Abstract
The degree of somatic mutation of immunoglobulin variable (Ig V) region genes is an important prognostic indicator of clinical course and outcome in B-cell chronic lymphocytic leukemia (B-CLL), although the reason for this association remains unclear. Furthermore, some B-CLL cells continue to acquire Ig V gene mutations after the transforming event. Because activation-induced cytidine deaminase (AID) is an essential component of the canonical somatic hypermutation process in healthy B cells, its expression in B-CLL is potentially relevant to the disease. We detected full-length AID transcripts and 3 splice variants by conventional reverse transcription polymerase chain reaction (RT-PCR) in approximately 40% of the cases examined. More sensitive real-time quantitative PCR detected AID transcripts in virtually all B-CLL samples tested, although the range of transcript levels was very large between different cases and varied within individual cases over time. Limiting dilution assays revealed that AID expression was restricted to a small fraction of the leukemic cells in the blood. However, this small fraction is not unique in its ability to express AID, because in vitro stimulation of B-CLL cells with appropriate stimuli significantly increased the fraction of AID-expressing cells. These data suggest that AID-mediated DNA alterations may occur in a variably sized, minor subset of B-CLL cells at any given time.
Introduction
B-cell chronic lymphocytic leukemia (B-CLL) results from an accumulation of monoclonal B lymphocytes that express CD19, CD5, CD23, and a reduced density of surface membrane immunoglobulin.1 The immunoglobulin variable (Ig V) region genes of B-CLL clones from approximately 50% of patients contain somatic mutations2 and the presence of Ig V gene mutations in the leukemic clone heralds a significantly better prognosis for the patient.3,4 Most B-CLL clones express IgM antibodies, but approximately 8% express switched isotypes,5 with IgG being the most prevalent switched class. In addition to somatic mutations and class switching at the clonal level, some individual leukemic B cells within the clones of certain patients continue these processes. Ongoing class switching in vivo and in vitro6-9 has been documented in the B-CLL cells of some patients, as has ongoing Ig VH gene diversification.10 It is not clear whether these ongoing processes are clinically significant, either by themselves or as reflections of other in vivo events.
Both class switch recombination (CSR) and somatic hypermutation (SHM) are dependent on the action of activation-induced cytidine deaminase (AID).11-14 The deamination activity of AID is an essential component of its function,15,16 and current data suggest that this acts on single-stranded DNA exposed by transcription.17-19 AID is both necessary and sufficient to initiate canonical SHM and CSR in mammalian cells20,21 with some requirement for protein synthesis.22 Not surprisingly, AID is preferentially expressed in secondary lymphoid organs, specifically in germinal center (GC) B cells13 and GC founder cells.9 AID is also induced in murine spleen B cells12 and human peripheral blood mononuclear cells (PBMCs)11 by in vitro stimulation with interleukin-4 (IL-4) and lipopolysaccharide (LPS) or CD40L.
Some B-CLL cells express AID.9,23-25 Patients whose B-CLL cells express AID can also evidence ongoing CSR by way of circle transcripts and switch circles as well as switched immunoglobulin proteins. The phenomenon can be mimicked in vitro in B-CLL cells in which AID is initially undetectable by stimulation with IL-4 and CD40L.9 This report confirms and extends those findings by demonstrating that in cases of B-CLL, AID is expressed as a native transcript and as several splice variant forms, that the detectable message is limited to a small population of leukemic cells, and that the size of this population varies from patient to patient and within a given patient over time.
Materials and methods
Patients and healthy donors
The Institutional Review Boards of North Shore University Hospital (Manhasset, NY) and Long Island Jewish Medical Center (New Hyde Park, NY) approved these studies. Informed consent was provided by all participants in accordance with the Declaration of Helsinki. The 99 patients in this study were diagnosed with typical B-CLL on the basis of clinical criteria and laboratory features. Patients were selected on the basis of the availability of DNA sequences for both the Ig VH and Ig VL genes in each case. As a source of healthy lymphocytes, leukocyte-enriched fractions from the blood of healthy volunteers matched for age were purchased from Long Island Blood Service (Melville, NY). These samples did not contain detectable titers of human immunodeficiency virus or hepatitis B virus antibodies.
Isolation of PBMCs and B cells
PBMCs were separated from heparinized venous blood of patients with B-CLL and from leukocyte fractions of healthy donors by density gradient centrifugation using Ficoll-Paque (Amersham Biosciences, Piscataway, NJ). PBMCs were cryopreserved with a programmable cell-freezing machine (Cryomed, Mt Clemens, MI) and thawed at the time of analysis. A negative selection isolation technique using a B-cell purification kit (Miltenyi Biotec, Auburn, CA) was used to enrich B lymphocytes from PBMCs. The purity of the collected fraction was analyzed by flow cytometry, and only fractions with more than 95% CD19+, CD5+ cells were used.
Reverse transcription-polymerase chain reaction
Total RNA was isolated from PBMCs and purified B cells from CLL samples and donors using Triazol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed in 20 μL by using the M-MLV RT and a poly(dT)12-18 primer (Invitrogen). Reactions were carried out at 42°C for 1 hour, heated at 65°C for 10 minutes, and then diluted to a final volume of 100 μL. Aliquots of 1 μL were used in 25 μL for each reverse transcription-polymerase chain reaction (RT-PCR). The complete coding region of AID message (GenBank no. AB04043126 ) was amplified with primers 63F (5′-CAGGGAGGCAAGAAGACAC-3′) and 1137R (5′-GCCTGAGGAAGAGTTTG-3′) for 33 cycles of denaturation for 30 seconds at 94°C, annealing for 45 seconds at 56°C, and extension for 90 seconds at 72°C. A nested PCR for this region was also used with the reaction described earlier followed by amplification of 1μL with primers 68F (5′-GAGGCAAGAAGACACTCTGG-3′) and 676R (5′-GTGACATTCCTGGAAGTTGC-3′) for 15 cycles (30 seconds at 94°C, 45 seconds at 56°C, 90 seconds at 72°C). Exons 1, 2, and 3 were amplified with primers 63F and 415R (5′-GCGGTCCTCACAGAAGTAG-3′) for 33 cycles (30 seconds at 94°C, 45 seconds at 56°C, 45 seconds at 72°C). The region between exon 3 and the end of the coding region in exon 5 was amplified using primers 392F (5′-GGAACCCCAACCTCAGTC-3′) and 676R for 33 cycles (30 seconds at 94°C, 60 seconds at 58°C, 90 seconds at 72°C). β-actin was always amplified simultaneously on the same samples for 25 cycles (15 seconds at 94°C, 30 seconds at 60°C, 45 seconds at 72°C) by using the primers β-F (5′-ATCTGGCACCACACCTTCTACAATGAG-3′) and β-R (5′-CGTCATACTCCTGCTTGCTGATCCAC-3′).
DNA sequencing
The multiple bands generated by the amplification with primers 63F and 1137R were run on a 2% agarose gel, excised, purified with the QIAEX II Gel Extraction kit (QIAGEN, Valencia, CA), and directly sequenced. The individual sequences were then aligned to the AID genomic DNA and/or mRNA sequence as appropriate (GenBank no. AB040430, AB04043126 ). Because all of the variants shared sequence between exon 1 and exon 3, the region between exon 3 and the coding end of exon 5 was amplified with primers 392F and 676R and cloned into a TA vector (Invitrogen). A total of 96 clones were screened and processed using Wizard minipreps (Promega, Madison, WI). The clones were a mixture of 5 products different in length. At least 5 clones for each variant were sequenced using M13 forward and reverse primers.
Real-time quantitative RT-PCR
We used the TaqMan 5′ nuclease assay in these studies. Total RNA from purified B-CLL cells was isolated by Rneasy Mini Kit and digested with DNase I (Qiagen). RNA (1 μg) was reverse transcribed in 20 μL by using Superscript II RT and a poly(dT)12-18 primer (Invitrogen). After adding 80 μL water and mixing, 5-μL aliquots were used for each TaqMan reaction. TaqMan primers and probes were designed using Primer Express software version 1.5 (Applied Biosystems, Foster City, CA). The probe was labeled with FAM at the 5′ end and TAMRA at the 3′ end. Real-time PCR for AID and glyceraldehyde phosphate dehydrogenase (GAPDH) was performed using the TaqMan PCR core reagent kit (Applied Biosystems) and ABI Prism 7700 Sequence Detection System (PE-Applied Biosystems). The PCR reaction mixture contained 3.5 mM MgCl2; 0.2 mM each of deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), and deoxyguanosine triphosphate (dGTP); 0.4 mM deoxyuridine triphosphate (dUTP); 0.5 μM forward primer; 0.5 μM reverse primer; 0.1 μM TaqMan probe; 0.25 U uracil DNA glycosylase; and 0.625 U AmpliTaq Gold polymerase in 1 × TaqMan PCR buffer. cDNA (5 μL) was added to the PCR mix, and the final volume of PCR reaction was 25 μL. All samples were run in duplicates. GAPDH was used as endogenous control. The thermal cycler conditions were 50°C for 2 minutes, 95°C for 10 minutes, and 45 cycles of 95°C for 0.30 minute and 60°C for 1 minute. Data were analyzed using Sequence Detection System (SDS) software version 1.6.3. Results were obtained as CT (threshold cycle) values. The software determines a threshold line on the basis of the baseline fluorescent signal, and the data point that meets the threshold is given as threshold cycle (CT). CT is inversely proportional to the starting template copy number. The difference in CT values between the AID and GAPDH reactions (ΔCT) was converted into AID expression units by setting GAPDH equal to 10 000 and dividing by 2ΔCT. All measurements were performed in duplicate. TaqMan Sequences include the following: GAPDH, forward primer (GAAGGTGAAGGTCGGAGTC) and reverse primer (GAAGATGGTGATGGGATTTC); TaqMan probe, (FAM-caagcttcccgttctcagcc-TAMRA); AID, forward primer (cctcctaatgagagtatctgggtgat) and reverse primer (ttaaaacatacagcgcatgattgg); and TaqMan probe (FAM-tgaccccaaaccatctctccaaagca-TAMRA).
Limiting dilution assay
Purified B-CLL cells were distributed into multiple aliquots at each of several dilutions, ranging from 50 cells to 6400 cells per aliquot. The cells were directly deposited in 96 PCR plates, each well of which contained the lysis solution (1 μL5 × first-strand RT buffer, 0.5 μL 0.01M dithiothreitol [DTT], 10% Nonidet-P40, 0.4 μL 10 mM mix of deoxynucleotide triphosphates (dNTPs), 0.05 μg oligo d(T)12-18), RNase Guard (Amersham Biosciences), and Prime RNase Inhibitor (Eppendorf, Hamburg, Germany). cDNA was prepared in the same plate by adding the Superscript II Reverse Transcriptase (Invitrogen). cDNA (3 μL) was amplified in 25 μL for 2 rounds of PCR for either AID or the VH gene of the particular B-CLL clone. The first AID PCR was performed using the primer pair 63F and 415R for 35 cycles at the same conditions listed earlier. A volume of 1 μL of the first PCR was used for the seminested PCR, in which the primer 415R was coupled with the new primer 127F (5′-TGTCCGCTGGGCTAAGG-3′); the PCR conditions were the same as the previous PCR. The first VH PCR was performed using the VH family-specific framework region 1 forward primer in conjunction with the appropriate reverse constant region (CH) primer.2 A volume of 1 μL of the first PCR was used for a seminested PCR, in which a more internal CH primer (hMi7 ) was used; the PCR conditions were previously described. PCR products were run on 2% agarose gels stained with ethidium bromide. A band at approximately 310 base pair (bp) indicated a positive result. The fraction of AID PCR-positive (AID+) aliquots at each dilution was plotted on a log scale versus the total number of cells assayed at each dilution. The interpolated dilution at which 67% of the tested aliquots are positive is roughly equal to the reciprocal frequency of positive cells in the sample.27 If there were insufficient numbers of positive wells to properly interpolate, or no positive wells, the upper 95% confidence bound of the Poisson distribution was divided by the total number of cells (Σ number of wells × cells/well) assayed to establish an estimated upper threshold of the positive fraction.
In vitro B-CLL cell activation
Purified B cells were suspended as 1 × 106/mL in RPMI-1640 (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS) and antibiotics. B cells (4 million) were incubated with phorbol 12-myristate 13-acetate (PMA) and ionomycin (both from Sigma, St Louis, MO); both reagents were used at a final concentration of 10 ng/mL. In parallel, 4 × 106 cells were also incubated with CD32-transfected murine L cells (purchased from the ATCC, Manassas, VA) and anti-CD40 (200 ng/mL, monoclonal antibody [mAb] 89; Beckman Coulter, Miami, FL) and IL-4 (10 ng/mL; Sigma). Cells were harvested approximately 84 hours after culture set-up (day 3.5) and washed twice with cold phosphate-buffered saline (PBS). The activation status of the cells was confirmed by phenotypic analysis. Cells were suspended in PBS containing 1% bovine serum albumin and 0.1% sodium azide and subjected to immunofluorescent analyses for surface membrane expression of CD5, CD19, and either CD23, CD25, CD38, CD69, CD40, CD71, CD79b, CD95, or HLA-DR by standard methods.
Results
Detection of AID expression by RT-PCR
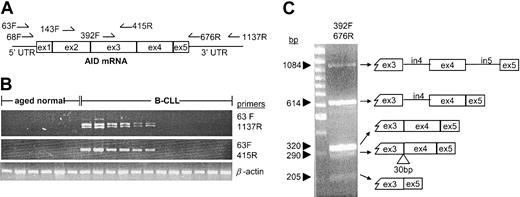
AID gene expression was initially detected by RT-PCR using primers that amplified various regions of the expressed sequence (Figure 1A). When a primer pair that amplified the entire coding region was used (63F and 1137R), multiple bands were detected in some B-CLL samples (Figure 1B, upper panel). However, a primer pair that amplified only the first 3 exons (63F and 415R) produced a single band from the same samples (Figure 1B, middle panel), suggesting that variants existed that shared the sequence between exons 1 and 3. This PCR was performed on cDNA made from mononuclear cells (MNCs) from the blood of 99 patients with B-CLL and 20 healthy individuals, as well as from human tonsil; 2 RAMOS subclones known to have varying levels of AID activity were analyzed as controls.28 As expected, AID was readily detected in the RAMOS cell line and in tonsil MNCs (not shown), but it was never detected in healthy controls (Figure 1B, middle panel). In contrast, transcripts for AID were observed in 39 of the 99 patients with B-CLL studied (∼40%). When the B-CLL samples were classified by Ig V gene mutation status using the standard 2% mutation threshold,2 AID message was detected in 60% (26 of 43) of unmutated cases and 23% (13 of 56) of mutated cases. AID expression also was tested in purified B cells from 19 patients with B-CLL and 20 healthy individuals matched for age. Seven of these B-CLL samples yielded positive results (37%), whereas all healthy B-cell samples were negative (Figure 1B).
Some B-CLL samples express the AID full-length transcript and its splice variants. (A) Map of the PCR primers used showing their relative position on the AID mRNA sequence (GenBank no. AB040431). Exons are indicated by boxes and noncoding regions by lines. (B) Representative RT-PCR results from purified B cells from healthy individuals of similar age and a series of B-CLL cases. Each vertical lane represents one patient or subject. Full coding sequence using primer pair 63F and 1137R (upper panel). First 3 exons using primer pair 63F and 415R (middle panel). β-actin (lower panel). (C) The region between exon 3 and exon 5 contains multiple variants. PCR products from CLL332 using the primer pair 392F and 676R were run on a 2% agarose gel (left). The total PCR product was cloned, and individual clones representing each band were sequenced using M13 primers. The sequences are diagrammed at right. From the bottom: variant missing exon 4 (205 bp), variant lacking the first 30 bp of exon 4 (290 bp), full-length transcript (320 bp), variant retaining the intron 4 sequence (614 bp), variant containing intron 4 and intron 5 (1084 bp). The last variant is identical to the genomic sequence of AID and most likely arose from genomic DNA contaminating the RNA used for the RT-PCR.
Some B-CLL samples express the AID full-length transcript and its splice variants. (A) Map of the PCR primers used showing their relative position on the AID mRNA sequence (GenBank no. AB040431). Exons are indicated by boxes and noncoding regions by lines. (B) Representative RT-PCR results from purified B cells from healthy individuals of similar age and a series of B-CLL cases. Each vertical lane represents one patient or subject. Full coding sequence using primer pair 63F and 1137R (upper panel). First 3 exons using primer pair 63F and 415R (middle panel). β-actin (lower panel). (C) The region between exon 3 and exon 5 contains multiple variants. PCR products from CLL332 using the primer pair 392F and 676R were run on a 2% agarose gel (left). The total PCR product was cloned, and individual clones representing each band were sequenced using M13 primers. The sequences are diagrammed at right. From the bottom: variant missing exon 4 (205 bp), variant lacking the first 30 bp of exon 4 (290 bp), full-length transcript (320 bp), variant retaining the intron 4 sequence (614 bp), variant containing intron 4 and intron 5 (1084 bp). The last variant is identical to the genomic sequence of AID and most likely arose from genomic DNA contaminating the RNA used for the RT-PCR.
The multiple bands observed with the PCR that amplified the entire coding region were then analyzed in more detail. PCR performed with primers that amplified the region between exon 3 and the 3′ end of exon 5 (392F and 676R; Figure 1A) resolved 5 bands (Figure 1C). The total PCR product was cloned, and individual clones representing each band were sequenced. All sequences derived from the canonical AID sequence are diagrammed in Figure 1C. The shortest variant was missing the sequence of exon 4, and an intermediate variant was lacking the first 30 bp of exon 4. A longer variant retained the intron 4 sequence. The variant that contained intron 4 and intron 5 may have resulted from DNA contamination of the RNA used for the RT-PCR because it matches the genomic sequence, although it cannot be excluded as a novel mRNA variant. The unequivocable mRNA variants were observed in B lymphocytes from allergic individuals29 and patients with B-CLL.24,30
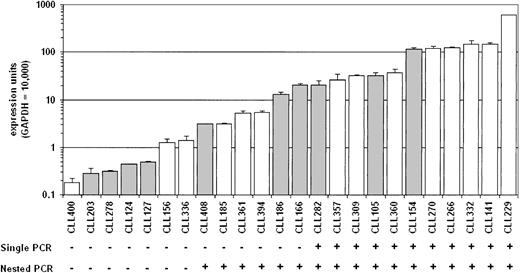
The RT-PCR products from the analyses mentioned earlier displayed a broad range of intensities, suggesting quantitative differences in AID expression between patients. Therefore, real-time quantitative PCR was performed on cDNA from purified B-CLL cells. AID message was detectable in all B-CLL samples tested, even those that had been negative by standard RT-PCR or more sensitive nested RT-PCR (Figure 2). The range of expression among the B-CLL samples tested was considerable, with a 1000-fold range from the highest value to the lowest. There was a continuous distribution of expression levels. Interestingly, there was no significant association between AID expression level and Ig V gene mutation status when these quantitative values were considered (P > .01).
Quantitation of AID message in B-CLL cells. Quantitative real-time PCR was performed on cDNA from purified B-CLL cells, and the relative expression units are plotted on a log scale. The open bars represent the B-CLL cases with less than 2% nucleotide difference from the corresponding germ line Ig V region gene, and the filled bars are B-CLL cases with 2% or more differences. The results of either single or nested AID-specific RT-PCR for the same samples are shown below the graph. The single PCR used the primers 63F and 1137R for 33 cycles. Nested PCR was performed on the single PCR products using the primers 68F and 676R for 15 cycles.
Quantitation of AID message in B-CLL cells. Quantitative real-time PCR was performed on cDNA from purified B-CLL cells, and the relative expression units are plotted on a log scale. The open bars represent the B-CLL cases with less than 2% nucleotide difference from the corresponding germ line Ig V region gene, and the filled bars are B-CLL cases with 2% or more differences. The results of either single or nested AID-specific RT-PCR for the same samples are shown below the graph. The single PCR used the primers 63F and 1137R for 33 cycles. Nested PCR was performed on the single PCR products using the primers 68F and 676R for 15 cycles.
Expression of AID in B-CLL cells is variable
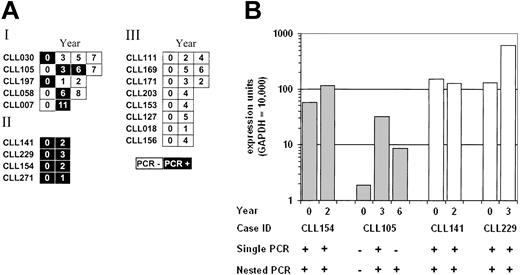
Because the results described in “Detection of AID expression by RT-PCR” indicate that RT-PCR can distinguish substantial variations in AID expression levels, 17 cases were analyzed at time points separated by 1 year or more (Figure 3A). Four of these cases were positive at all time points tested (II), 8 were consistently negative (III), and 5 changed over time (I). Of those cases that changed, 2 were initially positive, 1 was initially negative, and 2 were positive at intermediate time points only. Quantitative PCR was performed on 4 of the 17 samples, and the levels mirrored the RT-PCR results (Figure 3B). In particular, quantitative PCR expression values from patient CLL105 fluctuated across the detection limit of both the single and nested RT-PCR assays, illustrating the subjective and arbitrary nature of PCR positivity as a correlative parameter.
AID expression can change over time in some B-CLL cases. (A) Schematic representation of the AID RT-PCR results on 17 B-CLL cases at different time points. AID amplification was performed by using the primers 63F and 415R for 33 cycles. The open boxes represent PCRs without a detectable product; the filled boxes are a detectable product. The numbers in the boxes correspond to the number of years counted from the first tested sample date. (B) Real-time quantitative PCR results on B-CLL cases at different time points. The labeling of the graph and PCR conditions are as for Figure 2.
AID expression can change over time in some B-CLL cases. (A) Schematic representation of the AID RT-PCR results on 17 B-CLL cases at different time points. AID amplification was performed by using the primers 63F and 415R for 33 cycles. The open boxes represent PCRs without a detectable product; the filled boxes are a detectable product. The numbers in the boxes correspond to the number of years counted from the first tested sample date. (B) Real-time quantitative PCR results on B-CLL cases at different time points. The labeling of the graph and PCR conditions are as for Figure 2.
A subset of B-CLL cells express AID
PCR-based measurements of gene expression on populations of cells represent an averaged total of the expression in the individual cells. As such, these methods do not distinguish between homogeneous expression of the gene of interest by the entire population versus selective, strong expression of the gene by a minority of the population. Single cell-based PCR methods could resolve this issue conveniently if the fraction of expressing cells is a few percent or more; however, detection of very small numbers of expressing cells is more difficult and laborious because of the need to analyze a large number of individual cells. Alternatively, limiting dilution assays can identify positive fractions well below the convenient limit of single cell PCR, while avoiding the technical difficulties associated with single cell deposition.
Therefore, purified B-CLL cells were diluted to concentrations ranging from 50 to 6400 cells per well. A minimum of 32 wells was tested at each dilution. Nested PCR for AID and the specific VH gene used by each B-CLL case were independently performed on each well, and the ratio of AID+ to VH+ wells was determined. Because each well containing B-CLL cells yielded a PCR product for the VH gene, this served both as an experimental control for sample distribution and PCR efficiency. A representative experiment is shown in Figure 4A with analysis shown in Figure 4B; the results with purified B cells from 9 B-CLL cases and from 2 elderly healthy individuals are summarized in Tables 1 and 2. The highest fraction observed was 0.2% for patient CLL332, whereas 3 other cases had positive fractions between 0.01% and 0.1%. Four of the 5 remaining cases had some positive wells, but not enough to accurately calculate a positive fraction. In these cases, the likelihood that the positive wells contained more than one AID+ cell was minimal, and the Poisson distribution statistics applied to assign an upper threshold. No positive wells were observed with peripheral blood B cells from 2 healthy elderly individuals, even at 6400 cells per well. The fraction of AID+ cells correlates with the expression levels measured by quantitative PCR, suggesting that the AID message level per cell is similar across samples, and the differences seen in the quantitative measurements are thus a reflection of the size of the AID+ fraction. However, there may still be variation in the amount of AID message within the positive cells of a given patient. Surprisingly, even in samples with relatively high AID expression levels, the fraction of cells expressing AID is small (< 1%), indicating that the positive cells most likely express considerable levels of AID message.
AID expression is limited to a fraction of the B-CLL clone. (A) A representative (CLL 266) limiting dilution experiment is shown. Each gel lane contains the PCR product derived from a well containing the indicated number of cells. The PCR used primers specific for (left panel) the VH gene of the clone or (right panel) AID. Thirty-two wells were tested at each dilution. The last lane at the highest cell density (*) did not contain reverse transcriptase. The anticipated size of the VH band was approximately 360 bp and approximately 310 bp for AID. (B) Representative analysis of the data generated by the limiting dilution experiments. The fraction of AID+ aliquots at each dilution was plotted on a log scale versus the total number of cells assayed at each dilution. The interpolated dilution at which 67% of the tested aliquots are positive is approximately equal to the reciprocal frequency of positive cells in the sample.
AID expression is limited to a fraction of the B-CLL clone. (A) A representative (CLL 266) limiting dilution experiment is shown. Each gel lane contains the PCR product derived from a well containing the indicated number of cells. The PCR used primers specific for (left panel) the VH gene of the clone or (right panel) AID. Thirty-two wells were tested at each dilution. The last lane at the highest cell density (*) did not contain reverse transcriptase. The anticipated size of the VH band was approximately 360 bp and approximately 310 bp for AID. (B) Representative analysis of the data generated by the limiting dilution experiments. The fraction of AID+ aliquots at each dilution was plotted on a log scale versus the total number of cells assayed at each dilution. The interpolated dilution at which 67% of the tested aliquots are positive is approximately equal to the reciprocal frequency of positive cells in the sample.
Quantitation of AID+ fraction of high AID-expressing samples
. | Cells/well . | . | . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | 50 . | 100 . | 200 . | 400 . | 1600 . | AID+ fraction, % . | QPCR value* . | ||||
| CLL332 | 5/95 | 15/94 | 36/94 | 51/93 | — | 0.210 | 147 | ||||
| CLL105 | — | 0/32 | — | 3/32 | 0.012 | 32 | |||||
| CLL266 | — | 3/32 | — | 10/32 | 0.062 | 125 | |||||
| CLL357 | — | 1/30 | — | 4/32 | 0.024 | 26 | |||||
. | Cells/well . | . | . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | 50 . | 100 . | 200 . | 400 . | 1600 . | AID+ fraction, % . | QPCR value* . | ||||
| CLL332 | 5/95 | 15/94 | 36/94 | 51/93 | — | 0.210 | 147 | ||||
| CLL105 | — | 0/32 | — | 3/32 | 0.012 | 32 | |||||
| CLL266 | — | 3/32 | — | 10/32 | 0.062 | 125 | |||||
| CLL357 | — | 1/30 | — | 4/32 | 0.024 | 26 | |||||
Summary of the results of the limiting dilution experiments performed on 8 CLL cases and 2 elderly healthy individuals (Figure 4A-B). The results are shown as the ratio of the AID+ wells to VH-positive wells at indicated dilutions for each sample. In the last 2 columns on the right, the AID+ fractions are expressed as a percentage, and the real-time quantitative PCR values are reported for each sample, respectively. — indicates that no experiment was performed at those conditions.
Expression units, as per Figure 2.
Quantitation of AID+ fraction of low AID-expressing samples
. | Cells/well . | . | . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | 400 . | 800 . | 1600 . | 3200 . | 6400 . | AID+ fraction, % . | QPCR value* . | ||||
| CLL203 | 1/93 | — | 2/92 | — | — | <0.0048 | 0.28 | ||||
| CLL127 | — | 0/32 | 1/32 | 0/32 | — | <0.0031 | 0.50 | ||||
| CLL400 | — | 0/28 | 0/29 | 0/25 | — | <0.0024 | 0.18 | ||||
| CLL361 | — | 1/32 | 0/32 | 1/31 | — | <0.0058 | 5.20 | ||||
| CLL156 | — | 0/32 | 0/32 | — | 1/28 | <0.0022 | 1.27 | ||||
| Normal A | 0/31 | — | 0/31 | — | 0/31 | <0.0014 | — | ||||
| Normal B | 0/32 | — | 0/32 | — | 0/30 | <0.0008 | — | ||||
. | Cells/well . | . | . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | 400 . | 800 . | 1600 . | 3200 . | 6400 . | AID+ fraction, % . | QPCR value* . | ||||
| CLL203 | 1/93 | — | 2/92 | — | — | <0.0048 | 0.28 | ||||
| CLL127 | — | 0/32 | 1/32 | 0/32 | — | <0.0031 | 0.50 | ||||
| CLL400 | — | 0/28 | 0/29 | 0/25 | — | <0.0024 | 0.18 | ||||
| CLL361 | — | 1/32 | 0/32 | 1/31 | — | <0.0058 | 5.20 | ||||
| CLL156 | — | 0/32 | 0/32 | — | 1/28 | <0.0022 | 1.27 | ||||
| Normal A | 0/31 | — | 0/31 | — | 0/31 | <0.0014 | — | ||||
| Normal B | 0/32 | — | 0/32 | — | 0/30 | <0.0008 | — | ||||
Summary of the results of the limiting dilution experiments performed on 8 CLL cases and 2 elderly healthy individuals (Figure 4A-B). The results are shown as the ratio of the AID+ wells to VH-positive wells at indicated dilutions for each sample. In the last 2 columns on the right, the AID+ fractions are expressed as a percentage, and the real time quantitative PCR values are reported for each sample, respectively. If there were insufficient numbers of positive cells to properly interpolate as in Figure 4B, a Poisson distribution was used to establish the upper bound of the 95% confidence interval. — indicates that no experiment was performed at those conditions.
Expression units, as per Figure 2.
B-CLL cells up-regulate AID on stimulation in vitro
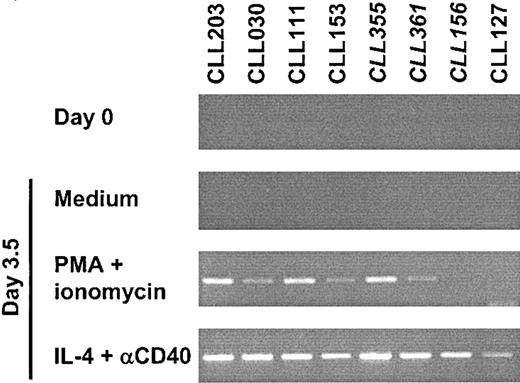
The preceding data raised the possibility that the small fraction of AID+ cells is unique in its ability to express AID. To test this hypothesis, in vitro stimulation assays were performed on 8 B-CLL samples that had very small fractions of AID-expressing cells as evidenced by a negative RT-PCR. Purified B-CLL cells were cultured for 3.5 days in the presence of either PMA and ionomycin or CD32-transfected L cells with anti-CD40 mAb and IL-4. At the end of the culture period, AID expression was readily detected in all the samples that were stimulated by the latter culture conditions, whereas AID message was detected in 6 of the 8 samples stimulated by the former culture conditions (Figure 5). The efficiency of cellular activation was corroborated for each sample by changes in surface membrane expression of activation markers (HLA-DR, CD69, CD62L, CD71, and CD79b) measured by direct immunofluorescence (data not shown). Cells cultured in medium alone (Figure 5) or in the presence of only CD32-transfected L cells (data not shown) remained RT-PCR-negative throughout the culture period. Real-time PCR was performed for 3 of these experiments and confirmed substantial, although varied, increases in AID message levels in response to the stimuli. The increased AID expression levels suggested that the fraction of cells expressing AID had increased markedly, and this increase was confirmed by limiting dilution analysis (Table 3). Culturing with PMA plus ionomycin caused fewer cells to become AID+ than did culturing with CD32-transfected L cells with anti-CD40 mAb and IL-4, consistent with the qualitatively weaker PCR results (Figure 5). The results indicate that the AID+ fraction detected prior to culture is not a uniquely competent subclone, but is instead a small subset of the B-CLL cells that are capable of expressing AID when appropriately stimulated.
B-CLL can up-regulate AID on appropriate stimulation in vitro. RT-PCR results from purified B cells from 8 B-CLL samples, 5 mutated and 3 unmutated (italicized). RNA was extracted from purified B-CLL cases before (day 0, first panel from the top) and after 3.5 days of stimulation with either PMA and ionomycin (third panel) or CD32-transfected L cells with anti-CD40 mAb and IL-4 (fourth panel). Cells cultured in medium alone (second panel) or with only CD32-transfected L cells (not shown) remained RT-PCR negative throughout the culture period. AID amplification was performed using the primers 63F and 415R for 33 cycles. β-actin was positive and qualitatively similar in intensity for all tested samples (data not shown).
B-CLL can up-regulate AID on appropriate stimulation in vitro. RT-PCR results from purified B cells from 8 B-CLL samples, 5 mutated and 3 unmutated (italicized). RNA was extracted from purified B-CLL cases before (day 0, first panel from the top) and after 3.5 days of stimulation with either PMA and ionomycin (third panel) or CD32-transfected L cells with anti-CD40 mAb and IL-4 (fourth panel). Cells cultured in medium alone (second panel) or with only CD32-transfected L cells (not shown) remained RT-PCR negative throughout the culture period. AID amplification was performed using the primers 63F and 415R for 33 cycles. β-actin was positive and qualitatively similar in intensity for all tested samples (data not shown).
AID+ fraction and expression following in vitro stimulation
. | Cells/well . | . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | 50 . | 100 . | 400 . | 800 . | 1600 . | 3200 . | QPCR value* . | |||||
| CLL203 | ||||||||||||
| Before | — | — | 1/93 | — | — | 0.28 | ||||||
| PMA + ionomycin | 3/95 | 7/95 | — | — | — | — | 4.78 | |||||
| αCD40 + IL-4 | 93/93 | 90/95 | — | — | — | — | 2398 | |||||
| CLL361 | ||||||||||||
| Before | — | — | — | 1/32 | 0/32 | 1/31 | 5.2 | |||||
| PMA + ionomycin | — | 0/32 | 1/32 | — | 1/30 | — | 11.8 | |||||
| αCD40 + IL-4 | — | 10/32 | 23/32 | — | 30/31 | — | 235 | |||||
| CLL127 | ||||||||||||
| Before | — | — | — | 0/32 | 1/32 | 0/31 | 0.5 | |||||
| PMA + ionomycin | — | 0/32 | 0/31 | — | 1/28 | — | 7.7 | |||||
| αCD40 + IL-4 | — | 2/32 | 2/32 | — | 5/31 | — | 116.8 | |||||
. | Cells/well . | . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample . | 50 . | 100 . | 400 . | 800 . | 1600 . | 3200 . | QPCR value* . | |||||
| CLL203 | ||||||||||||
| Before | — | — | 1/93 | — | — | 0.28 | ||||||
| PMA + ionomycin | 3/95 | 7/95 | — | — | — | — | 4.78 | |||||
| αCD40 + IL-4 | 93/93 | 90/95 | — | — | — | — | 2398 | |||||
| CLL361 | ||||||||||||
| Before | — | — | — | 1/32 | 0/32 | 1/31 | 5.2 | |||||
| PMA + ionomycin | — | 0/32 | 1/32 | — | 1/30 | — | 11.8 | |||||
| αCD40 + IL-4 | — | 10/32 | 23/32 | — | 30/31 | — | 235 | |||||
| CLL127 | ||||||||||||
| Before | — | — | — | 0/32 | 1/32 | 0/31 | 0.5 | |||||
| PMA + ionomycin | — | 0/32 | 0/31 | — | 1/28 | — | 7.7 | |||||
| αCD40 + IL-4 | — | 2/32 | 2/32 | — | 5/31 | — | 116.8 | |||||
Summary of the results of the limiting dilution experiments performed on 3 CLL cases before and after 3.5 days of culture in the presence of either PMA and ionomycin or CD32-transfected L cells with anti-CD40 mAb and IL-4. The results are shown as a ratio of the AID+ wells to VH+ wells at indicated dilutions for each sample. In the last column, the real-time quantitative PCR values are reported. — indicates that no experiment was performed at those conditions.
Expression units, as per Figure 2.
Discussion
Our study demonstrates that AID is expressed in the leukemic cells of most, if not all, patients with B-CLL. The surprising result of this study is that AID is expressed in a very small fraction of the B-CLL clone and that this fraction can vary from patient to patient and from time to time. Nevertheless, it is clear that many of the cells that are AID-negative are competent to express this mRNA when appropriately stimulated in vitro.
Previous reports suggested that AID is preferentially expressed in B-CLL cases with unmutated Ig V genes.23-25 The results presented here do not support this contention, although they suggest an explanation for the observation. When PCR assays are used, the sensitivity of the PCR establishes an arbitrary limit of detection that can vary from protocol to protocol. Indeed our data, generated by standard PCR, suggested enriched AID expression in B-CLL cases with unmutated Ig VH genes (Figure 2 and data not shown). However, more precise real-time quantitative PCR analyses that transcend arbitrary expression thresholds reveal that AID expression is not black or white across patients with B-CLL, but instead covers a wide range of total expression levels regardless of VH genotype. When the actual expression values for AID in mutated and unmutated cases are compared on a log scale, there is no significant difference between them. Furthermore, it is tenuous to correlate fixed parameters, such as Ig V gene mutation status, with markers or parameters that can fluctuate significantly unless such fluctuation can be controlled for or averaged. Most important, population-based assays such as PCR can miss or confound the contribution of small minority populations, which is critical for AID expression in B-CLL (Figure 4, Tables 1, 2).
Why does only a small subset of B-CLL cells express AID? Either the cells that express AID are genetically distinct, AID-competent subclones or they represent inducible cells that recently encountered a putative AID-inducing stimulus. The former suggestion is undermined by the results of the stimulation studies, indicating that many more cells can be induced to become AID expression-positive in vitro than are detected in vivo (Figure 5, Table 3). However, even with inducing stimuli, not all leukemic cells expressed AID, leaving open the possibility that a subset of the B-CLL cells are genetically advantaged in their ability to express AID.
Why then does only a subset of the AID-competent cells actually express AID in vivo? One explanation is that there are specific signals and/or microenvironments that can induce AID and that access to these elements is limited. In vitro, B-CLL cells rapidly down-regulate putative products of AID activity, mainly circle transcripts and switch circles.9 If the same is true in vivo, then the AID+ cells detected in the blood would have recently experienced an AID-inducing stimulus. An intriguing possibility is that this stimulus occurs in a specific microenvironment. It has been suggested that B-CLL cells are sustained in vivo by intermittent and repetitive engagement of their B-cell antigen receptor (BCR) by autoantigens31 to which B-CLL cells are known to react.32-34 In addition, stromal elements found in select microenvironments35,36 may maintain viability by aborting apoptosis through interactions with cell surface or secreted molecules.37 These 2 stimuli, engagement of the BCR with consequent signal transduction and stromal cell interaction, may be coupled to the proliferation centers observed in the solid lymphoid tissues of patients with B-CLL.38 As such, AID-expressing cells might represent the most recent emigrants from solid tissue compartments. If true, then the functional characteristics of AID+ cells would give insight into the nature of events occurring in these proliferation areas and offer a view of the events normally obscured in the bulk population of cells in the peripheral blood. Limiting dilution assays of AID expression like those in Table 1, using B-CLL cells derived from bone marrow, lymph node, or spleen, may be informative in this regard.
It is unclear whether AID mRNA expression documented here translates into functional AID protein or to what extent it leads to DNA alterations in the cells in which it is expressed. Indeed, conventional analyses of protein levels and function may be challenging in light of the selective expression of AID message in such a small subset of the leukemic clone. Furthermore, the presence of multiple splice variants whose roles have not been defined makes predictions of in vivo functionality difficult. However, the association between significant AID expression and ongoing CSR9,30 suggests that AID is functionally active in some B-CLL cells.
It has been demonstrated that the SHM machinery can target other loci besides the Ig genes in healthy B cells during the GC reaction39-41 with implications for lymphoid malignancies.42 Because AID is necessary and sufficient for induction of somatic mutations, its expression in a subpopulation of B-CLL cells suggests a mechanism by which ongoing mutation of both the Ig V and other loci could occur. It is difficult to definitively associate AID expression with the presence of ongoing novel Ig V or other gene mutations, because the time of their occurrence in relation to the time of AID detection is not readily determinable. Nevertheless, it is tempting to link the 2. Furthermore, ongoing Ig V gene mutations10 and CSR8 have been noted in both mutated and unmutated B-CLL cases, consistent with our observations that both subgroups of B-CLL can express AID.
B-CLL is not characterized by a consistent and distinct cytogenetic abnormality, and translocations are rare even among those leukemic cells that do acquire chromosomal aberrations.43 This suggests that accumulated point or other small mutations or deletions might play a role in the transition from quiescent to aggressive disease. Indeed, transgenic mice that constitutively express AID rapidly acquire T-cell lymphomas without translocations.44 Deletions appear to be especially relevant in B-CLL,45 and AID plays a role in DNA deletions associated with CSR as well as point mutations involved in the SHM process. The pool of AID-expressing B-CLL cells may be the wellspring of increasingly dangerous clonal variants. This pool is fractionally small, with 0.2% being the largest observed here. However, even if the positive fraction is as low as 0.001%, at any given time there would still be several million individual AID-expressing cells in a typical patient with B-CLL, because patients usually have leukemic tumor burdens of 10 000 to 100 000 cells/mm3 in the blood. Furthermore, it is not clear whether the cells that express AID at a given time are the same and only cells that express it at earlier or later time points. This appears unlikely if AID can be up-regulated in most B-CLL cells, as our in vitro stimulation data suggest (Figure 5, Table 3), and if there is any significant cell turnover within the clone, as our preliminary in vivo measurements of B-CLL cell kinetics suggest.46 Instead, it seems more likely that there is a temporally moving “window” of AID expression among members of the clone, and that the size of that window may change as the clone evolves. This situation would expose a broader swath of the clone to the potential consequences of AID-mediated actions, expanding the opportunities for evolution of clonal variants. If AID expression frequencies are higher in the bone marrow or other solid lymphoid tissues, the window of AID-expressing cells could be even larger. In addition, there could be a selection for subclones that have enhanced capacity to express AID, either because of direct changes in the AID expression pathway or indirectly through modulations of the encounters with AID-inducing stimuli. This could bootstrap a system for increasing levels of genetic diversity by selecting for ever larger windows of AID expression concurrent with any adventitious genetic changes.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-05-1585.
Supported in part by grants from the National Institutes of Health, National Cancer Institute (CA 81554 and CA 87956), the Jean Walton Fund for Lymphoma and Myeloma Research, the Joseph Eletto Leukemia Research Fund, The S.L.E. Foundation, Inc, and the Laurie Strauss Leukemia Research Foundation.
E.A. and B.T.M. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ”advertisement” in accordance with 18 U.S.C. section 1734.
We thank Taraneh Banapour, Alamelu Chandrasekaren, and Craig Gawel for technical assistance, as well as past members of the Laboratory of Experimental Immunology for Ig V gene sequence data on the B-CLL cases described here. We thank Davorka Messmer and Charles Chu for helpful discussions.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal