Abstract
The tyrosine kinase receptor fetal liver kinase 1 (Flk-1) plays a crucial role in vasculogenesis and angiogenesis, but its target genes remain elusive. Comparing Flk-1+/+ with Flk-1-/- embryonic stem (ES) cells, we identified transcripts regulated by the vascular endothelial growth factor A (VEGF-A)/Flk-1 pathway at an early stage of their differentiation to endothelial and mural precursors. Further analysis of a number of these genes (Nm23-M1, Nm23-M2, Slug, Set, pp32, Cbp, Ship-1, Btk, and Pim-1) showed that their products were transiently up-regulated in vivo in endothelial cells (ECs) during angiogenesis of the ovary, and their mRNA was rapidly induced in vitro by VEGF-A in human umbilical cord vein endothelial cells (HUVECs). Functional analysis by RNA interference (RNAi) in ES cells induced to differentiate demonstrated that Pim-1 is required for their differentiation into ECs and smooth muscle cells (SMCs). In HUVECs, RNAi showed that Pim-1 is required in ECs for VEGF-A-dependent proliferation and migration. The identification of Flk-1 target genes should help in elucidating the molecular pathways that govern the vasculogenesis and angiogenesis processes. (Blood. 2004;103:4536-4544)
Introduction
The formation of the vascular system begins with vasculogenesis, the aggregation of endothelial precursors into a primitive network of tubes both in the yolk sac and in the embryo. Further vessel development, known as angiogenesis, consists of the remodeling and sprouting of the primitive capillary network in the embryo. This process also characterizes the formation of new vessels in adult tissues.1
Vascular endothelial growth factor A (VEGF-A) is a critical player both in vasculogenesis and in physiologic and pathologic angiogenesis. Targeted inactivation of VEGF-A showed an embryonal lethal phenotype even in heterozygotes, indicating that a tight dose-dependent regulation of this factor is required for vascular development.2,3 VEGF-A binds to 2 tyrosine kinase receptors, fms-like tyrosine kinase-1 (Flt-1; also known as VEGF-R1) and fetal liver kinase 1 (Flk-1; also known as VEGF-R2 and kinase domain receptor [KDR]).4 During vessel development, Flt-1 functions as a VEGF-A decoy receptor, while Flk-1 is the major mediator of the mitogenic, motogenic, and differentiation effects of VEGF-A.5-7 Flk-1 is the earliest marker of developing endothelial cells (ECs), as it appears prior to the distinction between endothelial and hematopoietic cells.8,9 Embryos lacking Flk-1 die with no sign of endothelial or hematopoietic cells, and chimeric embryo with Flk-1 null embryonic stem (ES) cells demonstrated that this receptor is also required for the migration of hematopoietic and endothelial progenitors to the yolk sac.5,6 Differentiation of ES cells in vitro showed that Flk-1-positive cells can differentiate into not only endothelial and hematopoietic cells but also mural cells.10-12 Thus, Flk-1 marks the hemangioblast that has been described as the common progenitor of hematopoietic, mural, and endothelial cells, and its expression persists in the angioblasts that will give rise to ECs and smooth muscle cells (SMCs).
In the adult, Flk-1 expression is restricted to endothelial cells and transiently up-regulated during angiogenesis.13,14 Flk-1 signaling upon VEGF-A binding acts as an early event to induce quiescent endothelial cells of veins and arteries to proliferate and migrate. A good example of VEGF-A-dependent angiogenesis is the cyclic corpus luteum of the ovary, which represents the organ site with the strongest angiogenesis.15 VEGF-A is a key mediator in angiogenesis of the ovary, as it is expressed in the granulosa cell layer of the corpus luteum, and the administration of VEGF-A inhibitors suppress luteal angiogenesis.16-20
In spite of the importance of this receptor for endothelial specification and in angiogenesis, little is known about its downstream targets. To gain a molecular understanding of the Flk-1 pathway signaling on vascular development, we differentiated in vitro murine embryonic stem (ES) cells into ECs and smooth muscle cells (SMCs) with VEGF-A165 (further referred to as VEGF-A) in chemically defined serum-free conditions. The differential screening between Flk-1+/+ and Flk-1-/- ES cells identified Flk-1-regulated transcripts expressed in angioblasts. Their endothelial expression was further demonstrated in hormonally induced mice ovaries as a model of angiogenesis in vivo, and the response to VEGF-A was measured in human umbilical cord vein endothelial cells (HUVECs). Functional in vitro analysis of Pim-1, identified in this screening, suggests that this gene is a downstream Flk-1 effector in vasculogenesis.
Materials and methods
ES differentiation in vitro
R1 ES cells, Flk-1 knock-out (KO) clones 5547 and 5548, and Flk-1+/- clones 5539 and 5540,6 a gift from Bill Stanford (Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario, Canada), were initially maintained on mouse inactivated embryo fibroblast layers in Dulbecco modified essential medium (DMEM; Gibco, Grand Island, NY) containing 15% fetal calf serum (FCS; Hyclone, Logan, UT), 1500 U/mL leukemia inhibitory factor (LIF), and 50 μM 2-mercaptoethanol. ES cell wild-type (WT) and mutant clones were grown on 0.1% gelatin for 2 weeks before embryoid body (EB) formation: 50 to 2000 cells/mL were aggregated in a nonadherent 96-well plate (Cellstar, Campbell, CA) for 4 days in α-modified essential medium (α-MEM) with 15% FCS and 50 μM 2-mercaptoethanol. EBs were then plated on a vitronectin-coated 24-well plate for another 4 days in α-MEM with 15% KnockOut SR (Gibco) and 50 μM 2-mercaptoethanol. ES cells were induced to differentiate to ECs in the presence of 50 ng/mL VEGF-A165 (Sigma, St Louis, MO). The nonadherent cells were removed by washing 1 day after plating on vitronectin-coated dishes. As ES clones behaved similarly, the data reported are of the heterozygous clone 5539 and the knock-out clone 5548.
In vitro and in vivo angiogenesis
To induce angiogenesis in the ovary, C57BL6 mice were stimulated with 10 U/mouse of pregnant mare's serum gonadotropin (PMS-G) to induce follicular maturation, and after 44 hours, with 10 U/mouse of human chorionic gonadotropin (hCG) to induce the luteic phase (LP) as previously described.21
FACS analysis
Cultured cells were harvested after incubation with dissociation buffer (Gibco) under gentle agitation and incubated for 30 minutes in phosphate-buffered saline (PBS)-5% FBS on ice to block unspecific binding. Cells were then incubated with primary antibodies for 30 minutes in PBS-1% bovine serum albumin (BSA) and after 3 washes were stained with conjugated secondary antibodies. Cells were analyzed by fluorescence-activated cell-sorter (FACS) scan, and dead cells were excluded by staining them with propidiumiodide.
Immunostaining
Cells were plated on coated cover glasses, fixed with 4% paraformaldehyde in PBS for 20 minutes at room temperature (rt), washed twice, and incubated with the blocking buffer (PBS + 1% BSA + 0.1% Triton X-100). The fixed cells were incubated with primary antibody in blocking buffer for 1 hour at rt followed by 3 washes and incubation with conjugated secondary antibodies in blocking buffer for 1 hour at rt. For nuclear staining, cells were incubated for 5 minutes at rt with DAPI (4,6 diamidino-2-phenylindole) reagent. To investigate the acetylated low-density lipoprotein (Ac-LDL) uptake, the cultured cells were incubated with 10 μg/mL Alexa Fluor 488 Ac-LDL (Molecular Probes, Eugene, OR) for 4 hours and washed twice with medium. The samples were examined with a Nikon E600 microscope (Melville, NY). Colocalizations were performed with Leica TCS SP2 confocal microscopy (Heidelberg, Germany). We used a monoclonal antibody (mAb) for murine E-cadherin ECCD2 (Zymed, South San Francisco, CA); murine Flk-1 AVAS12, murine platelet endothelial cell adhesion molecule 1 (PECAM-1) MEC13.1, murine vascular endothelial (VE)-cadherin 11D4.1 (Pharmingen, San Diego, CA); smooth muscle α-actin (α-SMA) 1A4 (Sigma); platelet-derived growth factor receptor β (PDGFR-β28; Transduction Laboratories, Lexington, KY); CBP (C-1), Ship-1 (P1C1), and anti-Pim-1 (N-16) polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); and BTK (C-20), pp32 (C-18), SET (E-15), Slug (D-19), NM23-H1 (L-14), and NM23-H2 (L-16) (Santa Cruz Biotechnology). As secondary antibodies, we used R-phycoerythrin-conjugated or fluorescein isothiocyanate (FITC)-conjugated anti-rat immunoglobulin G (IgG; H + L), FITC-conjugated anti-goat IgG (H + L), R-phycoerythrin-conjugated or FITC-conjugated anti-mouse IgG (H + L), and FITC-conjugated anti-rabbit IgG (H + L) in 1:100 dilution.
RNA extraction
RNA samples were extracted directly from cultured cells or frozen tissues using Trizol reagent (Gibco) followed by isopropanol alcohol precipitation. For isolation of poly A+ mRNA we used the Qiagen Oligotex direct mRNA Poly A extraction kit as indicated by the manufacturer (Valencia, CA).
SSH cDNA library generation
Suppression subtractive hybridization (SSH) methods22 were used to construct a subtractive library using the Clontech (Palo Alto, CA) polymerase chain reaction (PCR)-Select cDNA Subtraction Kit. Briefly, 2 μg poly A+ RNA samples from EBs was purified, and after cDNA synthesis the tester cDNA was subtracted twice to the driver cDNA and nested PCR was amplified. The PCR products were then cloned into the pGEM-T vector (Promega, Madison, WI). Using this method, we generated 4 different libraries: a forward unsubtracted and a subtracted library from cDNA derived from EBs Flk-1+/+ at 5.5 days of differentiation; and a reverse unsubtracted and a subtracted library from cDNA derived from EBs Flk-1-/- at 5.5 days of differentiation.
Differential colony screening
To identify the differentially expressed clones in the forward subtracted library, 7000 single colonies on nitrocellulose filters were screened. Colonies were plated in 4 replicas and were grown overnight on the filters, which were exposed for 5 minutes in a denaturing solution (0.5 M NaOH, 1.5 M NaCl) followed by a neutralizing solution (0.5 M Tris [tris(hydroxymethyl)aminomethane]-HCl, 1.5 M NaCl) and baked at 80°C for 2 hours.
The differential colony screening was performed by hybridization with the forward subtracted probes, forward unsubtracted probes, reverse unsubtracted probes, and reverse subtracted probes, which were derived from the radiolabeled corresponding cDNAs. Using this intercross analysis we identified the differentially expressed clones.
Macroarray analysis
To evaluate the difference in the gene expression levels, we performed a macroarray analysis. Nested PCR amplification of each clone identified was performed with rTaq DNA Polymerase (Takara, Kyoto, Japan), purified, and analyzed on agarose gel. Of each DNA product, 8 replicas were spotted on nitrocellulose filters and hybridized separately with the full-length cDNA probes obtained from the EBs mRNA WT and KO using the SMART PCR cDNA Synthesis Kit (Clontech). The single spot signals were quantified using the 1D Image Analysis Software (Kodak, Rochester, NY), and the values were normalized against the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene signal on each filter.
Real-time reverse transcriptase (RT)-PCR
The gene expression analysis was performed with the Light Cycler RNA Amplification Kit SYBR Green I (Roche, Mannheim, Germany); we designed specific primers for each gene using the OLIGO 4.0 PCC software (Hayward, CA); each experiment was repeated 4 times.
Design and cloning siRNA cassettes
The U6 promoter (-315 to +1) was PCR amplified from mouse genomic DNA with an XhoI restriction site at the transcriptional initiation site and cloned into pGEM-T vector. Oligonucleotides coding for mouse Pim-1 small interfering RNA (siRNA) were designed to contain a sense 21 nucleotide strand (5′-GTCTCTTCAGAGTGTCAGCAC-3′) sequence followed by a spacer (5′-TTCAAGAGA-3′) and its reverse complementary strand followed by 5 thymidines as an RNA polymerase III transcriptional stop signal. The human PIM-1 sense oligonucleotide (5′-GTCTCTTCAGAATGTCAGCAT-3′) was cloned with the same spacer as for the mouse and followed by its reverse complementary sequence. The complementary oligonucleotides were phosphorylated, annealed, and subcloned into XhoI-PstI cloning sites pGEM-T/U6 promoter vector. The U6 promoter-siRNA constructs were then subcloned into ClaI-SalI lentiviral vector, eliminating the protein G kinase (PGK) promoter and the green fluorescent protein (GFP) cassette.
Lentiviral vector production and titration
The lentiviral vectors (pCCLsin.PTT.PGK.EGFP.Wpre, pMDLg/pRRE, pRSV-Rev, and pMD2.VSVG) were a kind gift from L. Naldini (San Raffaele Telethon Institute for Gene Therapy, Milan, Italy), and the recombinant lentiviruses were produced as previously described.23 Briefly, 293FT cells were transiently transfected using the calcium-phosphate method for 16 hours, after which the lentiviruses were harvested 24 and 48 hours later and filtered through 0.22-μm pore cellulose acetate filters. Recombinant lentiviruses were concentrated by ultracentrifugation for 2 hours at 50 000g. Vector infectivity was evaluated by infecting cells with a GFP vector and titrating siRNA-expressing virus by real-time quantitative RT-PCR of a common lentiviral genome region compared with the GFP vector.
Proliferation, chemotaxis, and in vitro angiogenesis assay
Mitogenic activity was performed in HUVECs. Cells were seeded in 96-well plates (6 × 103 cells/well) and grown for 16 hours in complete M199 medium. The cells were then starved for 24 hours in M199 medium containing 2% FBS and 0.5% BSA, incubated with VEGF-A in the presence of 3H-thymidine for 24 hours at 37°C, and trypsinized; 3H-thymidine incorporation was measured using a γ-counter (Hewlett Packard, Palo Alto, CA). Chemotaxis analysis was performed as described24 with Boyden chamber using 48-well plates. Migrated cells were fixed in methanol stained with Giemsa solution and counted. Sprouts were performed as described.25 Sprouting was quantified by measuring the length (ocular grid at × 20-× 40 magnification) of the capillary sprouts. In vitro angiogenic assay was performed using Matrigel (BD Biosciences, Bedford, MA) in 96-well tissue culture as described.24
Results
Flk-1+/+ ES cells fully differentiate into endothelial and mural cells, while Flk-1-/- ES cells fail to differentiate
It was previously demonstrated that sorted Flk-1-positive mouse ES cells can differentiate into both ECs and SMCs, and that EC differentiation is dependent on VEGF-A in a dose-dependent manner.12,26 We introduced few variations on these protocols to identify genes relevant for endothelial and mural differentiation considering that the cross talk between endothelial and mural cell precursors is crucial for full development.27 EC and SMC precursors were selected by plating embryoid bodies (EBs) on vitronectin-coated dishes, thus selecting the precursors by adhesion of αvβ3 integrins.28 The subsequent differentiation was performed in serum-free medium supplemented with recombinant VEGF-A. In these conditions we obtained full EC and SMC differentiation. (FACS analysis at the various steps of differentiation with endothelial and mural markers is shown in Supplemental Figure S1; see the Supplemental Materials link at the top of the online article on the Blood website). Immunofluorescence analysis showed that PECAM1-positive cells tended to be at the center of the colony, while smooth muscle α-actin (α-SMA)-positive cells were dispersed at the periphery, forming a network on a different focal plane (Figure 1A-C). At the interface between the 2 cell types from day 5.5 onward, PECAM1-positive endothelial precursors began forming cordlike tubes, which within a few days were surrounded by α-SMA-positive cells in vessellike structures (Figure 1D-E). At day 8.5, differentiated ES cells were not only positive for VE-cadherin (Figure 1F) but were also able to uptake Ac-LDL particles and to form capillaries in vitro on matrigel (Figure 1G-H); SMCs were able to form a cytoskeletal network of α-SMA (Figure 1I). Thus ES cells were fully differentiated into ECs as determined not only by the expression of specific endothelial markers but also by their ability to undergo in vitro angiogenesis.
ES cells differentiate into mature ECs and SMCs by VEGF-A treatment. Indirect immunofluorescence analysis by double staining of endothelial and mural precursors. (A-C) At 5.5 days of ES cell differentiation, ECs stained with PECAM1 (green) and α-SMA-positive cells (red) form a network on different focal planes. Nuclei were stained with DAPI as indicated. Scale bar, 40 μm. (D-E) From day 5.5, EC precursors stained with PECAM1 (green) form primitive capillary tubes, which at 8.5 days are surrounded by SMCs stained with α-SMA (red). Scale bar, 16 μm. (F-G) At 8.5 days of differentiation, ECs are positive for VE-cadherin, which is a late marker of differentiation, and are able to uptake the acetylated low-density lipoprotein (Ac-LDL) particles. Green fluorescent Ac-LDL particles were added directly to the medium of differentiated ECs and analyzed at the microscope. Scale bar, 200 μm. (H) Differentiated ECs at 8.5 days plated on matrigel form capillary-like structure in vitro. Differentiated ECs from VEGF-A-induced EBs were disaggregated and plated on Matrigel. (I) SMCs showed fully differentiated α-SMA fibers. The field was at the periphery of the EBs to observe SMCs. Scale bar, 24 μm.
ES cells differentiate into mature ECs and SMCs by VEGF-A treatment. Indirect immunofluorescence analysis by double staining of endothelial and mural precursors. (A-C) At 5.5 days of ES cell differentiation, ECs stained with PECAM1 (green) and α-SMA-positive cells (red) form a network on different focal planes. Nuclei were stained with DAPI as indicated. Scale bar, 40 μm. (D-E) From day 5.5, EC precursors stained with PECAM1 (green) form primitive capillary tubes, which at 8.5 days are surrounded by SMCs stained with α-SMA (red). Scale bar, 16 μm. (F-G) At 8.5 days of differentiation, ECs are positive for VE-cadherin, which is a late marker of differentiation, and are able to uptake the acetylated low-density lipoprotein (Ac-LDL) particles. Green fluorescent Ac-LDL particles were added directly to the medium of differentiated ECs and analyzed at the microscope. Scale bar, 200 μm. (H) Differentiated ECs at 8.5 days plated on matrigel form capillary-like structure in vitro. Differentiated ECs from VEGF-A-induced EBs were disaggregated and plated on Matrigel. (I) SMCs showed fully differentiated α-SMA fibers. The field was at the periphery of the EBs to observe SMCs. Scale bar, 24 μm.
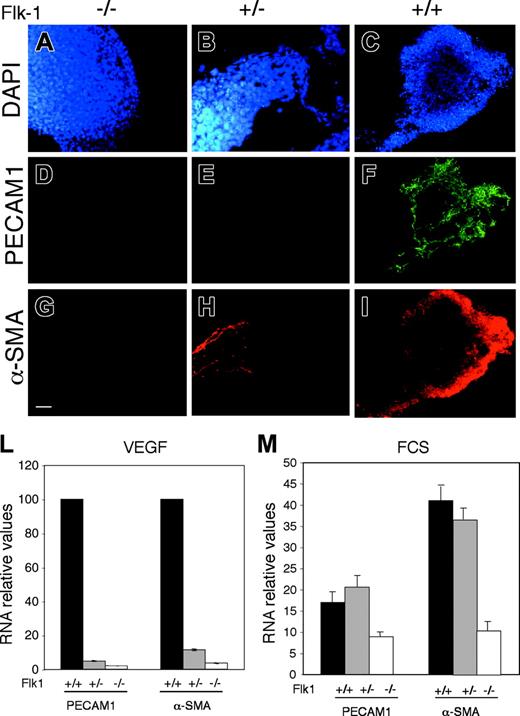
The in vitro differentiation protocol of ES cells was performed to differentiate Flk-1+/+, Flk-1+/-, and Flk-1-/- cells. Although wild-type and mutant ES cells were equally viable (data not shown), immunofluorescence analysis of PECAM1 and α-SMA at 5.5 days revealed that Flk-1-/- could not differentiate into endothelial and mural cells, while only a limited number of α-SMA-positive cells were observed in Flk+/- (Figure 2A-I). Accordingly, RT-PCR analysis of RNA extracted from ES cells treated with VEGF-A at day 5.5 showed a strong reduction of PECAM-1 and α-SMA both in Flk-1+/- and Flk-1-/- with respect to the wild-type cells (Figure 2L). Thus in chemically defined conditions, only Flk-1 wild-type ES cells could differentiate. This result is in contrast with previous observations demonstrating that Flk-1-/- ES cells could differentiate, albeit at low efficiency, into hematopoietic cells and ECs.29,30 In fact, performing the ES differentiation with FCS we observed that Flk-1-/- could differentiate into endothelial cells as they expressed PECAM1 mRNA (Figure 2M). However, in the presence of serum the endothelial differentiation was less efficient, as, at day 8.5, we could not obtain more than 40% of PECAM1-positive cells (data not shown). This suggests that FCS can induce the differentiation of ECs via the activation of receptors other than Flk-1. Taken together, these data demonstrate that it is possible to fully differentiate ES cells to ECs and SMCs strictly dependent on Flk-1 signaling and, therefore, identify Flk-1 target genes involved in these differentiation pathways.
Flk-1 mutant ES cells do not differentiate into ECs and SMCs in chemically defined medium. Immunostaining of Flk-1 knock-out (Flk-1-/-), heterozygous (Flk-1+/-), and wild type (Flk-1+/+) cells at 5.5 days of differentiation with VEGF-A as indicated. (A-C) DAPI staining of cell nuclei. (D-F) PECAM1 staining of EC precursors. (G-I) α-SMA staining for mural precursors. Scale bar, 800 μm. (L) Quantitative RT-PCR of PECAM1 and α-SMA mRNAs and at 8.5 days of differentiation with VEGF-A in serum-free medium of Flk-1 wild-type, heterozygous, and knock-out cells as indicated. (M) Quantitative RT-PCR of PECAM1 and α-SMA mRNAs at 8.5 days of differentiation in FCS of Flk-1 wild-type, heterozygous, and knock-out cells as indicated. The data shown in panels L-M are the results of 3 independent experiments. Error bars indicate standard deviation.
Flk-1 mutant ES cells do not differentiate into ECs and SMCs in chemically defined medium. Immunostaining of Flk-1 knock-out (Flk-1-/-), heterozygous (Flk-1+/-), and wild type (Flk-1+/+) cells at 5.5 days of differentiation with VEGF-A as indicated. (A-C) DAPI staining of cell nuclei. (D-F) PECAM1 staining of EC precursors. (G-I) α-SMA staining for mural precursors. Scale bar, 800 μm. (L) Quantitative RT-PCR of PECAM1 and α-SMA mRNAs and at 8.5 days of differentiation with VEGF-A in serum-free medium of Flk-1 wild-type, heterozygous, and knock-out cells as indicated. (M) Quantitative RT-PCR of PECAM1 and α-SMA mRNAs at 8.5 days of differentiation in FCS of Flk-1 wild-type, heterozygous, and knock-out cells as indicated. The data shown in panels L-M are the results of 3 independent experiments. Error bars indicate standard deviation.
Identification of Flk-1 target genes
To identify new genes coding for endothelial and pericyte regulators, we extracted RNA from Flk-1+/+ and Flk-1-/- ES cells at 5.5 days of differentiation, as their precursors, harvested at this stage, represented the earliest time point to identify Flk-1 target genes. We then performed cDNA suppression subtractive hybridization to identify the differentially expressed genes.22 We screened about 7000 colonies, which revealed 134 differentially expressed (≥ 2-fold) transcripts (Supplemental Table S1). The differential expression of the identified clones was evaluated by macroarray hybridization probed with RNA extracted from ES Flk-1+/+ or Flk-1-/- cells at 5.5 days of differentiation. Present among the differentially expressed genes were genes known to be expressed in endothelial and endothelial precursors such as EphB3, endothelial differentiation gene 5 (Edg5), and Sfrp1 (also known as FrzA) Sry-related high mobility group box 18 (Sox18)—as well as mural markers such as αactin 2, insulin-like growth factor II (IGFII), and fibulin-5. This confirms that the subtraction library identified genes expressed in precursors of these cell types. It is worth noting that the mural markers overall were less represented, as the mural cells were less represented due to the in vitro condition adopted (Table S1 and Figure 1).
The differential expression of putative regulatory genes was further evaluated by quantitative RT-PCR. As reported in Table 1, only minor variations with respect to the macroarray analysis were observed. Interestingly, the RT-PCR analysis confirmed the differential expression in EC precursors of genes that were previously described as expressed and/or regulating hematopoietic cells such as Slug (also known as Snai2), Nm23-M1, Nm23-M2, Src homology 2-containing inositol-5-phosphatase 1 (Ship-1), Burton tyrosine kinase (Btk), and provirus integration site of Moloney murine leukemia virus 1 (MoMuLV1; Pim-1). In addition, this screening identified genes involved in chromatin remodeling and in general transcription regulation such as Set, pp32, and CREB-binding protein (Cbp).
Comparison of the expression levels of transcripts differentially expressed in Flk1+/+ versus Flk1−/− ES cells at 5.5 days of differentiation in vitro by macroarray analysis and real-time quantitative RT-PCR
Category/gene . | Fold increase by macroarray . | Fold increase by RT-PCR . |
|---|---|---|
| Nuclear factor | ||
| Cbp | 4.3 ± 0.6 | 7.6 ± 0.5 |
| Hec | 6.4 ± 1.2 | 3.4 ± 0.2 |
| Nm23M1 | 3.1 ± 0.8 | 3.8 ± 0.7 |
| Nm23M2 | 3.9 ± 0.6 | 2.1 ± 0.3 |
| Pim-1 | 2.9 ± 0.9 | 3.2 ± 0.2 |
| pp32 | 3.7 ± 0.8 | 3.2 ± 0.5 |
| Set | 5.2 ± 0.8 | 2.4 ± 0.2 |
| Slug | 3.4 ± 0.3 | 5.9 ± 0.5 |
| Sox18 | 5.7 ± 0.2 | 4.4 ± 0.7 |
| Growth factor | ||
| Sfrp1 (FrzA) | 4.9 ± 0.3 | 11.3 ± 1.2 |
| Receptors | ||
| Edg5 | 4.6 ± 0.3 | 3.1 ± 0.5 |
| EphB3 (Sek4) | 3.5 ± 0.2 | 2.2 ± 0.3 |
| Signal transduction | ||
| Ship-1 | 9.9 ± 0.8 | 7.0 ± 0.4 |
| Btk | 3.0 ± 0.3 | 3.2 ± 0.2 |
| Unknown | ||
| Mesdc2 | > 20 | 13.6 ± 3.1 |
Category/gene . | Fold increase by macroarray . | Fold increase by RT-PCR . |
|---|---|---|
| Nuclear factor | ||
| Cbp | 4.3 ± 0.6 | 7.6 ± 0.5 |
| Hec | 6.4 ± 1.2 | 3.4 ± 0.2 |
| Nm23M1 | 3.1 ± 0.8 | 3.8 ± 0.7 |
| Nm23M2 | 3.9 ± 0.6 | 2.1 ± 0.3 |
| Pim-1 | 2.9 ± 0.9 | 3.2 ± 0.2 |
| pp32 | 3.7 ± 0.8 | 3.2 ± 0.5 |
| Set | 5.2 ± 0.8 | 2.4 ± 0.2 |
| Slug | 3.4 ± 0.3 | 5.9 ± 0.5 |
| Sox18 | 5.7 ± 0.2 | 4.4 ± 0.7 |
| Growth factor | ||
| Sfrp1 (FrzA) | 4.9 ± 0.3 | 11.3 ± 1.2 |
| Receptors | ||
| Edg5 | 4.6 ± 0.3 | 3.1 ± 0.5 |
| EphB3 (Sek4) | 3.5 ± 0.2 | 2.2 ± 0.3 |
| Signal transduction | ||
| Ship-1 | 9.9 ± 0.8 | 7.0 ± 0.4 |
| Btk | 3.0 ± 0.3 | 3.2 ± 0.2 |
| Unknown | ||
| Mesdc2 | > 20 | 13.6 ± 3.1 |
Data are presented as mean ± SEM. The fold change detected by quantitative RT-PCR represents the average between the results of 3 independent experiments.
Identification of genes induced in angiogenesis of the ovary
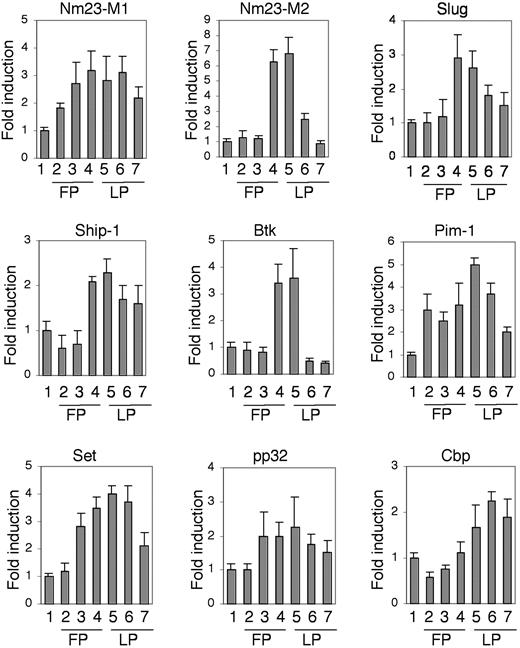
To understand whether the above identified genes were expressed and up-regulated in ECs during the angiogenic process, we further analyzed their expression in vivo in hormonally induced angiogenesis of the ovary. This model was chosen because the cyclic corpus luteum (CL) is a site with pronounced angiogenesis where vessel growth and maturation can easily be observed, and VEGF-A is essential for this process.14,15,17 C57BL6 mice were stimulated with pregnant mare's serum gonadotropin (PMS-G) to induce the follicular phase (FP), and successively with human chorionic gonadotropin (hCG) to induce the luteic phase (LP).21 Because ECs represent more than 50% of all cells of the growing CL,15 gene induction could be analyzed by quantitative RT-PCR. This was confirmed by the analysis of Flk-1 and Sfrp1/FrzA, which were known to be up-regulated in ECs during the angiogenesis of the ovary.13,21 Indeed, the RT-PCR analysis showed that their mRNAs were maximally induced from the late FP to the early LP, which corresponds to the most intense formation of the neovessels, and decreased at later time points (data not shown). Importantly, this analysis revealed that most of the identified genes were up-regulated during the angiogenesis of the ovary, although to varying levels and kinetics (Figure 3). Nm23-M2, Set, Btk, and Pim-1 showed the highest induction levels, while Nm23-M1, Slug, Ship-1, pp32, and Cbp showed minor, but still significant, induction. As could be predicted, Sox18, EphB3, and Edg5 were also induced with similar kinetics (data not shown).
Genes expressed in Flk-1-positive endothelial precursors are transiently up-regulated during angiogenesis in vivo. Time-course analysis of transcripts in hormonally induced ovaries. Vertical bars indicate the fold induction of each transcript as indicated from RNA collected from untreated (1) or hormonally induced ovaries during the follicular phase (FP) at 5 hours (2), 20 hours (3), and 44 hours (4) after pregnant mare's serum gonadotropin (PMS-G) stimulation and luteic phase (LP) at 8 hours (5), 16 hours (6), and 32 hours (7) after human chorionic gonadotropin (hCG) stimulation. Super ovulation in mice was induced as previously described,21 and the induction was quantified by real time RT-PCR and normalized on GAPDH expression. The data shown are the results of 3 independent experiments.
Genes expressed in Flk-1-positive endothelial precursors are transiently up-regulated during angiogenesis in vivo. Time-course analysis of transcripts in hormonally induced ovaries. Vertical bars indicate the fold induction of each transcript as indicated from RNA collected from untreated (1) or hormonally induced ovaries during the follicular phase (FP) at 5 hours (2), 20 hours (3), and 44 hours (4) after pregnant mare's serum gonadotropin (PMS-G) stimulation and luteic phase (LP) at 8 hours (5), 16 hours (6), and 32 hours (7) after human chorionic gonadotropin (hCG) stimulation. Super ovulation in mice was induced as previously described,21 and the induction was quantified by real time RT-PCR and normalized on GAPDH expression. The data shown are the results of 3 independent experiments.
New genes expressed in ECs during angiogenesis
To verify whether the newly identified genes were actually expressed in activated ECs, we performed immunofluorescence analysis. The ovaries collected at various time points after hormone induction were double stained with each specific antibody and with PECAM1 to label ECs. The immunofluorescence analysis performed at 8 hours of LP demonstrates that all genes analyzed were indeed expressed in endothelial cells (Figure 4). Importantly, no gene products analyzed were expressed in ECs of the ovary before active angiogenesis, but were up-regulated during the FP reaching maximal signal levels during the early LP, thus mirroring the RNA expression measured by RT-PCR. This expression pattern was evident for Pim-1, which was strongly induced in ECs during angiogenesis (Figure 5), while Cbp behaved differently. In fact, Cbp could be detected before angiogenic induction, and at later time points its expression was increased and not restricted to ECs (Supplemental Figure S2).
Genes expressed in Flk-1-positive endothelial precursors are expressed in endothelial cells of the ovary during angiogenesis. Immunofluorescence analysis on frozen ovary sections at 8 hours of the LP. Endothelial-positive cells were identified by staining with PECAM1 (green) antibody, and specific colocalization was valued by double staining with specific antibodies (red) as indicated. Colocalization analysis was performed with a confocal microscope. CL indicates corpus luteum. Scale bar, 40 μm.
Genes expressed in Flk-1-positive endothelial precursors are expressed in endothelial cells of the ovary during angiogenesis. Immunofluorescence analysis on frozen ovary sections at 8 hours of the LP. Endothelial-positive cells were identified by staining with PECAM1 (green) antibody, and specific colocalization was valued by double staining with specific antibodies (red) as indicated. Colocalization analysis was performed with a confocal microscope. CL indicates corpus luteum. Scale bar, 40 μm.
Pim-1 is transiently expressed in ECs of the ovary during angiogenesis. Immunofluorescence analysis on frozen ovary sections at ovaries during the follicular phase (FP) and luteic phase (LP) at the time indicated. Endothelial-positive cells were identified by staining with PECAM1 (green) antibody, and specific colocalization was valued by double staining with Pim-1 antibodies (red) as indicated. Colocalization analysis was performed with a confocal microscope. Scale bar, 40 μm.
Pim-1 is transiently expressed in ECs of the ovary during angiogenesis. Immunofluorescence analysis on frozen ovary sections at ovaries during the follicular phase (FP) and luteic phase (LP) at the time indicated. Endothelial-positive cells were identified by staining with PECAM1 (green) antibody, and specific colocalization was valued by double staining with Pim-1 antibodies (red) as indicated. Colocalization analysis was performed with a confocal microscope. Scale bar, 40 μm.
The identified genes respond to VEGF-A stimulation in HUVECs
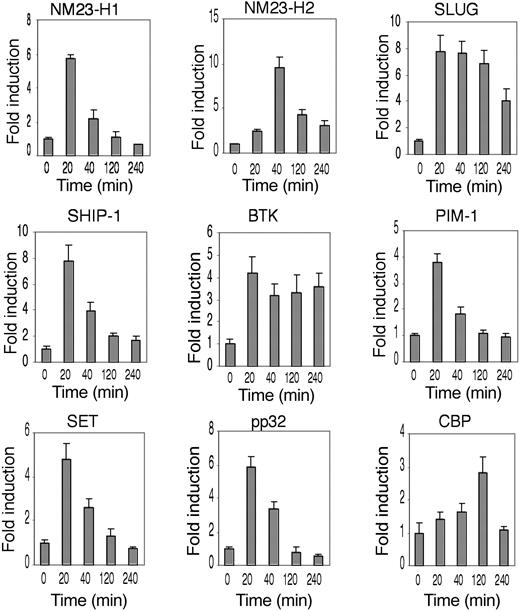
To verify whether the genes identified are downstream of Flk-1 signaling, we analyzed the kinetics of transcript induction by VEGF-A treatment of HUVECs of the human orthologues of each gene considered. RT-PCR analysis of mRNA extracted from HUVECs stimulated with VEGF-A was performed using specific primers. The results of this analysis are reported in Figure 6. NM23-H1, MESDC2, SLUG, pp32, and SHIP-1 mRNAs were strongly induced (≥ 6-fold), with a peak at 20 minutes. Similar kinetics, although with lower induction levels (3- to 6-fold), were observed for SET, BTK, and PIM-1 mRNAs. NM23-H2 mRNA was induced maximally at 40 minutes, while CBP induction was delayed, reaching a peak at 120 minutes.
Genes expressed in Flk-1-positive endothelial precursors are induced by VEGF-A in HUVECs. Time-course analysis of the human orthologues of the mouse genes as in Figure 3, as indicated. Vertical bars for each transcript, as indicated, measure the fold induction at the time indicated. Induction analysis was performed with RT-PCR and normalized on GAPDH expression. The data shown are the results of 3 independent experiments.
Genes expressed in Flk-1-positive endothelial precursors are induced by VEGF-A in HUVECs. Time-course analysis of the human orthologues of the mouse genes as in Figure 3, as indicated. Vertical bars for each transcript, as indicated, measure the fold induction at the time indicated. Induction analysis was performed with RT-PCR and normalized on GAPDH expression. The data shown are the results of 3 independent experiments.
Pim-1 is required for endothelial and mural cell differentiation
Pim-1 was further selected for analysis of its functional role in ES cell differentiation and in VEGF-A-stimulated HUVECs. This gene was chosen because no role in vasculogenesis or angiogenesis has been previously demonstrated. By contrast, our analysis showed that Pim-1 was specifically expressed in Flk-1-positive precursors. It was transiently expressed in vivo in ECs during angiogenesis and induced by VEGF-A in HUVECs. In addition, Pim-1 codes for a serine/threonine kinase, which in the nucleus could regulate gene expression by direct phosphorylation of transcription factors.31-33 To analyze the role of Pim-1 in vasculogenesis in vitro, we generated a lentiviral vector expressing a small interfering RNA (siRNA) recognizing the mouse Pim-1 mRNA, under the control of the RNA Polymerase III promoter U6. This vector was chosen because in contrast to other retroviral vectors analyzed, it was expressed in all ES cells and was not silenced during the differentiation protocol (data not shown). The optimal sequence to silence Pim-1 mRNA was identified using an RNA accessibility test.34 Infection with Pim-1 siRNA lentivirus showed a strong reduction of Pim-1 product in ES cells (Figure 7A). The lentiviral-infected ES cells silenced for Pim-1 were then induced to differentiate in vitro. Pim-1 siRNA did not inhibit cell growth or differentiation toward other cell types as shown by the expression of E-cadherin, which decreased both in the mock-infected cells and in cells expressing Pim-1 siRNA (Figure 7B), and by the muscle marker Myogenin and the endodermal marker α-1 fetoprotein, which were strongly increased in these cells (Table 2). As shown in Figure 7B, FACS analysis of endothelial markers revealed a significant reduction of PECAM1 and Flk-1 from day 5.5, while reduction of VE-cadherin was clearly observed at 8.5 day of ES cell differentiation. The expression of VE-cadherin could be appreciated only later, as this membrane protein is a late marker in the EC differentiation process.36 Similar reduction was observed for the mural markers PDGFR-β and α-SMA, thus suggesting that Pim-1 is required for the establishment of ECs and SMCs from their precursor.
Pim-1 is required for vasculogenesis and angiogenesis in vitro. (A) Western blot analysis of Pim-1 silencing obtained by infecting ES cells with a lentivirus vector expressing Pim-1 siRNA. (B) FACS analysis of differentiated ES cells at 5.5 and 8.5 days using specific antibodies for each marker as indicated. Green lines indicate the positive cell population, and numbers above each curve indicate the percentage of positive cells compared with red lines, which represent negative controls. ES cells were infected with a lentivirus vector generating, under the control of the U6 promoter, siRNA for renilla luciferase gene (control) or the murine Pim-1. (C) HUVECs were infected with the lentiviral vector expressing siRNA to silence the renilla luciferase (control) or human PIM-1 under the control of the U6 polymerase promoter. Pim-1 expression in HUVECs was valued by Western blot at 2 hours after VEGF-A induction. (D) PIM-1 silencing affects both proliferation and migration of HUVECs in vitro. Vertical bars represent the fold induction as indicated. Proliferation and migration tests were performed as described.24 The data are the results of 3 independent experiments. (E) Pim-1 silencing inhibited the angiogenesis assay in vitro. A representative experiment is shown. The assay was performed by plating HUVECs either infected with the mock vector or infected with the siRNA lentiviral vector on Matrigel, and HUVECs were induced with VEGF-A as described.24 (F) PIM-1 silencing inhibited sprouting of ECs. A representative experiment is shown. The sprouting assay was performed as described.35 Original magnification × 40.
Pim-1 is required for vasculogenesis and angiogenesis in vitro. (A) Western blot analysis of Pim-1 silencing obtained by infecting ES cells with a lentivirus vector expressing Pim-1 siRNA. (B) FACS analysis of differentiated ES cells at 5.5 and 8.5 days using specific antibodies for each marker as indicated. Green lines indicate the positive cell population, and numbers above each curve indicate the percentage of positive cells compared with red lines, which represent negative controls. ES cells were infected with a lentivirus vector generating, under the control of the U6 promoter, siRNA for renilla luciferase gene (control) or the murine Pim-1. (C) HUVECs were infected with the lentiviral vector expressing siRNA to silence the renilla luciferase (control) or human PIM-1 under the control of the U6 polymerase promoter. Pim-1 expression in HUVECs was valued by Western blot at 2 hours after VEGF-A induction. (D) PIM-1 silencing affects both proliferation and migration of HUVECs in vitro. Vertical bars represent the fold induction as indicated. Proliferation and migration tests were performed as described.24 The data are the results of 3 independent experiments. (E) Pim-1 silencing inhibited the angiogenesis assay in vitro. A representative experiment is shown. The assay was performed by plating HUVECs either infected with the mock vector or infected with the siRNA lentiviral vector on Matrigel, and HUVECs were induced with VEGF-A as described.24 (F) PIM-1 silencing inhibited sprouting of ECs. A representative experiment is shown. The sprouting assay was performed as described.35 Original magnification × 40.
Expression levels in ES cells infected with Pim-1 RNAi versus Flk1+/+ ES cells at 5.5 and 8.5 days of differentiation in vitro
. | Fold increase in Pim-1 RNAi . | . | |
|---|---|---|---|
| Gene . | Day 5.5 . | Day 8.5 . | |
| Flk-1 | −2.7 ± 0.4 | −14.2 ± 21 | |
| PECAM1 | −2.8 ± 0.3 | −4.5 ± 0.5 | |
| VE-cadherin | −8.8 ± 1.2 | −42 ± 6.3 | |
| Tie-2 | −2.9 ± 0.3 | −35.0 ± 4.4 | |
| Flt4 | −5.9 ± 1.0 | −1.4 ± 0.1 | |
| VEGF-A | −12.3 ± 1.1 | −1.3 ± 0.1 | |
| VEGF-D | −1.6 ± 0.2 | −2.5 ± 0.4 | |
| α-SMA | −4.6 ± 0.6 | −2.5 ± 0.3 | |
| SM22α | −4.5 ± 0.5 | −2.6 ± 0.4 | |
| Calponin-h1 | −7.3 ± 0.9 | −1.7 ± 0.2 | |
| SM-MHC | −3.7 ± 0.3 | +1.7 ± 0.1 | |
| Brachyury | −37.3 ± 4.9 | +1.0 ± 0.1 | |
| α 1-fetoprotein | −1.1 ± 0.1 | +143 ± 18 | |
| Nestin | +1.4 ± 0.2 | −3.5 ± 0.5 | |
| Myogenin | +81.2 ± 11 | +129.0 ± 15 | |
. | Fold increase in Pim-1 RNAi . | . | |
|---|---|---|---|
| Gene . | Day 5.5 . | Day 8.5 . | |
| Flk-1 | −2.7 ± 0.4 | −14.2 ± 21 | |
| PECAM1 | −2.8 ± 0.3 | −4.5 ± 0.5 | |
| VE-cadherin | −8.8 ± 1.2 | −42 ± 6.3 | |
| Tie-2 | −2.9 ± 0.3 | −35.0 ± 4.4 | |
| Flt4 | −5.9 ± 1.0 | −1.4 ± 0.1 | |
| VEGF-A | −12.3 ± 1.1 | −1.3 ± 0.1 | |
| VEGF-D | −1.6 ± 0.2 | −2.5 ± 0.4 | |
| α-SMA | −4.6 ± 0.6 | −2.5 ± 0.3 | |
| SM22α | −4.5 ± 0.5 | −2.6 ± 0.4 | |
| Calponin-h1 | −7.3 ± 0.9 | −1.7 ± 0.2 | |
| SM-MHC | −3.7 ± 0.3 | +1.7 ± 0.1 | |
| Brachyury | −37.3 ± 4.9 | +1.0 ± 0.1 | |
| α 1-fetoprotein | −1.1 ± 0.1 | +143 ± 18 | |
| Nestin | +1.4 ± 0.2 | −3.5 ± 0.5 | |
| Myogenin | +81.2 ± 11 | +129.0 ± 15 | |
Data are presented as mean ± SEM. The fold change detected by quantitative RT-PCR represents the average among the results of 4 independent measurements.
To verify the role of PIM-1 in ECs, we performed functional tests by silencing PIM-1 in HUVECs, which were then induced with VEGF-A (Figure 7C). Gene silencing was obtained by expressing PIM-1 siRNA under the control of the U6 promoter in lentiviral vector. The mock-infected HUVECs responded to VEGF-A with proliferation and migration, while we observed a significant inhibition of both VEGF-A-dependent proliferation and migration in cells expressing PIM-1 siRNA (Figure 7D). In addition, PIM-1 knock down inhibited capillary formation on matrigel and endothelial cell sprouting (Figure 7E-F). These data strongly suggest that PIM-1 is a bona fide downstream KDR effector in HUVECs.
Discussion
VEGF-A/Flk-1 signaling plays a crucial role in the vasculogenic and angiogenic processes, however little was known about the genes regulated by this signaling. The present study reports, for the first time, the identification of VEGF-A/Flk-1-regulated genes during vasculogenesis and angiogenesis and demonstrates the functional role of Pim-1 in these processes. To identify Flk-1 target genes, we screened for differentially expressed genes in Flk-1 wild type versus Flk-1 knock out at the early stages of EC differentiation. This screening identified several new genes previously not known to be involved in angiogenesis.
The in vitro model of ES cell differentiation adopted in this study is based on the induction of ES cells with VEGF-A in chemically controlled medium isolating the differentiating precursors on a selective substrate. This approach permits full differentiation between ECs and SMCs, as it allows the cross-talk between their precursors. Moreover, under these stringent conditions EC and SMC differentiation is strictly dependent on the expression of Flk-1. This was shown by the fact that in these conditions Flk-1 mutant ES cells could not differentiate in vitro toward ECs and SMCs. On the contrary, we (this work) and others29,30 observed that with FCS, Flk-1 mutant ES cells could still differentiate, albeit at low efficiency, into ECs. This implies that in the presence of other growth factors, endothelial precursors can, at least in part, bypass the Flk-1 requirement for endothelial differentiation.
The transcription profile of Flk-1 wild-type endothelial precursors, performed with a subtraction library, identified 134 transcripts. The expression of a group of potential regulators was confirmed by quantitative RT-PCR and further analyzed. The analysis of expression, at the RNA and protein level, demonstrated their specific induction in ECs in vivo in the angiogenesis of the ovary, while the analysis in HUVECs revealed that these genes were induced by VEGF-A with fast kinetics. To our knowledge, none of the genes identified in this screening was previously known to be an Flk-1 target gene. Several genes identified were previously described to play a role in endothelial differentiation and angiogenesis, thus confirming that the screening was giving the right genes. For the remaining genes, the relation with vasculogenesis and/or angiogenesis was less obvious. Several genes, previously identified on diverse cell types, were selectively up-regulated in ECs in angiogenesis of the ovary and induced by the KDR signaling in HUVECs. It is worth noting that most of these genes have been described as expressed and/or as playing a role in cells of hematopoietic origins. As Flk-1 marks the common progenitor of vascular and hematopoietic cells, it is possible that the abovementioned genes are regulated by Flk-1 signaling in the hemangioblast. However, we have no evidence to support this hypothesis, which should be further investigated. In fact, the gene expression analysis reported in this work has been performed in Flk-1-positive endothelial precursors expressing endothelial markers and demonstrated their expression in adult ECs—after the separation from the hematopoietic lineage. As Flk-1 expression persists in the angioblast that will originate both ECs and SMCs, one must assume that in hematopoietic precursors these genes are regulated by other pathways. This is the case with Slug, which was specifically expressed in our analysis under the direct signaling of Flk-1 and has been shown to be induced in primitive hematopoietic cells by stem cell factor (SCF)/c-kit signaling.37
As hematopoietic and vascular cells have a common precursor, the expression of this group of genes might unveil similar functions and provide hints of their role in ECs. All of these genes are expressed at early stages of hematopoietic cell differentiation and all together contribute to different functions of these cells, including cell proliferation, survival, differentiation, and migration. The nuclear factors Nm23-M1 and Nm23-M2 appear to play a role at early stages of hematopoietic differentiation, although their precise function remains elusive.38 The nonreceptor tyrosine kinase Btk is a component of the B-cell receptor signaling involved in B-cell development and antibody production.39 Ship-1 is a negative modulator of the signaling in hematopoietic cells.40 Pim-1 is expressed in immature hematopoietic cells and was first identified as a common integration site in MoMuLV-induced murine T-cell lymphomas.41,42 Pim-1 is a serine/threonine kinase able to phosphorylate and regulate the activity of transcription factors.33,43
Finally, the general transcription regulators Set, pp32, and Cbp were regulated by the Flk-1 signaling. Set and pp32 may play a role in the establishment and maintenance of the “histone code” of the chromatin, as both were found to be subunits of the inhibitor of acetyltransferases (INHAT),44 while Cbp codes for a general transcription coactivator that also has acetyltransferase activity.45 It must be pointed out that during the induction of the angiogenesis of the ovary, Cbp up-regulation was not limited to the ECs. This expression pattern, per se, does not exclude a role of this gene in endothelial precursors during vasculogenesis or in ECs during angiogenesis. In fact, it has been observed that Cbp knock outs, which express a truncated form of the protein, die at about day 9.5 with hematopoietic and vascular defects.46
Functional analysis of Pim-1 in vasculogenesis and angiogenesis was also performed. Pim-1 represented a good candidate as it was specifically expressed in ECs during angiogenesis, it codes for a nuclear kinase able to phosphorylate different substrates, and no previous role in vasculogenesis or angiogenesis for this gene was found. We performed the functional analysis by expressing Pim-1 RNAi in ES cells and HUVECs. These experiments demonstrated that Pim-1 is indeed required for the developmental differentiation of endothelial precursors as well as mural cells. For the latter, however, it is possible that their differentiation inhibition is due to the lack of inductive signals due to the reduction of ECs. This seems to be confirmed by the specific expression of Pim-1 in endothelial cells in vivo, although further experiments are needed as, during vasculogenesis, Pim-1 could play a role in the common precursor. Pim-1 depletion in ES cells also caused a reduction of Flk-1 expression on the cell surface. This regulation is likely to be indirect due to the reduction of ECs, although this point needs further investigation. In HUVECs, PIM-1 was shown to be required for VEGF-A-dependent proliferation and migration, thus demonstrated to be a bona fide effector of VEGF-A/Flk-1 not only in vasculogenesis but also in angiogenesis. These results in vitro contrast with Pim-1 knock out, which did not show major vasculogenic defects.31,32 This discrepancy could be explained by gene compensations in the knock-out animals, similar to the behavior of the Rb gene where it has been shown that the conditional inactivation of the gene in adult mice induces different phenotypic consequences when compared with its germ-line loss.47 Pim-1 regulates by phosphorylation proteins implicated in cell cycle progression, such as CDC25, and cell differentiation, such as c-Myb.33,43,48 It is therefore likely that the VEGF-A/Flk-1 pathway induces Pim-1, which then phosphorylates yet unknown targets involved in amplification and differentiation of endothelial cells.
The inhibition of VEGF-A/Flk-1 signaling has proved to be effective in limiting the angiogenesis implicated in tumor progression.4,49 In this respect, Pim-1 may represent a new candidate for inhibition of angiogenesis in adults. Given that Pim-1 is a serine/threonine kinase,42 specific small molecule kinase inhibitors could be developed for use as selective angiogenesis blockers.50
Taken together, the genes identified in this screening support the idea that Flk-1 is required for the early stages of vasculogenesis, which consist of the expansion and differentiation of endothelial progenitors as well as the reprogramming of quiescent ECs of vessels that happens during angiogenesis. To activate these cellular functions, Flk-1 expressed in ECs and their precursors induces the expression of a series of genes that could be cell-type specific but could also be not exclusive to endothelial cells.
As VEGF-A/Flk-1 plays a central role in vasculogenesis and angiogenesis inducing different functions in activated cells, the genes identified in this study should provide a valuable resource to better understand these processes.
Supported by grants from Associazione Italiana Ricerca sul Cancro (AIRC), Ministero Italiano dell' Istruzione, dell'Università e della Ricerca (MIUR), and Fondazione Monte Paschi Siena (MPS).
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Prepublished online as Blood First Edition Paper, February 24, 2004; DOI 10.1182/blood-2003-11-3827.
The authors thank F. Bussolino, I. Delany, and N. Valiante for critical reading of the manuscript; B. Grandi for technical help; Chiron SpA for the DNA sequence and FACS facilities; W. L. Stanford for Flk-1-/- ES cells; and L. Naldini for the lentiviral vector.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal