Abstract
Interleukin 6 (IL-6) is a growth and survival factor for multiple myeloma cells. As we report here, the IL-6–dependent human myeloma cell line INA-6 responds with a remarkably rapid and complete apoptosis to cytokine withdrawal. Among the antiapoptotic members of the B-cell lymphoma-2 (Bcl-2) family of apoptosis regulators, only myeloid cell factor-1 (Mcl-1) was slightly induced by IL-6. Overexpression studies demonstrated, however, that IL-6 does not exert its survival effect primarily through this pathway. The IL-6 signal transduction pathways required for survival and the target genes controlled by them were analyzed by using mutated receptor chimeras. The activation of signal transducer and activator of transcription 3 (Stat3) turned out to be obligatory for the survival of INA-6 cells. The same held true for survival and growth of XG-1 myeloma cells. Gene expression profiling of INA-6 cells by using oligonucleotide microarrays revealed many novel IL-6 target genes, among them several genes coding for transcriptional regulators involved in B-lymphocyte differentiation as well as for growth factors and receptors potentially implicated in autocrine or paracrine growth control. Regulation of most IL-6 target genes required the activation of Stat3, underscoring its central role for IL-6 signal transduction. Taken together, our data provide evidence for the existence of an as yet unknown Stat3-dependent survival pathway in myeloma cells.
Introduction
Multiple myeloma (MM)/plasmacytoma is a B-cell neoplasm characterized by the accumulation of clonal malignant plasma cells in the bone marrow. Although the pathogenesis of the disease still remains unclear, it is well established that interleukin-6 (IL-6), derived from either autocrine or paracrine sources, plays an essential role in the malignant progression of MM.1 IL-6 binds to a plasma membrane receptor complex whose signal-transducing component is the glycoprotein 130 (gp130). Binding of IL-6 activates gp130-associated protein tyrosine kinases of the Janus kinase (JAK) family.2 The JAKs phosphorylate gp130 tyrosine residues, which then serve as docking sites for src homology 2 (SH2) domain-containing signaling molecules such as the signal transducers and activators of transcription 3 and 1 (Stat3, Stat1),3,4 the SH2-containing phosphatase-2 (SHP-2), and the suppressor of cytokine signaling-3 (SOCS3).5,6 Among 5 tyrosine residues phosphorylated in gp130, tyrosine 759 recruits SHP-2 and SOCS3. SHP-2 has been implicated in the activation of the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3-K) pathways,7-10 whereas SOCS3 has been identified as a feedback inhibitor of JAK activity.11 Tyrosines 905 and 915 associate both Stat3 and Stat1, whereas tyrosines 767 and 814 preferentially mediate Stat3 activation.4 After tyrosine phosphorylation, Stat3 and Stat1 form homodimers or heterodimers, enter the nucleus, and regulate the transcription of target genes.12,13
Stat3 is found constitutively activated in a growing number of tumor-derived cell lines as well as samples from human cancers, including lymphomas and leukemias.14 Furthermore, a constitutively active Stat3 mutant causes cellular transformation in fibroblasts, revealing its oncogenic potential.15 In MM and plasmacytoma cells, Stat3 signaling was also reported to play an important role.16-19
Other studies, however, demonstrated the PI3-K/Akt pathway to be critical for IL-6–dependent growth and survival of MM cells. This pathway is frequently found activated in these cells, and inhibiting either PI3-K or Akt activity curtails IL-6–dependent or independent tumor cell expansion.20-23 In contrast, Xu et al24 reported that inhibition of apoptosis by IL-6 might be due to a down-regulation of the jun kinase (JNK) pathway in MM cells.24 Furthermore, IL-6 was shown to trigger MM cell proliferation by way of induction of the Ras-dependent MAPK pathway.25
Members of the B-cell lymphoma-2 (Bcl-2) family of apoptosis regulators were shown to play an important role in IL-6–dependent survival of MM cells.17,26,27 For the MM cell line U266, resistance to Fas-induced apoptosis correlates with an up-regulation of the antiapoptotic protein Bcl-xL.17 In other MM cell lines, the antiapoptotic action of IL-6 seems to correlate best with the induction of myeloid cell factor-1 (Mcl-1).26,28,29 Mcl-1 induction was observed to be mediated through the Stat3 pathway,19,30 the PI3-K pathway,31,32 or in a cooperative manner by both.33 More recent data question a central role of Mcl-1 for myeloma survival.34
To further elucidate the role of different IL-6–induced signaling pathways and genes in MM, we chose the human IL-6–dependent cell line INA-635 as a model system. As we demonstrate, this cell line depends strictly on IL-6 with regard to survival but not proliferation. Although IL-6 slightly induced the expression of Mcl-1, overexpression of this antiapoptotic protein did not suffice to protect the cells from apoptotic death. By microarray technology, genes differentially expressed in response to IL-6 in INA-6 cells were identified. INA-6 cells engineered to express chimericmutated receptors impairing the activation of particular signaling pathways allowed us to correlate expression patterns and survival. The results support the pivotal role of Stat3 activation for the antiapoptotic effect of IL-6. Therefore, our data provide evidence that IL-6–dependent survival of INA-6 MM cells relies on an additional Stat3-dependent antiapoptotic pathway.
Materials and methods
Cell culture
INA-6 and XG-1 cells were maintained in RPMI 1640 medium with Glutamax-I supplemented with 10% fetal calf serum, antibiotics (all from Life Technologies, Karlsruhe, Germany), 50 μM 2-β-mercaptoethanol and 1 ng/mL IL-6. Recombinant human IL-6 was a generous gift from S. Rose-John (Kiel, Germany).
Apoptosis assay
Apoptosis was assessed by using the annexin V–fluorescein isothiocyanate (FITC) apoptosis detection kit II (BD Biosciences, Heidelberg, Germany). For transfectants carrying receptor-enhanced green fluorescent protein (EGFP) fusion proteins, annexin V–FITC and propidium iodide (PI) were replaced by annexin V–phycoerythrin (PE) and 7-amino-actinomycin D (7-AAD), respectively. Flow cytometric analysis was done with a FACScan flow cytometer with the use of the CellQuest software (BD Biosciences).
Cell cycle analysis
Cell cycle distribution was measured by flow cytometric analysis by using PI staining. Cells (3 × 104) were washed with phosphate-buffered saline and fixed in 80% ethanol for 20 minutes at –20°C. After rehydrating in phosphate-buffered saline for 15 minutes, cells were incubated with the same solution containing 100 μg/mL RNase, 0.05% Tween 20, and 10 μg/mL PI. Fluorescence of FL2-A was analyzed on a flow cytometer as described earlier.
RNA isolation
Total RNA was prepared from 5 × 106 cultured cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA quality was examined by the RNA 6000 LabChip Kit on the 2100 bioanalyzer (Agilent Technologies, Waldbronn, Germany).
Gene expression analysis by DNA-oligonucleotide arrays
Double-stranded DNA was synthesized from total RNA, amplified as cRNA, labeled, and hybridized to Affymetrix U95A chips (Affymetrix, Santa Clara, CA), which were washed and scanned according to procedures developed by the manufacturer.
Two independent sets of experiments were performed to assess IL-6–dependent expression patterns in INA-6 cells. Normalization of the individual arrays on the basis of total intensity and comparative data analyses were performed with the Affymetrix software (MicroArray Suite 5.0.1/MicroDB 2.0/DMT 2.0). Significance criteria for selecting genes differentially expressed in response to IL-6 were chosen as follows: the microarrays hybridized with RNA from INA-6 cells withdrawn from IL-6 for 12 hours were set as reference chips to which microarrays derived from INA-6 cells treated with IL-6 for 1 hour, 4 hours, or permanently were compared. Genes were regarded differentially expressed if in both experimental sets the change P values were below .0025 and beyond .9975 for increased and decreased probe set signal intensities, respectively. Similarly, 2 sets of experiments were carried out with the INA-6 cells engineered to express mutated chimeric receptors.
Real-time PCR
RNA quantification by real-time polymerase chain reaction (PCR) detection was done with a LightCycler (Roche Diagnostics, Mannheim, Germany). Reverse transcription (RT) was performed by using SuperScript II (Life Technologies). An optimal PCR reaction was established by using the FastStart DNA Master SYBR Green I Kit (Roche Diagnostics). The nucleotide sequences of the primer pairs are available on request. The identity of the PCR products was confirmed by sequencing. Signals for the genes of interest from each RNA sample were normalized to that sample's signal for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The fold change was calculated relative to the signal observed for samples from cytokine-starved cells.
Ribonuclease protection assay
Gene fragments serving as probe template were cloned by RT-PCR into pGEM-T (Promega, Mannheim, Germany) except for HB-EGF and SGK. Probes for the latter were created from plasmids kindly provided by G. Raab (Boston, MA) and F. Lang (Tübingen, Germany), respectively. Probe sequences are available on request. Probes were labeled with α-32[P]–uridine triphosphate (UTP) (3000 Ci/mmol [11.1 × 1013 Bq/mol]) by means of RiboQuant In Vitro Transcription Kit (BD Biosciences/Pharmingen, San Diego, CA). Total RNA (5 μg) was hybridized with probe sets (4 × 104 cpm/μL for each probe) at 56°C overnight. Samples were digested with RNases A/RNase and inactivated, and the RNA was precipitated. The protected fragments were separated on a denaturing gel containing 5% polyacrylamide and 8 M urea. Bands were quantified by using a PhosphorImager (FLA-3000; Fujifilm, Japan). GAPDH was used for normalization.
Immunoblot analysis
Cell lysis and immunoprecipitation were performed as described by Bellido et al.36 Proteins were separated by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis, blotted to polyvinyl diflouride membrane, and detected by chemiluminescence (Pierce, Rockford, IL). Antibodies to Stat3 (S21320), Mcl-1 (sc-819), and β-actin (Clone AC-74) were from Transduction Laboratories (Lexington, KY), Santa Cruz Biotechnology (Santa Cruz, CA), and Sigma (Taufkirchen, Germany), respectively. Antityrosine-phosphorylated Stat3 (no. 9131), anti-Bad (no. 9292), and antiphospho-Bad (no. 9291) were from Cell Signaling Technology (Beverly, MA). Bcl-X (AF800), Bcl-2 (AF810), and Bax (AF820) antibodies were from R&D Systems (Wiesbaden, Germany).
Construction of expression vectors
Standard cloning procedures were performed as outlined by Sambrook et al.37 Lentiviral vector constructs VigΔSBΔNde, pHIT-G and SpΔ2 were a generous gift from K. Überla (Bochum, Germany).
The expression vector pSVL-Eg-Flag containing the chimeric erythropoietin receptor Eg (erythropoietin receptor [EpoR]/gp130 receptor)38 was taken as source for constructing the Eg-EGFP fusion protein. The Eg-Flag construct was amplified by PCR by using a primer introducing a SpeI site upstream and a BamHI site downstream of the gene (5′-GAT CCA CTA GTG CTA CCG GTC GCC ACC ATG GAC AAA CTC AGG GTG-3′ and 5′-GGT GGATCC CGT TTA TCG TCT TTG-3′), digested, and ligated into the NheI/BamHI opened EGFP-N1 vector (BD Biosciences/Clontech, Palo Alto, CA). To transfer the resulting Eg-Flag-EGFP construct into the lentiviral expression vector VigΔSBΔNde a PCR was performed with a primer introducing an ApaI site upstream and an EclXI site downstream of the gene (5′-CCG GTC GGGCCC ATG GAC AAA CTC AGG GTG C-3′ and CTA GAG TCGCGGCCG CTT TAC-3′). The PCR product was subjected to an ApaI/EclXI digest and inserted into the corresponding sites of VigΔSBΔNde by replacing GFP behind the MLV promoter finally leading to the plasmid MLV-Eg/Flag/EGFP.
The lentiviral expression vector MLV-Eg/Flag/EGFP encoding the wild-type domain of gp130 was used to generate mutants carrying tyrosine-phenylalanine substitutions removing docking sites for Stat1, Stat3, or SHP2. For substitution of tyrosines 905 and 915, a PCR-based mutagenesis was used by using one primer introducing both substitutions (5′-TTG TAG TCG ACC TGA GGC ATG AAG CCG CCT TGC CGT ACA GTC TGT GGT AAG AAA CTT TTA GG-3′) and the other situated within the MLV promoter (5′-GCC AGT CCT CCG ATT GAC TGA G-3′). The PCR product was EspI/SalI digested and subcloned into MLV-Eg/Flag/EGFP by replacing the corresponding wild-type gp130 fragment. The QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was applied to substitute tyrosines 759, 767, and 814 by using primer pairs 5′-TCG AGC ACT GTC CAG TTT TCT ACC GTG GTA CAC-3′, 5′-TCT ACC GTG GTA CAC TCC GGA TTC AGA CAC CAA GTT CCG T-3′, and 5′-TGC CCA GGC AAC AGT TCT TCA AAC AGA ACT GCA G-3′, respectively.
To express Mcl-1 and Bcl-XL genes in INA-6 cells, the vector MLV-IRES-EGFP was constructed by inserting an internal ribosome entry site (IRES) and the EGFP cDNA into VigΔSBΔNde by way of BamHI and EclXI. The IRES-EGFP PCR product was generated from pExp3EGFP39 (gift from Z. McIvor and M. Cross, Leipzig, Germany) by using primers introducing BamHI, SphI, XbaI, SpeI sites upstream (5′-GTGGATCCGCATGCC TCTAGA AACTAGTTA TCA AGC TTA TCG ATA CC-3′) and an EclXI site downstream (AAA CGGCCG CTT TAC TTG TAC AGC TCG-3′). Mcl-1 and Bcl-xL cDNAs were from the Deutsches Ressourcenzentrum für Genomforschung GmbH (Berlin, Germany). Open reading frames were cloned into the multiple cloning site of MLV-IRES-EGFP by PCR.
Lentiviral infection
The VigΔSBΔNde-derived lentiviral vectors were packaged into lentiviruses by cotransfecting HEK 293T cells with packaging plasmids pHIT/G40 and SpΔ241 by using the calcium phosphate coprecipitation method. Medium was replaced after 24 hours. The next day, culture supernatants were centrifuged at 16g for 10 minutes, passed through 0.45-μm filter, and kept at –75°C until use. For lentiviral infection, 100 μL medium containing 105 cells was mixed with 200 μL lentiviral culture supernatant in a 24-well plate and incubated for 2 to 4 hours. After adding 1 mL medium the incubation was continued for 2 days. The cells were then placed in fresh medium. High-speed cell sorting was performed by using a FACS-Vantage (BD Biosciences) on the basis of green fluorescence.
Results
IL-6 protects from apoptosis but does not directly stimulate proliferation in INA-6 cells
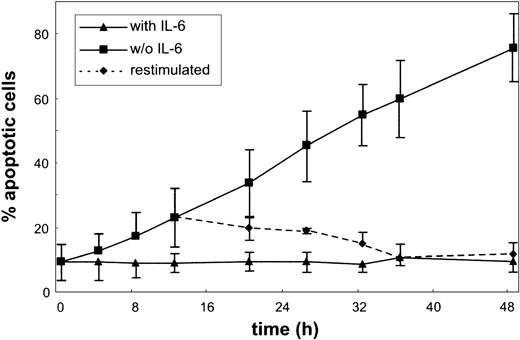
The effect of IL-6 on apoptosis and cell cycle of the MM cell line INA-6 was studied. As shown in Figure 1, INA-6 cells responded to IL-6 withdrawal with a remarkably rapid onset of apoptosis. Cells grown in the presence of IL-6 exhibited a low spontaneous apoptosis rate as quantified by the annexin V assay. As early as 4 hours after IL-6 withdrawal, the number of apoptotic cells started to rise, until after about 2 days almost all cells were dead. The process of apoptosis was still reversible after 12 hours of IL-6 deprivation as shown by re-addition of IL-6.
Apoptosis in the MM cell line INA-6. Cells were cultured without or with IL-6 for the times indicated, and aliquots were subjected to annexin V and PI staining. Apoptosis ratio was calculated as the percentage of annexin V–positive cells of all viable (PI-negative) cells. The dotted line represents a culture restimulated with IL-6 after 12 hours of IL-6 starvation. Data are mean values ± SD of 3 independent experiments.
Apoptosis in the MM cell line INA-6. Cells were cultured without or with IL-6 for the times indicated, and aliquots were subjected to annexin V and PI staining. Apoptosis ratio was calculated as the percentage of annexin V–positive cells of all viable (PI-negative) cells. The dotted line represents a culture restimulated with IL-6 after 12 hours of IL-6 starvation. Data are mean values ± SD of 3 independent experiments.
Analysis of the INA-6 cell cycle revealed a high portion of S-phase cells (more than 40%, data not shown), demonstrating the high proliferative potential of these cells. Withdrawal of IL-6 did not alter the cell cycle profile compared with cells grown in the presence of the cytokine, until after about 24 hours reliable cell cycle analysis was no longer possible because of high rates of apoptosis. We cannot rule out, however, that the cytokine exerts a delayed proliferative effect that is concealed by the rapid onset of apoptosis on IL-6 withdrawal. In contrast, serum deprivation yielded a rapid partial G1 block in INA-6 cells (data not shown), indicating that serum factors are required to sustain the growth of these cells.
The data demonstrate that IL-6 acts primarily as a survival factor on INA-6 cells. Therefore, these cells provide an excellent cellular system to study the molecular mechanisms of IL-6–mediated prevention of apoptosis.
Bcl-2 family apoptosis regulators do not play a major role in the IL-6–dependent survival of INA-6 cells
To analyze the mechanism of IL-6–mediated survival of INA-6 cells, we studied the expression of Bcl-2 family proteins in these cells. Among them Bcl-xL, Bcl-2, and Mcl-1 have previously been reported as being involved in the antiapoptotic effect of the cytokine.17,26,27
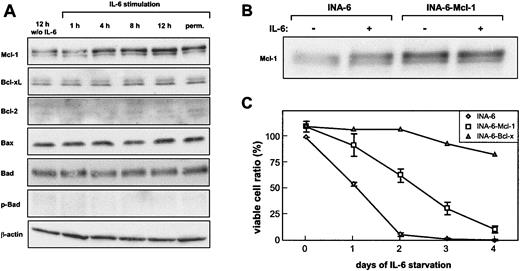
In INA-6 cells, the only expression change observed in response to IL-6 was a slight induction of Mcl-1 (Figure 2A). Bcl-xL, Bax, and Bad expression did not change, whereas Bcl-2 expression could not be detected in INA-6 cells. Furthermore, no induction of Bad phosphorylation by IL-6 was observed.
Mcl-1 induction does not sufficiently explain the IL-6–dependent survival of INA-6. (A) Cells were deprived of IL-6 for 12 hours and were restimulated for the times indicated. INA-6 cells permanently cultured in the presence of IL-6 were taken as a reference. Cell lysates were prepared, and equal amounts of protein were subjected to immunoblot analysis. (B) Mcl-1 and EGFP were coexpressed in INA-6 cells by lentiviral infection (INA-6–Mcl-1).As a control, infection was performed with a corresponding vector carrying EGFP cDNA only (INA-6). Infection efficiency was about 75% as measured by green fluorescence. The infected cells were cultured in the presence of IL-6 (+) or were withdrawn from IL-6 for 12 hours (–). Mcl-1 expression was analyzed by immunoblotting. (C) Control, Mcl-1–, and Bcl-xL–expressing INA-6 cells were grown with or without IL-6 for the times indicated and were subjected to an annexin V apoptosis assay. Viable cells are referred to as annexin V–negative cells. INA-6 cells grown in the presence of IL-6 were set 100%. Data for control and INA-6–Mcl-1 cells represent mean values ± SD of 3 independent experiments.
Mcl-1 induction does not sufficiently explain the IL-6–dependent survival of INA-6. (A) Cells were deprived of IL-6 for 12 hours and were restimulated for the times indicated. INA-6 cells permanently cultured in the presence of IL-6 were taken as a reference. Cell lysates were prepared, and equal amounts of protein were subjected to immunoblot analysis. (B) Mcl-1 and EGFP were coexpressed in INA-6 cells by lentiviral infection (INA-6–Mcl-1).As a control, infection was performed with a corresponding vector carrying EGFP cDNA only (INA-6). Infection efficiency was about 75% as measured by green fluorescence. The infected cells were cultured in the presence of IL-6 (+) or were withdrawn from IL-6 for 12 hours (–). Mcl-1 expression was analyzed by immunoblotting. (C) Control, Mcl-1–, and Bcl-xL–expressing INA-6 cells were grown with or without IL-6 for the times indicated and were subjected to an annexin V apoptosis assay. Viable cells are referred to as annexin V–negative cells. INA-6 cells grown in the presence of IL-6 were set 100%. Data for control and INA-6–Mcl-1 cells represent mean values ± SD of 3 independent experiments.
We next addressed the question whether the induction of Mcl-1 can account for IL-6–dependent survival of INA-6 cells. To this purpose, Mcl-1 was stably overexpressed by using a lentiviral vector that allowed coexpression with EGFP through an IRES sequence from the same expression cassette. Apoptosis of EGFP-positive cells was analyzed by flow cytometry. Ectopic expression of Mcl-1 reduced the number of apoptotic cells and prolonged the survival of INA-6 cells on IL-6 withdrawal (Figure 2C). Despite the fact, however, that Mcl-1 overexpression yielded higher protein levels than IL-6 stimulation of unmanipulated cells (Figure 2B), it was not sufficient to protect the cells from apoptotic death. Furthermore, overexpression of Bcl-xL by the same approach resulted in an efficient protection of INA-6 cells from apoptosis (Figure 2C), although Bcl-xL expression was not altered in response to IL-6, as described earlier. This finding demonstrates that disturbing the balance of proapoptotic and antiapoptotic Bcl-2 members can potentially block apoptosis in INA-6 cells, but that the induction of Mcl-1 does not suffice to do so. Therefore, the survival pathway initiated by IL-6 in INA-6 cells remains unresolved.
Dissection of signaling pathways mediating survival of myeloma cells
To obtain insight into the mechanism of IL-6–dependent INA-6 cell survival, we next asked which IL-6–initiated signal transduction pathway mediates this effect and which target genes are controlled by it. To this purpose a chimeric EpoR/gp130 receptor system established previously to define the signaling specificity through gp130 was used.4,38 Point mutations affecting recruitment of SHP-2, Stat1, and Stat3 to gp130 were introduced as outlined in Figure 3A. EpoR/gp130 variants were expressed in INA-6 cells by lentiviral infection. A carboxy-terminal EGFP fusion served to sort cells expressing the receptor chimeras by FACS. EGFP-positive cells were enriched to at least 80% and subjected to apoptosis, cell cycle, and immunoblot analyses.
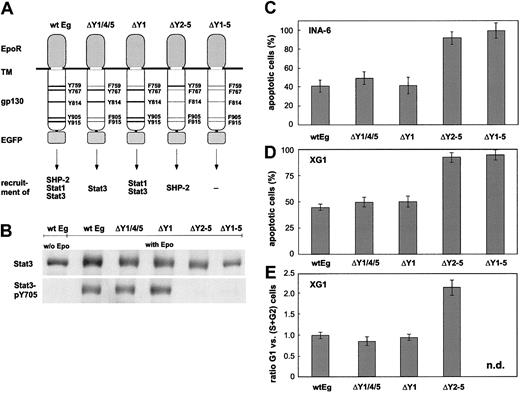
Ability of mutated EpoR/gp130 chimeras to mediate survival and proliferation. (A) Schematic representation of the mutations introduced into EpoR/gp130 (Eg). Tyrosine (Y) to phenylalanine (F) substitutions are printed in light face. The ability of the gp130 mutants to recruit downstream effector molecules is outlined. (B) INA-6 strains expressing the EpoR/gp130 chimeras as indicated were deprived of IL-6 for 12 hours before stimulation with Epo (7 U/mL) for another 8 hours. Thereafter, cell lysates were applied to immunoblot analysis by using antibodies against Stat3 or tyrosinephosphorylated Stat3. Cytokine-starved wild-type Eg strain was taken as a reference. (C-D) INA-6 and XG-1 strains were cultured without or with Epo for 24 hours (INA-6) and 72 hours (XG-1) in the absence of IL-6 and subsequently subjected to an apoptosis assay. The graph represents the percentage of annexin V–positive cells (mean values ± SD of 3 experiments) in the Epo-treated cultures as compared with those cultured without cytokine. (E) XG-1 strains were cultured without or with Epo for 72 hours in the absence of IL-6. Then, a flow cytometric cell cycle analysis was performed. The ratio of cells in G1 to those in S + G2 phases is shown (mean values ± SD of 3 experiments). n.d. indicates not determined.
Ability of mutated EpoR/gp130 chimeras to mediate survival and proliferation. (A) Schematic representation of the mutations introduced into EpoR/gp130 (Eg). Tyrosine (Y) to phenylalanine (F) substitutions are printed in light face. The ability of the gp130 mutants to recruit downstream effector molecules is outlined. (B) INA-6 strains expressing the EpoR/gp130 chimeras as indicated were deprived of IL-6 for 12 hours before stimulation with Epo (7 U/mL) for another 8 hours. Thereafter, cell lysates were applied to immunoblot analysis by using antibodies against Stat3 or tyrosinephosphorylated Stat3. Cytokine-starved wild-type Eg strain was taken as a reference. (C-D) INA-6 and XG-1 strains were cultured without or with Epo for 24 hours (INA-6) and 72 hours (XG-1) in the absence of IL-6 and subsequently subjected to an apoptosis assay. The graph represents the percentage of annexin V–positive cells (mean values ± SD of 3 experiments) in the Epo-treated cultures as compared with those cultured without cytokine. (E) XG-1 strains were cultured without or with Epo for 72 hours in the absence of IL-6. Then, a flow cytometric cell cycle analysis was performed. The ratio of cells in G1 to those in S + G2 phases is shown (mean values ± SD of 3 experiments). n.d. indicates not determined.
A similar approach by French et al42 that used epidermal growth factor receptor/gp130 chimeras was hampered by the known interaction of epidermal growth factor receptors with endogenous gp130.43 We did not observe a comparable phenomenon for the EpoR/gp130 hybrids.
INA-6 cells expressing EpoR/gp130 could be permanently propagated with Epo, demonstrating that the receptor chimera is fully functional. Consistent with previous observations,3,38,44 mutation of all 4 Stat3-recruiting tyrosine residues (Eg-Δ2-5 and Eg-Δ1-5) abrogated Stat3 activation through the hybrid receptors entirely (Figure 3B).
To compare the receptor variants with respect to their capability to provide an antiapoptotic signal, the corresponding cell populations were cultured without or with Epo in the absence of IL-6 for 24 hours. As shown in Figure 3C, only EpoR/gp130 variants able to activate Stat3 protected INA-6 cells from apoptosis. Mutating the tyrosine motifs 905 and 915 reduced the antiapoptotic potency only slightly, indicating that Stat1 activation that is mediated primarily through these motifs38 is not essential. Furthermore, tyrosine 759 known to associate SHP-2 and, hence, the signaling pathways initiated through it turned out to be fully dispensable for the IL-6–mediated survival effect. Hence, our results emphasize the importance of Stat3 for the gp130-mediated antiapoptotic signal in INA-6 cells.
To evaluate whether the pivotal role of Stat3 holds true for other IL-6–dependent myeloma cell lines as well, we chose to study the cell line XG-1. Like INA-6 cells, this cell line undergoes apoptosis on IL-6 withdrawal, but the onset of apoptosis is considerably slower in XG-1 as compared with INA-6 cells (data not shown). Furthermore, IL-6 exerts a significant proliferative effect on XG-1 cells.45 XG-1 strains expressing the EpoR/gp130 chimeric receptor variants were obtained by lentiviral infection as above for INA-6 cells. Analysis of apoptosis in these cells after 72 hours of Epo treatment in the absence of IL-6 revealed a highly similar situation as for the corresponding INA-6 strains (Figure 3C). Again, only those cells expressing receptor variants capable of activating Stat3 were protected from apoptosis. Furthermore, the same held true for the proliferative stimulus: only Stat3-activating receptor variants gave rise to an increased S/G1 phase ratio and, hence, to increased DNA synthesis (Figure 4D). We conclude from these results that for XG-1 cells, Stat3 represents a key regulator for both apoptosis and cell cycle control.
Microarray analysis of IL-6–regulated genes in INA-6 cells. INA-6 cells were cultured either in the presence of 1 ng/mL IL-6 (perm.) or deprived of IL-6 for 12 hours before restimulation with IL-6 for 1 hour or 4 hours, and expression patterns were studied using the Affymetrix U95A array. Two independent series of the experiment were carried out. Microarrays representing cells withdrawn from IL-6 for 12 hours were taken as baseline. Expression changes are indicated by the colors (red for induced and green for reduced expression) as shown.
Microarray analysis of IL-6–regulated genes in INA-6 cells. INA-6 cells were cultured either in the presence of 1 ng/mL IL-6 (perm.) or deprived of IL-6 for 12 hours before restimulation with IL-6 for 1 hour or 4 hours, and expression patterns were studied using the Affymetrix U95A array. Two independent series of the experiment were carried out. Microarrays representing cells withdrawn from IL-6 for 12 hours were taken as baseline. Expression changes are indicated by the colors (red for induced and green for reduced expression) as shown.
IL-6–regulated gene expression patterns in INA-6 cells
To identify IL-6 target genes whose regulation correlates with Stat3 activation and survival of INA-6 cells, gene expression profiling studies were performed. First, differential expression patterns were defined in INA-6 cells in response to IL-6. For this purpose, Affymetrix U95A oligonucleotide microarrays comprising approximately 12 000 probe sets specific for human genes were used. The cells were IL-6–starved for 12 hours, then restimulated with IL-6 for 1 hour or 4 hours, or continuously grown in the presence of IL-6.
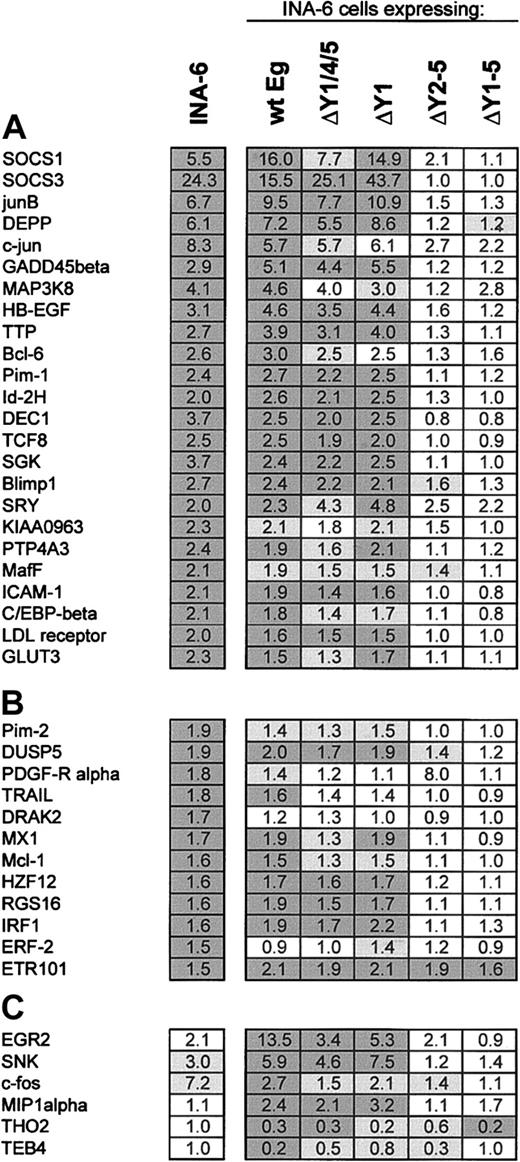
For a total of 174 probe sets representing approximately 150 different genes the data obtained indicate a differential expression in response to IL-6 by at least 1.5-fold at either time point. See the Supplemental Table link at the top of the online article on the Blood website. Forty-four genes yielded an at least 2-fold expression change, as shown in Figure 4. Among them, 22, 29, and 16 genes were found differentially expressed 2-fold or more at 1 hour, 4 hours, and permanent IL-6 treatment compared with IL-6 withdrawal, respectively. It is remarkable that only a few genes were repressed by IL-6, whereas the majority of differentially expressed genes were induced by the cytokine.
Our data demonstrate differential expression of both known (eg, Pim-1, junB, SOCS3, ICAM1, C/EBPβ) genes and novel IL-6 target genes in INA-6 cells. To verify the reliability of the microarray data we determined the mRNA levels of 9 genes by alternative techniques. For this purpose, genes from both groups were chosen. As shown in Table 1, the induction by IL-6 could be confirmed by real-time PCR and/or RNAse protection analysis in all cases. However, values for expression changes as measured by these methods were commonly higher than predicted by the microarray analysis, suggesting that the latter might tend to underestimate the amplitude of induction.
Induction of selected genes by IL-6 in INA-6 cells as determined by microarray analysis, real-time PCR, and RNA protection assay
. | Microarray . | . | . | Real-time PCR . | . | . | RNA protection assay . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene . | 1 h . | 4 h . | Perm . | 1 h . | 4 h . | Perm . | 1 h . | 4 h . | Perm . | ||||||
| junB* | 6.7 | 2.2 | 1.9 | 12.1 ± 3.1 | 3.7 ± 1.4 | 3.0 ± 1.6 | 7.5 ± 0.5 | 2.1 ± 0.1 | 2.0 ± 0.4 | ||||||
| SOCS3 | 24.3 | 3.7 | 4.0 | 24.7 ± 1.3 | 4.6 ± 1.7 | 3.0 ± 1.7 | 19.9 ± 6.9 | 3.2 ± 0.3 | 2.6 ± 0.4 | ||||||
| HB-EGF | 3.1 | 2.2 | 1.9 | 5.3 ± 0.7 | 3.2 ± 0.4 | 3.2 ± 0.8 | 4.8 ± 0.7 | 2.2 ± 0.1 | 2.9 ± 0.3 | ||||||
| Pim-1* | 2.4 | 1.0 | 1.1 | 8.2 ± 0.7 | 2.3 ± 0.3 | 1.5 ± 0.1 | ND | ND | ND | ||||||
| Bcl-3 | 1.9 | 1.7 | 1.5 | 8.3 ± 2.4 | 4.7 ± 1.9 | 5.0 ± 0.7 | ND | ND | ND | ||||||
| SGK | 3.7 | 4.3 | 4.4 | 12.1 ± 3.1 | ND | ND | 10.3 ± 2.9 | 6.5 ± 0.9 | 8.8 ± 1.0 | ||||||
| GADD45β | 3.2 | 2.0 | 1.3 | 6.0 ± 2.6 | ND | ND | 2.6 ± 0.3 | 1.3 ± 0.3 | 1.1 ± 0.6 | ||||||
| DEC1 | 3.7 | 2.1 | 2.0 | ND | ND | ND | 5.3 ± 1.9 | 2.7 ± 0.4 | 2.7 ± 0.4 | ||||||
| DEPP | 6.1 | 2.5 | 1.6 | ND | ND | ND | 7.4 ± 1.9 | 1.9 ± 0.6 | 2.0 ± 0.4 | ||||||
. | Microarray . | . | . | Real-time PCR . | . | . | RNA protection assay . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene . | 1 h . | 4 h . | Perm . | 1 h . | 4 h . | Perm . | 1 h . | 4 h . | Perm . | ||||||
| junB* | 6.7 | 2.2 | 1.9 | 12.1 ± 3.1 | 3.7 ± 1.4 | 3.0 ± 1.6 | 7.5 ± 0.5 | 2.1 ± 0.1 | 2.0 ± 0.4 | ||||||
| SOCS3 | 24.3 | 3.7 | 4.0 | 24.7 ± 1.3 | 4.6 ± 1.7 | 3.0 ± 1.7 | 19.9 ± 6.9 | 3.2 ± 0.3 | 2.6 ± 0.4 | ||||||
| HB-EGF | 3.1 | 2.2 | 1.9 | 5.3 ± 0.7 | 3.2 ± 0.4 | 3.2 ± 0.8 | 4.8 ± 0.7 | 2.2 ± 0.1 | 2.9 ± 0.3 | ||||||
| Pim-1* | 2.4 | 1.0 | 1.1 | 8.2 ± 0.7 | 2.3 ± 0.3 | 1.5 ± 0.1 | ND | ND | ND | ||||||
| Bcl-3 | 1.9 | 1.7 | 1.5 | 8.3 ± 2.4 | 4.7 ± 1.9 | 5.0 ± 0.7 | ND | ND | ND | ||||||
| SGK | 3.7 | 4.3 | 4.4 | 12.1 ± 3.1 | ND | ND | 10.3 ± 2.9 | 6.5 ± 0.9 | 8.8 ± 1.0 | ||||||
| GADD45β | 3.2 | 2.0 | 1.3 | 6.0 ± 2.6 | ND | ND | 2.6 ± 0.3 | 1.3 ± 0.3 | 1.1 ± 0.6 | ||||||
| DEC1 | 3.7 | 2.1 | 2.0 | ND | ND | ND | 5.3 ± 1.9 | 2.7 ± 0.4 | 2.7 ± 0.4 | ||||||
| DEPP | 6.1 | 2.5 | 1.6 | ND | ND | ND | 7.4 ± 1.9 | 1.9 ± 0.6 | 2.0 ± 0.4 | ||||||
Real-time PCR data and RNA protection assay, expressed as mean fold induction ± standard deviation, were obtained from at least 3 independent experiments by using RNA isolated from INA-6 cells incubated for 1 hour, 4 hours, or permanently with IL-6, compared with IL-6-deprived cells. Microarray data represent the mean fold induction of 2 experiments. ND indicates not determined; Perm, permanent.
junB and Pim-1 both are represented by 2-probe sets on the microarray. The microarray data given are the mean of the 2-probe set data.
Among the genes differentially expressed in response to IL-6, many code for proteins involved in cellular signaling, including cytokines, receptors, adaptor proteins, or protein kinases. Another predominant group is the IL-6 target genes coding for proteins participating in gene expression control at the transcriptional or posttranscriptional levels (Figure 4).
In accordance with the protein expression data for Bcl-2 family members as shown earlier, the microarray analysis revealed a significant induction of the Mcl-1 gene by 1.4- to 1.6-fold at all time points, whereas the genes coding for Bcl-x, Bad, or Bax were not changed. Bcl-2 was absent in both microarray and immunoblot analyses. Furthermore, the data did not indicate the induction of other antiapoptotic or down-regulation of proapoptotic regulators of the Bcl-2 or inhibitor of apoptosis families.
Analysis of gene expression patterns induced through mutated receptors
Stat3 activation turned out to be of pivotal importance for INA-6 and XG-1 cell survival. Therefore, the question arises as to which of the genes differentially expressed in response to IL-6 are controlled through that pathway. To this purpose, the expression patterns induced through the various EpoR/gp130 chimeras were analyzed.
We first evaluated whether signaling through EpoR/gp130 could reproduce a similar differential expression pattern as the one observed on IL-6 stimulation. Comparison of the data obtained by microarray analysis demonstrated that all 24 genes that were induced by at least 2-fold in INA-6 cells on a 1-hour treatment with IL-6 responded to Epo in cells expressing EpoR/gp130 as well (Figure 5A). The amplitude of expression changes turned out to be highly comparable. The same holds true for most genes that were induced by IL-6 between 1.5- and 2-fold after 1 hour (Figure 5B).
Gene expression profiling of INA-6 cells expressing mutated EpoR/gp130 chimeras. INA-6 strains expressing the indicated EpoR/gp130 chimeras were deprived of IL-6 for 12 hours before stimulation with Epo for 1 hour. Gene expression was analyzed by using Affymetrix U95A arrays (5 columns at the right). Two independent experimental series were carried out. The values are compared with those obtained for unmanipulatied INA-6 cells after treatment with IL-6 for 1 hour (left column). (A) Expression change values (fold) are shown for the genes significantly responding to a 1-hour IL-6 treatment of INA-6 cells by at least 2-fold. Dark gray background indicates change values significant (described in “Materials and methods”) in both experiments, light gray values significant in only one of the 2 experiments. (B) Values are shown for the genes significantly responding to IL-6 treatment by 1.5- to 2-fold and (C) for the genes changed through EpoR/gp130 but not significantly changed by IL-6 in INA-6 cells. Unigene accession numbers for the genes not included in Figure 3 are dual specificity phosphatase 5 (DUSP5), Hs.2128; tumor necrosis factor-related apoptosis-inducing ligand (TRAIL),: Hs.83429; death-associated protein kinase-related apoptosis-inducing protein kinase 2 (DRAK2), Hs.120996; human zinc finger protein 12 (HZF12), Hs.164284; regulator of G-protein signaling 16 (RGS16), Hs.183601; interferon regulatory factor 1 (IRF1), Hs.80645; EGF-response factor 2 (ERF-2), Hs.78909; early growth response 2 (EGR2), Hs.1395; serum-inducible kinase (SNK): Hs.3838; c-fos, Hs.25647; macrophage inflammatory protein 1-α (MIP1α), Hs.73817; THO2, Hs.16411; and TEB4, Hs.20141.
Gene expression profiling of INA-6 cells expressing mutated EpoR/gp130 chimeras. INA-6 strains expressing the indicated EpoR/gp130 chimeras were deprived of IL-6 for 12 hours before stimulation with Epo for 1 hour. Gene expression was analyzed by using Affymetrix U95A arrays (5 columns at the right). Two independent experimental series were carried out. The values are compared with those obtained for unmanipulatied INA-6 cells after treatment with IL-6 for 1 hour (left column). (A) Expression change values (fold) are shown for the genes significantly responding to a 1-hour IL-6 treatment of INA-6 cells by at least 2-fold. Dark gray background indicates change values significant (described in “Materials and methods”) in both experiments, light gray values significant in only one of the 2 experiments. (B) Values are shown for the genes significantly responding to IL-6 treatment by 1.5- to 2-fold and (C) for the genes changed through EpoR/gp130 but not significantly changed by IL-6 in INA-6 cells. Unigene accession numbers for the genes not included in Figure 3 are dual specificity phosphatase 5 (DUSP5), Hs.2128; tumor necrosis factor-related apoptosis-inducing ligand (TRAIL),: Hs.83429; death-associated protein kinase-related apoptosis-inducing protein kinase 2 (DRAK2), Hs.120996; human zinc finger protein 12 (HZF12), Hs.164284; regulator of G-protein signaling 16 (RGS16), Hs.183601; interferon regulatory factor 1 (IRF1), Hs.80645; EGF-response factor 2 (ERF-2), Hs.78909; early growth response 2 (EGR2), Hs.1395; serum-inducible kinase (SNK): Hs.3838; c-fos, Hs.25647; macrophage inflammatory protein 1-α (MIP1α), Hs.73817; THO2, Hs.16411; and TEB4, Hs.20141.
For verification of the microarray analysis, we selected 8 genes and estimated their mRNA levels by real-time PCR. As shown in Table 2, the microarray data could be confirmed. The high variation observed for the values of SOCS3 gene induction was due to the extremely low expression level of this gene in the absence of cytokine stimulation.
Induction of selected genes in INA-6 and XG-1 strains expressing mutated EpoR/gp130 chimeras after 1-hour Epo stimulation as determined by real-time PCR
. | IL-6 . | wtEg . | ΔY1/4/5 . | ΔY1 . | ΔY2-5 . | ΔY1-5 . |
|---|---|---|---|---|---|---|
| INA-6 cells | ||||||
| junB | 12.1 ± 3.1 | 14.7 ± 1.1 | 6.6 ± 1.5 | 15.7 ± 1.8 | 2.5 ± 0.5 | 2.5 ± 0.9 |
| SOCS3 | 24.7 ± 1.3 | 281.8 ± 59.3 | 136.1 ± 41.7 | 277.5 ± 72.0 | 4.7 ± 1.4 | 5.8 ± 1.5 |
| Pim-1 | 8.2 ± 0.7 | 4.1 ± 0.2 | 2.9 ± 0.3 | 3.9 ± 0.4 | 1.1 ± 0.2 | 1.2 ± 0.1 |
| HB-EGF | 5.3 ± 0.7 | 5.3 ± 0.6 | 3.5 ± 0.5 | 4.8 ± 0.4 | 1.4 ± 0.3 | 1.4 ± 0.5 |
| SGK | 14.0 ± 0.6 | 10.5 ± 3.3 | 6.3 ± 1.1 | 7.1 ± 1.5 | 1.4 ± 0.4 | 0.9 ± 0.1 |
| GADD45β | 6.0 ± 2.6 | 9.0 ± 2.5 | 5.4 ± 0.8 | 11.3 ± 2.4 | 3.2 ± 1.4 | 3.1 ± 1.3 |
| ETR101 | 2.1 ± 0.5 | 3.3 ± 0.9 | 2.3 ± 0.6 | 2.0 ± 0.5 | 1.6 ± 0.1 | 1.0 ± 0.2 |
| c-fos | 1.8 ± 0.4 | 5.9 ± 1.2 | 4.7 ± 1.2 | 2.3 ± 0.7 | 4.7 ± 1.6 | 3.8 ± 0.9 |
| XG-1 cells | ||||||
| junB | 5.6 ± 0.4 | 7.7 ± 0.6 | 5.6 ± 1.1 | 6.9 ± 1.0 | 2.5 ± 0.5 | 1.6 ± 0.4 |
| SOCS3 | 29.8 ± 2.1 | 108.8 ± 41.2 | 27.3 ± 3.7 | 54.6 ± 23.2 | 0.5 ± 0.2 | 2.6 ± 0.1 |
| Pim-1 | 3.3 ± 0.3 | 3.1 ± 0.5 | 2.5 ± 0.5 | 4.3 ± 1.2 | 1.2 ± 0.5 | 1.5 ± 0.5 |
| HB-EGF | 5.7 ± 0.5 | 6.7 ± 1.7 | 4.4 ± 1.0 | 4.6 ± 0.4 | 1.7 ± 0.2 | 1.8 ± 0.1 |
| SGK | 2.3 ± 0.3 | 2.8 ± 0.6 | 3.0 ± 0.6 | 2.9 ± 0.4 | 1.8 ± 0.4 | 1.4 ± 0.1 |
| GADD45β | 6.8 ± 1.5 | 12.3 ± 3.1 | 4.4 ± 0.4 | 6.6 ± 1.0 | 1.9 ± 0.6 | 1.3 ± 0.3 |
| ETR101 | 1.7 ± 0.2 | 3.0 ± 0.5 | 2.0 ± 0.3 | 1.8 ± 0.3 | 3.0 ± 0.5 | 1.5 ± 0.2 |
| c-fos | 9.7 ± 1.6 | 5.0 ± 1.1 | 2.9 ± 0.5 | 3.3 ± 0.5 | 1.7 ± 0.6 | 1.6 ± 0.1 |
. | IL-6 . | wtEg . | ΔY1/4/5 . | ΔY1 . | ΔY2-5 . | ΔY1-5 . |
|---|---|---|---|---|---|---|
| INA-6 cells | ||||||
| junB | 12.1 ± 3.1 | 14.7 ± 1.1 | 6.6 ± 1.5 | 15.7 ± 1.8 | 2.5 ± 0.5 | 2.5 ± 0.9 |
| SOCS3 | 24.7 ± 1.3 | 281.8 ± 59.3 | 136.1 ± 41.7 | 277.5 ± 72.0 | 4.7 ± 1.4 | 5.8 ± 1.5 |
| Pim-1 | 8.2 ± 0.7 | 4.1 ± 0.2 | 2.9 ± 0.3 | 3.9 ± 0.4 | 1.1 ± 0.2 | 1.2 ± 0.1 |
| HB-EGF | 5.3 ± 0.7 | 5.3 ± 0.6 | 3.5 ± 0.5 | 4.8 ± 0.4 | 1.4 ± 0.3 | 1.4 ± 0.5 |
| SGK | 14.0 ± 0.6 | 10.5 ± 3.3 | 6.3 ± 1.1 | 7.1 ± 1.5 | 1.4 ± 0.4 | 0.9 ± 0.1 |
| GADD45β | 6.0 ± 2.6 | 9.0 ± 2.5 | 5.4 ± 0.8 | 11.3 ± 2.4 | 3.2 ± 1.4 | 3.1 ± 1.3 |
| ETR101 | 2.1 ± 0.5 | 3.3 ± 0.9 | 2.3 ± 0.6 | 2.0 ± 0.5 | 1.6 ± 0.1 | 1.0 ± 0.2 |
| c-fos | 1.8 ± 0.4 | 5.9 ± 1.2 | 4.7 ± 1.2 | 2.3 ± 0.7 | 4.7 ± 1.6 | 3.8 ± 0.9 |
| XG-1 cells | ||||||
| junB | 5.6 ± 0.4 | 7.7 ± 0.6 | 5.6 ± 1.1 | 6.9 ± 1.0 | 2.5 ± 0.5 | 1.6 ± 0.4 |
| SOCS3 | 29.8 ± 2.1 | 108.8 ± 41.2 | 27.3 ± 3.7 | 54.6 ± 23.2 | 0.5 ± 0.2 | 2.6 ± 0.1 |
| Pim-1 | 3.3 ± 0.3 | 3.1 ± 0.5 | 2.5 ± 0.5 | 4.3 ± 1.2 | 1.2 ± 0.5 | 1.5 ± 0.5 |
| HB-EGF | 5.7 ± 0.5 | 6.7 ± 1.7 | 4.4 ± 1.0 | 4.6 ± 0.4 | 1.7 ± 0.2 | 1.8 ± 0.1 |
| SGK | 2.3 ± 0.3 | 2.8 ± 0.6 | 3.0 ± 0.6 | 2.9 ± 0.4 | 1.8 ± 0.4 | 1.4 ± 0.1 |
| GADD45β | 6.8 ± 1.5 | 12.3 ± 3.1 | 4.4 ± 0.4 | 6.6 ± 1.0 | 1.9 ± 0.6 | 1.3 ± 0.3 |
| ETR101 | 1.7 ± 0.2 | 3.0 ± 0.5 | 2.0 ± 0.3 | 1.8 ± 0.3 | 3.0 ± 0.5 | 1.5 ± 0.2 |
| c-fos | 9.7 ± 1.6 | 5.0 ± 1.1 | 2.9 ± 0.5 | 3.3 ± 0.5 | 1.7 ± 0.6 | 1.6 ± 0.1 |
Real-time PCR data from 2 independent experiments were obtained from INA-6 and XG-1 strains expressing mutated EpoR/gp 130 chimeras treated as described in Figure 5. The values for 1-hour IL-6 stimulation of unmanipulated cells are given for comparison (IL-6). Data are expressed as mean fold induction ± standard deviation from at least 3 measurements. Values from cytokine-starved wild-type Eg (wtEg) were taken as baseline.
The study demonstrates that the vast majority of genes were regulated in a significant manner (gray boxes in Figure 5) only by those EpoR/gp130 chimeras that allowed Stat3 activation. Comparison of the 3 Stat3-activating chimeras shows that the expression changes transmitted by Eg-ΔY1,4,5 were in most cases slightly lower when compared with the 2 other chimeras. This finding correlates well with its somewhat lower antiapoptotic potency as observed earlier.
Six genes were differentially expressed by at least 2-fold in cells expressing EpoR/gp130 but not in IL-6–treated INA-6 cells (Figure 5C). For all but one of them, however, expression control relied on phosphorylation sites in the receptor chimera's cytoplasmic region, suggesting that their induction does not occur through an interaction of EpoR/gp130 with other receptors. It appears likely, therefore, that a signaling threshold required for induction of these genes is reached by the higher number of receptor chimeras expressed in the engineered cells compared with endogenous gp130 in control cells.
To compare the expression pattern observed in receptor-manipulated INA-6 cells with the situation in XG-1 cells, the same 8 genes studied earlier in INA-6 cells by real-time PCR were next analyzed in XG-1 cells expressing the different receptor variants. As shown in Table 2 (lower part), these genes all were induced in XG-1 cells as well, albeit with a different amplitude in some cases. Most interestingly, in XG-1 and INA-6 cells the chimeric receptor variants produced comparable expression pattern with the exception of the c-fos gene that required Stat3 activation for its induction in XG-1 cells but not in INA-6 cells.
Taken together, our data imply a central role for Stat3 not only for the survival of INA-6 and XG-1 cells but also for the proliferative response of the latter. Furthermore, the transcription factor emerges as the main switch controlling the vast majority of genes that are differentially expressed in response to IL-6 in these MM cells.
Discussion
IL-6 was originally characterized as a cytokine that causes the terminal differentiation of activated B cells into immunoglobulin-secreting cells46 and was then shown to be a major growth and survival factor for MM cells.47 More recently, IL-6 has been shown to stimulate the growth of plasmablasts48 and to protect early plasma cells from apoptosis, allowing their terminal differentiation into plasma cells.49
As we show here, INA-6 human myeloma cells require IL-6 primarily as a survival factor, whereas their cell cycle is not influenced by the cytokine. The latter might be due to an activating mutation in the N-Ras gene, causing a constitutively activated MAPK cascade in these cells.35 To gain insight into the signaling pathways and target genes required for the IL-6–dependent survival of INA-6 cells, we used hybrid receptors containing mutated gp130 tyrosine phosphorylation sites. Survival of INA-6 cells depended strictly on Stat3 activation. Furthermore, the differential expression of almost all IL-6 target genes in INA-6 cells required Stat3 activation as well. This finding indicates that pathways initiated through tyrosine 759, directly through JAKs, or other regions of gp130, including those by ways of hematopoietic cell kinase (Hck), Grb2-associated binder 1 (Gab1), or proline-rich tyrosine kinase 2 (Pyk2),50,51 have negligible effect on gene expression regulation in these cells. The PI3-K/Akt pathway has been implicated in MM survival or growth by several studies. However, no phosphorylation of Akt in response to IL-6 was observed in INA-6 cells (data not shown). This finding is in agreement with the lack of IL-6–induced Bad phosphorylation. It appears possible that INA-6 cells possess a degenerate IL-6 signal transduction machinery that is “streamlined” to provide solely survival signals through Stat3. However, similar observations were made with the myeloma cell line XG-1, and preliminary microarray data obtained for the latter cell line show patterns of differentially expressed genes that are comparable to the situation in INA-6 cells (data not shown). Of note, a recent gene profiling analysis of the myeloma cell line ANBL-6 produced a dramatically different IL-6–dependent expression pattern.52 These cells, however, respond to IL-6 withdrawal with a remarkably complete cell cycle arrest, whereas they do not require the cytokine for survival. Hence, the genes regulated by IL-6 in these cells reflect primarily the release from the cell cycle arrest.
In accordance with the importance of Stat3 signaling in INA-6 cells, our microarray studies revealed an induction of genes participating in the JAK/STAT signaling pathway by IL-6. Stat1, Stat3, as well as Nmi, which is thought to stabilize the interaction of STAT factors with coactivators53 were induced in a delayed but persistent manner. Similarly, IL-6 induced the expression of flotillin, phospholipid scramblase 1, and caveolin-2 that are part of phospholipid raft and caveolae structures Stat3 has been shown to be associated with.54 In fact, caveolae have been shown to play an important role in IL-6 signaling of myeloma cells.55
Of interest, the immediate-early gene induced by IL-6 with the highest amplitude codes for SOCS3, a feedback inhibitor of the JAK/STAT pathway. With respect to the strict dependence of the cells on Stat3 activation this might appear as a paradox. Recent publications, however, offer a plausible explanation: these studies demonstrated that Stat1 activation through gp130 is completely suppressed by SOCS3, whereas Stat3 activation is only partially blocked.56-58 Therefore, the SOCS3 induction might serve to ensure a Stat3-dominated signaling and to eliminate the known proapoptotic action of Stat1.
Despite the rapid onset of apoptosis on IL-6 withdrawal neither microarray nor immunoblot analyses revealed major changes in the expression of Bcl-2 family members that could explain the IL-6–initiated survival effect. The weak but persistent induction of Mcl-1 that we were able to detect by both techniques does not suffice to protect INA-6 cells from apoptotic death as shown by ectopically overexpressing the protein. Differential expression of Bcl-2, Bcl-xL, and/or Mcl-1 has been widely observed in various other MM cell lines and primary cells on IL-6 stimulation.16,19,26,27,29 It is well established that the balance of proapoptotic and antiapoptotic Bcl-2 members is a crucial parameter in apoptosis control. In fact, down-regulation of Mcl-1 by antisense oligonucleotide strategies increased, and ectopic overexpression of Mcl-1 decreased apoptosis in various MM cell lines.28,29,59 Whether the induction of antiapoptotic Bcl-2 members is sufficient or even required for IL-6–dependent survival of MM cells, however, remains to be established. It is questionable whether this can be proven by manipulating the expression levels of Bcl-2 members below and beyond their physiologic range as usually done by antisense and overexpression techniques, respectively. How careful such data need to be interpreted is demonstrated by our finding that INA-6 cells can be efficiently protected against apoptosis by overexpressing Bcl-xL, although this protein is not an IL-6 target in these cells.
Shirogane et al60 reported on a synergistic role for Pim-1 and c-myc in Stat3-mediated Bcl-2 induction and survival of pre-B cells. Yet neither c-myc nor Bcl-2 expression changed in INA-6 cells in response to IL-6 nor did an overexpression of Pim-1 decrease apoptosis (data not shown). Survivin, a member of the inhibitor of apoptosis family, was reported as another target of Stat3 in lymphoma and endothelial cells,61,62 but no member of this family was differentially expressed in INA-6 cells according to the microarray data.
Therefore, our data strongly suggest the existence of yet another IL-6–dependent and Stat3-mediated mechanism triggering survival of INA-6 cells. Because immediate apoptosis regulators seem not to be a target of IL-6 signaling, it seems likely that IL-6 controls survival of INA-6 cells at a higher hierarchy level.
It is noteworthy in this context that the microarray data reveal the induction of a number of genes coding for cytokines, growth factors, and receptors by IL-6 in INA-6 cells, including HB-EGF, growth-arrest specific 6 (Gas6), CD44, osteopontin, the platelet-derived growth factor (PDGF) receptor α chain, and others. HB-EGF, a member of the epidermal growth factor family, was shown to be produced by several MM cell lines63 and to cooperate in XG-1 cells with IL-6 to promote their growth and survival.64 Yet, HB-EGF by itself had no influence on INA-6 cell survival (data not shown). Gas6 is induced in plasmablasts compared with B cells,65 and expression of its receptor, the Tyro3 receptor tyrosine kinase, is elevated in MM compared with healthy plasma cells.63 The proliferative action of Gas6 on mesangial cells correlates with Stat3 activation.66 CD44 is important for the interaction of plasma and bone marrow stromal cells,67 and osteopontin, a CD44 ligand, is involved in proliferation and apoptosis control through the PI3-K pathway.68 Our data demonstrate for the first time the induction of these genes by IL-6 and, thereby, suggest a further mechanism by which the cytokine might be involved in MM pathogenesis. In fact, it is tempting to speculate that IL-6 drives autocrine growth regulatory loops or takes part in the intercellular communication between MM and bone marrow stroma cells by inducing such factors and receptors.
Among the genes expressed differentially in INA-6 cells in response to IL-6, many code for proteins involved in expression control at the transcriptional or posttranscriptional levels. Remarkably, IL-6 turned out to control the expression of a number of transcriptional regulator genes known to play prominent roles in the differentiation of activated B cells into plasma cells. Among them are the Bcl-6 and Blimp1 genes coding for transcriptional repressors known to mutually suppress each other's transcription,69 Oct2 and its coactivator OBF-1, which is essential for germinal center formation,70 and IRF-4/MUM1 that has been implicated in MM development.71,72 The finding that all these now emerge as targets of IL-6 action further corroborates the important role the cytokine plays in the transcriptional control during B-cell differentiation. Only Oct2 had previously been demonstrated to underlie a control by IL-6.73 Bcl-6 blocks terminal differentiation into plasma cells and protects germinal center cells from apoptosis.74-76 Conversely, Blimp1 is associated with commitment to plasma cell differentiation by extinguishing the expression of miscellaneous genes important for B-cell receptor signaling, germinal center B-cell function, and proliferation.77,78 According to our data Bcl-6 seems to eventually override Blimp1 expression in INA-6 cells. This situation might result in reprogramming the cells from a noncycling plasma cell-like status to an earlier differentiation stage. It will be interesting to address the question of whether the proliferative and survival effect of IL-6 on MM cells may at least in part be due to such a change of differentiation stage.
In conclusion, our data provide evidence that IL-6–dependent survival of INA-6 cells requires the Stat3 pathway but does not involve previously known IL-6– or Stat3-activated antiapoptotic signals. It will be the challenge of future studies to identify this survival pathway.
Prepublished online as Blood First Edition Paper, September 11, 2003; DOI 10.1182/blood-2003-04-1048.
Supported by the Bundesministerium für Bildung und Forschung (BMB+F), Interdisciplinary Centre for Clinical Research (IZKF) at the University of Leipzig (01KS9504/1, project A13) and by the Deutsche Forschungsgemeinschaft (Bonn, Germany), SFB610, project A6.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank M. Cross for help with cell sorting; S. Rose-John for the generous gift of IL-6; G. Raab (Boston, MA), F. Lang (Tübingen, Germany), K. Überla (Bochum, Germany), Z. McIvor, and M. Cross (Leipzig, Germany) for providing vectors; the Interdisciplinary Center for Bioinformatics (IZBI, Leipzig, Germany) for support with microarray data analysis; and Andreas Günther and Frank Bakker (Erlangen, Germany) for critical reading of the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal