patients with SMCD without associated eosinophilia carried the FIP1L1-PDGFRA rearrangement (data not shown). For 1 patient (case 3), the FIP1L1-PDGFRA fusion was detected in enriched eosinophil and neutrophil fractions from peripheral blood by both FISH and RT-PCR (Figure 3) and in mononuclear cells by FISH.
RT-PCR analysis of FIP1L1-PDGFRA expression in cell fractions enriched for eosinophils or neutrophils.FIP1L1-PDGFRA fusion transcripts were detected in both eosinophils and neutrophils. PCR products of different sizes are observed, due to alternative splicing in FIP1L1, as observed previously.10,23 Two alternatively spliced transcripts with the same intensity were detected in eosinophils, whereas 1 predominant splice variant is present in neutrophils.
RT-PCR analysis of FIP1L1-PDGFRA expression in cell fractions enriched for eosinophils or neutrophils.FIP1L1-PDGFRA fusion transcripts were detected in both eosinophils and neutrophils. PCR products of different sizes are observed, due to alternative splicing in FIP1L1, as observed previously.10,23 Two alternatively spliced transcripts with the same intensity were detected in eosinophils, whereas 1 predominant splice variant is present in neutrophils.
Discussion
The identification of the FIP1L1-PDGFRA fusion gene in SMCD-eos has important implications. First, it is a novel finding that reveals an overlap between HES and SMCD-eos, both in terms of their clinical presentation and molecular pathogenesis. These findings complement recent reports of HES patients harboring the FIP1L1-PDGFRA fusion that has elevated tryptase levels indicative of increased mast cells and suggests there may be clinical, morphologic, and genetic overlap between the syndromes SMCD-eos and HES.23 Second, FIP1L1-PDGFRA is the therapeutic target for imatinib mesylate in a subset of SMCD-eos cases. The rapid resolution of eosinophilia in these patients, at a dose lower than that required for the induction of hematologic and cytogenetic response in bcr-abl–positive chronic myeloid leukemia (CML; 1100 mg vs 400 mg/d; Table 1), is reminiscent of the dramatic clinical responses seen in HES patients who carry the FIP1L1-PDGFRA rearrangement.10 While the possibility that inhibition of wild-type c-kit may contribute to the observed responses cannot be entirely excluded, imatinib mesylate's efficacy at the lower dose in SMCD is consistent with the lower IC50 (50% inhibitory concentration) for FIP1L1-PDGFRA10 compared with c-kit24 (3.2 nM vs 100 nM). The posttreatment loss of the abnormal FIP1L1-PDGFRA signal in all responding patients further supports its central role in disease pathogenesis (Table 1). Third, identification of FIP1L1-PDGFRA fusion provides a rational basis for therapy of SMCD. Our data suggest that SMCD-eos patients that carry FIP1L1-PDGFRA can be predicted, before therapy, to be imatinib mesylate responsive. Conversely, consistent with previous in vitro data,25 patients who carry an enzymatic-site c-kit mutation (Asp816Val) can be predicted to be imatinib mesylate refractory. Demonstration of the FIP1L1-PDGFRA rearrangement in multiple cell lineages strongly suggests that this molecular lesion occurs in an early hematopoietic progenitor. Finally, a re-evaluation of the WHO classification for SMCD may be appropriate. SMCD patients who are FIP1L1-PDGFRA positive are not recognized in the current WHO classification.18 These patients constitute a unique subgroup on the basis of their clinical characteristics and response to imatinib mesylate therapy.
The issue of whether loss of the CHIC2 locus by FISH continues to be perfectly correlated with expression of the FIP1L1-PDGFRA fusion gene as well as imatinib mesylate responsiveness will require study of a larger cohort of patients.
Prepublished online as Blood First Edition Paper, July 3, 2003; DOI 10.1182/blood-2003-05-1627.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We have no conflict of interest to declare. We are grateful to Novartis Pharmaceuticals (Basel, Switzerland) for providing the drug free of charge for patients on this study. Novartis Pharmaceuticals had no role in study design; the collection, analysis, and interpretation of data; the writing of the report; or in the decision to submit the paper for publication.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal