Abstract
The autoimmune/lymphoproliferative syndrome (ALPS) displays defective function of Fas, autoimmunities, lymphadenopathy/splenomegaly, and expansion of CD4/CD8 double-negative (DN) T cells. Dianzani autoimmune/lymphoproliferative disease (DALD) is an ALPS variant lacking DN cells. Both forms have been ascribed to inherited mutations hitting the Fas system but other factors may be involved. A pilot cDNA array analysis on a DALD patient detected overexpression of the cytokine osteopontin (OPN). This observation was confirmed by enzyme-linked immunosorbent assay (ELISA) detection of higher OPN serum levels in DALD patients (n = 25) than in controls (n = 50). Analysis of the OPN cDNA identified 4 polymorphisms forming 3 haplotypes (A, B, and C). Their overall distribution and genotypic combinations were different in patients (N = 26) and controls (N = 158) (P < .01). Subjects carrying haplotype B and/or C had an 8-fold higher risk of developing DALD than haplotype A homozygotes. Several data suggest that these haplotypes influence OPN levels: (1) in DALD families, high levels cosegregated with haplotype B or C; (2) in healthy controls, haplotype B or C carriers displayed higher levels than haplotype A homozygotes; and (3) in AB and AC heterozygotes, mRNA for haplotype B or C was more abundant than that for haplotype A. In vitro, exogenous OPN decreased activation-induced T-cell death, which suggests that high OPN levels are involved in the apoptosis defect.
Introduction
The autoimmune lymphoproliferative syndrome (ALPS), previously described as Canale-Smith Syndrome,1 is characterized by (1) defective function of the death receptor Fas involved in switching off the immune response, (2) autoimmunities that predominantly involve blood cells (ie, thrombocytopenia, anemia, neutropenia), (3) polyclonal accumulation of lymphocytes in the spleen and lymph nodes, and (4) expansion of CD4/CD8 double-negative (DN) T cells in the peripheral blood.2-4 The classic form of ALPS is due to deleterious mutations of the Fas or FasL genes2-4 and corresponds to those displayed by MLRlpr/lpr and MLRgld/gld mice developing an ALPS-like pattern.5
Our group has since identified several unrelated patients with an ALPS-like clinical pattern that fulfills the first 3 criteria but lacks expansion of DN T cells. Moreover, they did not display Fas or FasL mutations. Most of their parents were Fas-resistant and we suggested involvement of inherited mutations hitting the Fas pathway downstream from the receptor. Since the complete paradigm of ALPS could not be demonstrated, we provisionally named this disease autoimmune lymphoproliferative disease (ALD).6-8 The possibility that mutations acting downstream from Fas may cause autoimmune/lymphoproliferative patterns has then been confirmed by Wang et al9 in 2 unrelated patients with typical ALPS who carried mutations of the caspase 10 gene. These authors coined the terms ALPS-Ia and -Ib for the disease with mutations of Fas and FasL, respectively, ALPS-II for the mutation of caspase 10, and ALPS-III for the unknown mutations of the signaling pathway. McKusick has proposed Dianzani ALD (DALD) as the name for the disease lacking Fas/FasL mutations and expansion of DN cells to avoid confusion with the ALD acronym used for other diseases (OMIM reference no. 605233; http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM).
ALPS-like disorders do not behave as classical monogenic diseases. This is true in lpr and gld mice and even more evident in ALPS and DALD. The lpr and gld mutations cause the disease in homozygosity but its expression greatly depends on the genetic background, since it is much milder in BALB/c than in MLR mice.10 Most ALPS-Ia patients are heterozygous for the Fas mutation, but the parent carrying the mutation is generally healthy. Other complementary factors may thus be required in function of the severity of the mutation.11,12 The same observation is true in DALD patients, since both parents display generally defective Fas function but are healthy.6-8
The aim of this work was to identify factors involved in the development of DALD. In a pilot study, cDNA arrays used to analyze the expression of genes involved in apoptosis and proliferation in T cells from a DALD patient detected a striking up-regulation of the gene of osteopontin (OPN), an arginine-glycine-aspartate (RGD)–containing cytokine binding to several integrins and the CD44v6-7 isoforms.13-17 OPN is constitutively expressed by bone and several epithelial tissues where it is involved in bone remodeling, tissue repair, and cell migration,14 whereas in endothelial cells, macrophages, and smooth muscle cells it is mainly expressed upon activation in inflammatory contexts.18 Moreover, it is abundantly expressed by natural killer (NK) cells and activated T cells where it has been described as Eta-1 (early T-cell activation-1) because of its early production upon cell activation and has been shown to enhance the T-cell helper 1 (TH1) and inhibit the TH2 responses.19,20 The high OPN levels in DALD were intriguing in light of previous works detecting OPN up-regulation in MLRlpr/lpr mice and showing that when they are crossed with OPN–/– mice they display delayed onset of polyclonal B-cell activation.21,22 Moreover, high levels of OPN have also been observed in systemic lupus erythematosus (SLE),18,23 rheumatoid arthritis (RA),24 and multiple sclerosis (MS).25 This background prompted our analysis of the OPN levels and gene in DALD patients. Increased OPN serum levels correlated with distinct gene polymorphisms. In T-cell cultures, exogenous OPN inhibited activation-induced cell death (AICD). These data together suggest that high OPN levels associated with OPN polymorphisms favor development of autoimmunity/lymphoproliferation.
Patients, materials, and methods
Patients
We analyzed 26 (17 males, 7 females) DALD patients observed at the Pediatric Department, University of Turin, Italy (some data has already been presented in Ramenghi et al7 ). Diagnosis was based on the presence of all of the following criteria: (1) presence of autoimmune cytopenia involving one or more hematologic lineage; (2) presence of chronic nonmalignant lymphadenopathy (2 or more lymph nodes enlarged over 2 cm in diameter) and/or splenomegaly; (3) defective Fas-induced apoptosis in vitro; (4) lack of mutations in the Fas, FasL, or caspase 10 genes; (5) lack of DN T-cell expansion in the peripheral blood. Fas function was evaluated as previously reported.6,7 The Fas, FasL, and caspase 10 genes were sequenced from genomic DNA to rule out known mutations causing ALPS. Clinical and serologic features of patients are summarized in Table 1. Peripheral blood specimens and serum were obtained from patients, their relatives, and healthy controls with written informed consent. The study was planned according to the guidelines of the local ethical committee.
Clinical data of DALD patients
Patient no. . | Sex . | Age, y* . | IgG, mg/dL† . | OPN, ng/mL† . | OPN genotype . | ANA . | AM . | HM . | SM . | Autoimmunities . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 6 | 966 | 332 | AB | + | + | + | + | AHA, AN, T, RF |
| 2 | M | 10 | 1100 | 254 | AA | - | + | - | + | AN, E |
| 3 | M | 16 | 506 | ND | CC | - | + | - | + | AN, T |
| 4 | M | 9 | 1143 | 286 | AC | + | - | - | + | AHA, A |
| 5 | F | 18 | 2775 | 426 | AB | + | - | - | + | AN, RF |
| 6 | M | 16 | 2810 | 405 | CC | - | + | - | + | T, IH |
| 7 | F | 24 | 1259 | 337 | AC | + | - | + | + | AHA, IH |
| 8 | F | 15 | 1019 | 222 | AC | + | - | - | + | AHA, T, A1 |
| 9 | M | 16 | 1132 | 340 | AB | + | - | + | + | AHA, T, IH |
| 10 | M | 11 | 679 | 90 | AA | + | + | + | + | AHA, T, V |
| 11 | F | 11 | 1542 | 347 | AB | + | + | - | + | AHA, |
| 12 | F | 17 | 836 | 211 | AC | +/- | - | - | + | AHA, T, E |
| 13 | M | 15 | 1153 | 298 | AC | - | + | - | - | T |
| 14 | M | 5 | 900 | 156 | AC | + | + | - | + | T |
| 15 | M | 10 | 1117 | 226 | AC | + | + | + | + | AN, T, IH |
| 16 | M | 33 | 984 | 494 | BC | ND | + | + | + | AN, T, A1 |
| 17 | M | 6 | 878 | 109 | CC | + | + | - | - | T |
| 18 | F | 7 | 1180 | 167 | AA | + | + | + | + | AHA, AN, T, E, A, C, SR, RF |
| 19 | M | 10 | 830 | 103 | AC | + | + | - | + | IDDM, T, A1, AN |
| 20 | M | 6 | ND | 37 | AC | + | + | - | + | AN, T |
| 21 | M | 13 | ND | 383 | AC | - | + | - | + | AN |
| 22 | F | 3 | ND | 187 | AB | + | + | + | + | SR |
| 23 | M | 17 | 2110 | 356 | AC | + | + | + | + | AHA, AN, T, RF |
| 24 | F | 9 | 842 | 147 | AC | + | - | - | + | T |
| 25 | M | 2 | ND | 512 | AC | + | + | - | + | AHA, IH, T |
| 26 | M | 11 | ND | 365 | AB | ND | + | + | + | AN, T |
Patient no. . | Sex . | Age, y* . | IgG, mg/dL† . | OPN, ng/mL† . | OPN genotype . | ANA . | AM . | HM . | SM . | Autoimmunities . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 6 | 966 | 332 | AB | + | + | + | + | AHA, AN, T, RF |
| 2 | M | 10 | 1100 | 254 | AA | - | + | - | + | AN, E |
| 3 | M | 16 | 506 | ND | CC | - | + | - | + | AN, T |
| 4 | M | 9 | 1143 | 286 | AC | + | - | - | + | AHA, A |
| 5 | F | 18 | 2775 | 426 | AB | + | - | - | + | AN, RF |
| 6 | M | 16 | 2810 | 405 | CC | - | + | - | + | T, IH |
| 7 | F | 24 | 1259 | 337 | AC | + | - | + | + | AHA, IH |
| 8 | F | 15 | 1019 | 222 | AC | + | - | - | + | AHA, T, A1 |
| 9 | M | 16 | 1132 | 340 | AB | + | - | + | + | AHA, T, IH |
| 10 | M | 11 | 679 | 90 | AA | + | + | + | + | AHA, T, V |
| 11 | F | 11 | 1542 | 347 | AB | + | + | - | + | AHA, |
| 12 | F | 17 | 836 | 211 | AC | +/- | - | - | + | AHA, T, E |
| 13 | M | 15 | 1153 | 298 | AC | - | + | - | - | T |
| 14 | M | 5 | 900 | 156 | AC | + | + | - | + | T |
| 15 | M | 10 | 1117 | 226 | AC | + | + | + | + | AN, T, IH |
| 16 | M | 33 | 984 | 494 | BC | ND | + | + | + | AN, T, A1 |
| 17 | M | 6 | 878 | 109 | CC | + | + | - | - | T |
| 18 | F | 7 | 1180 | 167 | AA | + | + | + | + | AHA, AN, T, E, A, C, SR, RF |
| 19 | M | 10 | 830 | 103 | AC | + | + | - | + | IDDM, T, A1, AN |
| 20 | M | 6 | ND | 37 | AC | + | + | - | + | AN, T |
| 21 | M | 13 | ND | 383 | AC | - | + | - | + | AN |
| 22 | F | 3 | ND | 187 | AB | + | + | + | + | SR |
| 23 | M | 17 | 2110 | 356 | AC | + | + | + | + | AHA, AN, T, RF |
| 24 | F | 9 | 842 | 147 | AC | + | - | - | + | T |
| 25 | M | 2 | ND | 512 | AC | + | + | - | + | AHA, IH, T |
| 26 | M | 11 | ND | 365 | AB | ND | + | + | + | AN, T |
ANA indicates antinuclear antibodies; AM, adenomegaly; HM, hepatomegaly; SM, splenomegaly; +, presence; -, absence; AHA, autoimmune hemolytic anemia; AN, autoimmune neutropenia; T, thrombocytopenia; RF, recurrent fever; E, eczema; ND, not determined; A, arthritis; IH, immune hepatitis; AI, alopecia; V, vitiligo; C, celiac disease; SR, skin rush; and IDDM, insulin-dependent diabetes mellitus.
Age at study.
Serum levels; 95th percentile of control OPN levels = 298 ng/mL.
T-cell isolation and immunophenotype analysis
Peripheral blood mononuclear cells (PBMCs) were prepared by gradient centrifugation. Expression of surface molecules was evaluated by direct immunofluorescence and cytofluorimetric analysis (FACScan; Becton Dickinson, San Jose, CA). The following monoclonal antibodies (mAbs) were used: anti-CD3 (Leu-4), anti-CD4 (Leu-3a), anti-CD8 (Leu-2a), anti–T-cell receptor αβ (anti-TCRαβ; Becton Dickinson), anti-Fas (Immunotech, Marseilles, France). CD4 and CD8 DN TCRαβ-positive cells were detected by 2-color immunofluorescence using fluorescein isothiocyanate (FITC)–conjugated anti-TCRαβ mAb and phycoerythrin (PE)–conjugated anti-CD4 and anti-CD8 mAbs.
Array hybridization, probe preparation, and macroarray analysis
Two sets of macroarray membranes were purchased from Sigma-Genosys (London, United Kingdom): Panorama Human Apoptosis Gene Arrays (PRAP0002) and Panorama Human Cytokine Gene Arrays (PRCK0002). For preparation of cDNA probes, 2 μg of total RNA was isolated from 6-day cultured PBMCs and reverse transcripted according to the manufacturer. Membranes were prehybridized for 2 hours at 65°C and denatured 32P-labeled probes were incubated at 65°C. Membranes were washed twice with 2× sodium chloride sodium citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) at 65°C for 15 minutes and then twice in 0.2× SSC/0.1% SDS at 65°C for 30 minutes and analyzed with a Phosphoimage scanner (BioRad, Hercules, CA).
OPN ELISA assay and cytokine measurements
Serum concentration of OPN was measured by capture enzyme-linked immunosorbent assay (ELISA) according to the protocol provided by the manufacturer (Calbiochem, San Diego, CA). The optical density was measured at 450 nm using a microplate reader (BioRad).
Serum levels of interleukin 2 (IL-2), interferon γ (IFN-γ), IL-4, IL-6, IL-10, and tumor necrosis factor α (TNF-α) levels were evaluated in a cytometric bead array kit (Becton Dickinson).
OPN haplotype determination
Primers used to analyze the OPN gene were as follows: OPN-1 (forward) 5′-AGACAGAGCATCGTCGGG-3′; OPN-2 (reverse) 5′-GGGGAGCTCCTTATTACATTCAAGATA-3′; OPN-3 (reverse) 5′-GGCTGTGGAATTCACGGC-3′; OPN-4 (forward) 5′-GCCGTGAATTCCACAGCC-3′; OPN-7 (forward) 5′-CCTCGGATAACCTAAAAGCCATGGT-3′; and OPN-8 (reverse) 5′-GCTCTAGACCACCAAATTCTTATTACATTCAAG-3′.
The cDNA was obtained from 10 μg of total RNA from PBMCs cultured for 6 days after activation with a Superscript II reverse transcriptase (RT) kit using oligo-dT following the manufacturer's manual (Invitrogen, Carlsbad, CA). Primers for polymerase chain reaction (PCR) amplification of all human OPN were OPN-1 and OPN-2. PCR products were purified and cloned into the pGEM-T vector (Promega, Madison, WI). Ten to 20 independent colonies were amplified with the primers set described above and sequenced with the primers OPN-1, -2, -3, and -4.
As the template for amplification of the genomic region comprising exon VI and exon VII, 200 ng of genomic DNA was used. The amplification products to evaluate polymorphisms at positions 282 and 750 were generated using the primers OPN-7 and -3; the amplification primers to evaluate polymorphisms at positions 1083 and 1239 were generated using the primers OPN-4 and -8. PCR products were sequenced with the primers OPN-7 and -3 for the first fragment and OPN-4 for the second fragment. Sequencing was performed with the ABI PRISM BigDye Terminator kit (Applied Biosystems, Foster City, CA) on an automatic sequencer (Applied Biosystems 3100 Genetic Analyser).
Functional assays
PBMCs (106) were grown in RPMI + 10% fetal calf serum (FCS) supplemented with IL-2 (10 U/mL) with or without recombinant OPN (rOPN; 500ng/mL; Calbiochem) and live cells were counted at days 4, 6, 11, and 14 after activation with phytohemagglutinin (PHA; 1 μg/mL).
Cell death assays were performed on these cells at day 6 after activation. Briefly, 5 × 104 cells were incubated with RPMI + 5% FCS + 1 U/mL IL-2 in the presence or absence of rOPN (500 ng/mL). Fas-induced cell death was evaluated by adding soluble CH11 anti-Fas mAb (1μg/mL), whereas AICD was evaluated by incubating cells in wells coated with the anti-CD3 OKT3 mAb (10 μg/mL). After a 16-hour incubation, live cells were counted with the trypan blue exclusion test or cytofluorimetric detection of apoptotic cells after staining with propidium iodide and FITC-conjugated annexin V. Assays were performed in triplicate and analyzed by a blind observer. Results are expressed as relative cell survival percentage calculated as follows: (total live cell count in the assay well/total live cell count in the respective control well) × 100. Spontaneous cell loss in the control wells was always less than 10% of the seeded cells.
Evaluation of cell staining with annexin V was performed using the Annexin V–Fluos kit (Boehringer Mannhein, Gmbh, Mannheim, Germany). Briefly, cells were stained with annexin V (1:50) and propidium iodide (1 μg/mL) in 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)/NaOH pH7.4, 140 mM NaCl, 5 mM CaCl2 and analyzed by flow cytometry. Live cells were those not displaying shrunken/hypergranular morphology and unstained by propidium iodide or annexin V.
Statistical analysis
For the ELISA experiments, the approximation of population distribution to normality was tested using statistics for kurtosis and symmetry. Results were asymmetrically distributed and hence presented as median values and percentiles. ELISA comparisons were performed with the nonparametric Mann-Whitney U test. Genotype and haplotype distributions were performed with the chi-square test. All P values are 2-tailed. The Pearson linear regression analysis was performed for correlation analysis between the OPN and immunoglobulin G (IgG) levels.
Maximum-likelihood estimation of haplotype frequency was calculated by the Arlequin software version 1.1 (Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland).
Results
Increased levels of OPN in DALD patients and their families
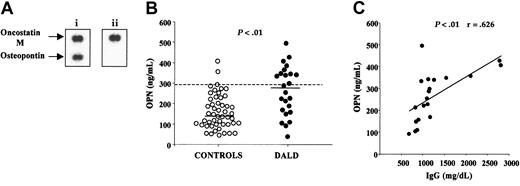
We used cDNA arrays to compare transcription of genes involved in cell apoptosis or proliferation in a DALD patient and her Fas-sensitive healthy brother. These arrays consisted of 573 selected cDNA fragments arrayed on nylon membranes that assessed expression of genes related to apoptosis and cell activation. The mRNA was obtained from T cells cultured for 6 days with PHA plus IL-2 to overcome differences due to the presence of higher counts of in vivo–activated cells in the patient. Expression of several genes was altered in the patient, but the most striking difference was up-regulation of the OPN gene (Figure 1A).
DALD patients display increased serum levels of OPN. (A) Autoradiographs of macroarray membranes hybridized with cDNA from a DALD patient (i) and from her healthy brother (ii). Bottom spots show OPN and top spots show oncostatin M as a control. (B) Serum concentrations of OPN (ng/mL) in 25 DALD patients and 50 healthy unrelated controls. The bold horizontal bars indicate the median values for each group; the dotted line, the 95th percentile of the controls. The P value of differences between patients and controls was calculated using the Mann-Whitney U test. (C) Correlation between OPN and IgG serum levels in DALD patients. The line is the best fitting regression line. The r and P values were evaluated using the Pearson correlation analysis. OPN levels were evaluated by ELISA.
DALD patients display increased serum levels of OPN. (A) Autoradiographs of macroarray membranes hybridized with cDNA from a DALD patient (i) and from her healthy brother (ii). Bottom spots show OPN and top spots show oncostatin M as a control. (B) Serum concentrations of OPN (ng/mL) in 25 DALD patients and 50 healthy unrelated controls. The bold horizontal bars indicate the median values for each group; the dotted line, the 95th percentile of the controls. The P value of differences between patients and controls was calculated using the Mann-Whitney U test. (C) Correlation between OPN and IgG serum levels in DALD patients. The line is the best fitting regression line. The r and P values were evaluated using the Pearson correlation analysis. OPN levels were evaluated by ELISA.
This observation prompted measurement of the OPN protein in the sera of 25 DALD patients and 50 healthy controls by ELISA. Figure 1B shows that DALD patients displayed significantly higher OPN serum levels (median, 286; range, 37-512 ng/mL) than the controls (median, 141; range, 45-405 ng/mL) (P < .001).
DALD patients often display altered levels of serum immunoglobulins (Table 1). Since OPN has been shown to modulate Ig production in mouse models, we compared the serum levels of OPN and IgG in DALD patients. Figure 1C shows a significant direct correlation between the 2 parameters: the higher the OPN levels, the higher the IgG levels (P < .01, r = 0.626).
OPN is produced during the immune response by several cell types and modulates T-cell differentiation. To assess whether the high levels of OPN in DALD patients are simply a marker of an activated immune response, we used a cytometric bead array kit to measure the serum levels of several cytokines (ie, IL-2, INF-γ, IL-4, IL-6, IL-10, and TNF-α) involved in the TH1 or TH2 response or macrophage activation in DALD patients and controls. Only IL-10 levels were significantly increased in the patients, but no correlation was found with their OPN levels (data not shown). Moreover, IL-10 levels were quite variable in different samples from the same patient, whereas OPN levels were consistent.
OPN polymorphisms are associated with DALD
Eleven single-nucleotide polymorphisms have been described in the OPN cDNA: one is in exon VI and 10 in exon VII (2 in the coding region and 8 in the 3′ untranslated region [UTR]; http://www.ncbi.nlm.nih.gov/SNP). To investigate the involvement of the OPN gene, we sequenced genomic DNA corresponding to exon VI and VII in 26 DALD patients and 158 ethnically matched healthy controls. Four known polymorphisms were identified in both patients and controls; they corresponded to positions +282T>C (exon VI), +750C>T (exon VII, coding region), +1083A>G, and +1239A>C (3′ UTR; numeration are referred to ATG =+1). Highly significant pairwise linkage disequilibrium was observed between alleles at these 4 polymorphism (P < 10–5). A maximum-likelihood haplotype frequency estimation yielded 3 likely haplotypic combinations: haplotype A (282T-750C-1083A-1239A), haplotype B (282C-750T-1083A-1239C), and haplotype C (282C-750T-1083G-1239C). All these haplotypes were directly observed in individuals homozygous at all, or all but one tested position and were confirmed by cloning and sequencing the entire OPN cDNA from 12 subjects heterozygotes at more than one position.
The genotypic distributions of the haplotypes did not deviate significantly from the Hardy-Weinberg equilibrium in both groups. Overall distributions of haplotypes and genotypes were significantly different in DALD patients and controls (P < .01; Table 2). Frequency of haplotype A was lower and haplotypes B and C were higher in DALD patients than in the controls. Haplotype B or C carriers had an 8-fold higher risk of developing DALD compared with homozygotes for haplotype A (odds ratio [OR] 7.86; 95% confidence interval (CI) 2.12 < 7.86 < 35).
Frequency distribution of OPN haplotypes and genotypes in 26 DALD patients and 158 controls
. | DALD . | . | Controls . | . | Statistic . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype/genotype* . | N† . | % . | N . | % . | P‡ . | OR . | 95% CI . | ||||
| A | 25 | 48 | 222 | 70 | .0027 | 0.39 | 0.21-0.74 | ||||
| B | 7 | 13.5 | 17 | 5 | .0595 | 2.74 | 0.90-7.40 | ||||
| C | 20 | 39.5 | 77 | 23 | .0491 | 1.94 | 0.99-3.73 | ||||
| AA | 3 | 11.5 | 80 | 51 | .0005 | 0.13 | 0.02-0.45 | ||||
| AB | 6 | 23 | 14 | 9 | NS | — | — | ||||
| AC | 13 | 50 | 48 | 30 | NS | — | — | ||||
| BC | 1 | 4 | 3 | 2 | NS | — | — | ||||
| CC | 3 | 11.5 | 13 | 8 | NS | — | — | ||||
. | DALD . | . | Controls . | . | Statistic . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype/genotype* . | N† . | % . | N . | % . | P‡ . | OR . | 95% CI . | ||||
| A | 25 | 48 | 222 | 70 | .0027 | 0.39 | 0.21-0.74 | ||||
| B | 7 | 13.5 | 17 | 5 | .0595 | 2.74 | 0.90-7.40 | ||||
| C | 20 | 39.5 | 77 | 23 | .0491 | 1.94 | 0.99-3.73 | ||||
| AA | 3 | 11.5 | 80 | 51 | .0005 | 0.13 | 0.02-0.45 | ||||
| AB | 6 | 23 | 14 | 9 | NS | — | — | ||||
| AC | 13 | 50 | 48 | 30 | NS | — | — | ||||
| BC | 1 | 4 | 3 | 2 | NS | — | — | ||||
| CC | 3 | 11.5 | 13 | 8 | NS | — | — | ||||
— indicates not evaluated; and NS, not significant.
Haplotype A (282T-750C-1083A-1239A), haplotype B (282C-750T-1083A-1239C), haplotype C (282C-750T-1083G-1239C); genotypes AA, AB, AC, BC, and CC; no subjects with the genotype BB were detected.
Number of chromosomes (for haplotypes) or number of subjects (for genotypes).
The haplotype overall P value = .0039; genotype overall P value = .0047.
OPN polymorphisms are associated with the OPN levels
These data suggest that haplotype B and C may be associated with production of high OPN levels, which in turn favor development of DALD. To assess this possibility we performed 3 sets of analyses.
First, we analyzed the serum levels and haplotypes in family members of DALD patients 1, 5, 11, and 21 (Table 1) who displayed OPN levels higher than the 95th percentile of the controls (OPNhigh phenotype). All the relatives were healthy and did not display signs of autoimmunity and/or lymphoproliferation, but their OPN levels (median, 286; range, 142-451 ng/mL) were higher than those of random controls (median, 141; range, 45-405 ng/mL) (P < .001). Moreover, the grandmother and the father of patient 1, the mother of patient 5, the father of patient 11, and the father of patient 21 displayed the OPNhigh phenotype with an inherited pattern compatible with a dominant transmission; OPN genotyping showed that this phenotype was cotransmitted with haplotype B or C in all (4/4) cases (data not shown).
Second, we compared the OPN serum levels of healthy subjects carrying or not carrying the haplotype B or C (ie, genotypes AB+BC+CC vs AA). We separately analyzed 2 groups of healthy subjects (ie, random controls and the relatives of DALD patients 1, 5, 11, and 21). In both groups, subjects carrying the haplotype B or C displayed significantly higher OPN levels than the AA homozygotes (Figure 2).
Healthy subjects carrying the haplotype B or C display higher serum levels of OPN than homozygotes for haplotype A. Two groups of subjects were evaluated (ie, random healthy subjects [n = 50] and the healthy relatives belonging to the families shown in Figure 3 [n = 12]). The genotypes are indicated with different symbols as shown in the figure. The bold horizontal lines indicate the median values for each group; the dotted line, the 95th percentile of the controls. The P values were calculated using the Mann-Whitney U test.
Healthy subjects carrying the haplotype B or C display higher serum levels of OPN than homozygotes for haplotype A. Two groups of subjects were evaluated (ie, random healthy subjects [n = 50] and the healthy relatives belonging to the families shown in Figure 3 [n = 12]). The genotypes are indicated with different symbols as shown in the figure. The bold horizontal lines indicate the median values for each group; the dotted line, the 95th percentile of the controls. The P values were calculated using the Mann-Whitney U test.
Third, we evaluated the mRNA level of each haplotype in 7 AC and 6 AB heterozygotes. Sequencing of 144 independent cDNA clones showed that haplotype A was always about 5-fold less expressed than haplotype B or C in each subject (Table 3).
mRNA level of each haplotype in 6 AB and 7 AC heterozygotes evaluated by sequencing of independent cDNA clones
. | . | . | Haplotype A clones† . | . | Haplotype B or C clones† . | . | ||
|---|---|---|---|---|---|---|---|---|
| Subject . | Genotype . | Total clones* . | n . | % . | n . | % . | ||
| 1 | AB | 8 | 0 | 0 | 8 | 100 | ||
| 2 | AB | 10 | 1 | 10 | 9 | 90 | ||
| 3 | AB | 15 | 3 | 20 | 12 | 80 | ||
| 4 | AB | 8 | 3 | 38 | 5 | 63 | ||
| 5 | AB | 8 | 1 | 13 | 7 | 88 | ||
| 6 | AB | 10 | 1 | 10 | 9 | 90 | ||
| 7 | AC | 10 | 2 | 10 | 18 | 90 | ||
| 8 | AC | 15 | 3 | 15 | 17 | 85 | ||
| 9 | AC | 36 | 4 | 36 | 7 | 64 | ||
| 10 | AC | 14 | 1 | 14 | 6 | 86 | ||
| 11 | AC | 20 | 2 | 20 | 8 | 80 | ||
| 12 | AC | 11 | 1 | 11 | 8 | 89 | ||
| 13 | AC | 25 | 2 | 25 | 6 | 75 | ||
| Total | — | 144 | 24 | 17 | 120 | 83 | ||
. | . | . | Haplotype A clones† . | . | Haplotype B or C clones† . | . | ||
|---|---|---|---|---|---|---|---|---|
| Subject . | Genotype . | Total clones* . | n . | % . | n . | % . | ||
| 1 | AB | 8 | 0 | 0 | 8 | 100 | ||
| 2 | AB | 10 | 1 | 10 | 9 | 90 | ||
| 3 | AB | 15 | 3 | 20 | 12 | 80 | ||
| 4 | AB | 8 | 3 | 38 | 5 | 63 | ||
| 5 | AB | 8 | 1 | 13 | 7 | 88 | ||
| 6 | AB | 10 | 1 | 10 | 9 | 90 | ||
| 7 | AC | 10 | 2 | 10 | 18 | 90 | ||
| 8 | AC | 15 | 3 | 15 | 17 | 85 | ||
| 9 | AC | 36 | 4 | 36 | 7 | 64 | ||
| 10 | AC | 14 | 1 | 14 | 6 | 86 | ||
| 11 | AC | 20 | 2 | 20 | 8 | 80 | ||
| 12 | AC | 11 | 1 | 11 | 8 | 89 | ||
| 13 | AC | 25 | 2 | 25 | 6 | 75 | ||
| Total | — | 144 | 24 | 17 | 120 | 83 | ||
Statistical analysis was performed on percentages using the Mann-Whitney U test and the proportions of haplotype A clones were significantly lower than that of haplotype B or C (P < .0001).
— indicates not applicable.
Total number of OPN cDNA clones analyzed in that subject.
Number (n) and percentage (%) of clones with haplotype A and B or C.
Effect of exogenous OPN on T-cell cultures
ALPS and DALD are due to an altered balance between lymphocyte proliferation and death that may cause both autoimmunity and accumulation of lymphocytes in the secondary lymphoid organs. To investigate the role of OPN in this balance, we evaluated the effect of addition of rOPN on the expansion and death response of cultured T cells.
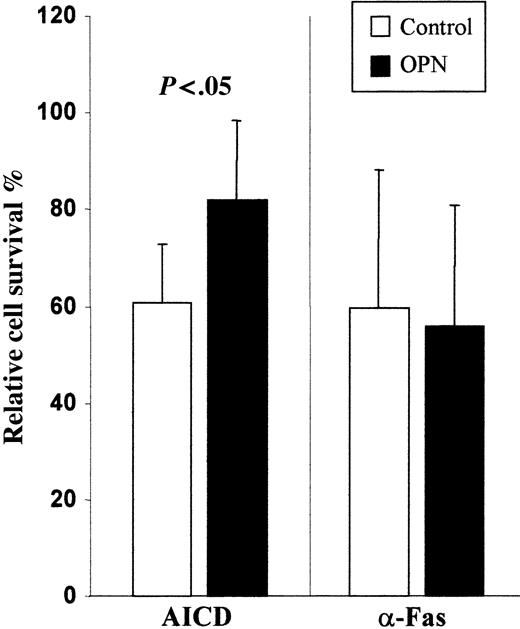
PBMCs obtained from healthy controls were activated with PHA + IL-2 and cultured for 14 days in the presence and absence of rOPN. Monitoring of T-cell expansion by counting live cells at days 4, 6, 11, and 14 showed that at each time rOPN slightly but significantly increased T-cell counts (data not shown). Moreover at day 6, we evaluated Fas-induced and AICD by triggering Fas or CD3, respectively, by mAb. Results showed that cells cultured with rOPN displayed significantly lower AICD than those cultured without it, whereas no significant difference was found in Fas-induced cell death (Figure 3). Phenotypic analysis of cells by immunofluorescence showed similar distribution of CD4+ and CD8+ cells and similar expression of Fas and FasL in cells cultured with or without rOPN. By contrast, rOPN did not display any substantial effect when it was only added at the time of the death assay to cells grown in its absence. Similar results were obtained by detecting apoptotic cells by annexin V staining (data not shown).
Recombinant OPN inhibits AICD but not Fas-induced cell death. T cells were grown for 6 days in the presence (▪) and absence (□) of rOPN. Then, Fas-induced cell death was triggered using a soluble anti-Fas mAb (CH11), whereas AICD was induced by incubating cells in wells coated with an anti-CD3 mAb (OKT3). The rOPN (500 ng/mL) was added in the assays involving the rOPN-grown cells. Cell survival was assessed after 18 hours. Results are expressed as relative survival percentage (ie, the proportion of surviving cells relative to the corresponding control, untreated or treated with rOPN). Results are the means ± SD of 5 experiments each performed in duplicate. The P value was calculated using the Mann-Whitney U test. Similar results were obtained in 2 experiments in which apoptotic cells were detected by propidium iodide and annexin V staining (data not shown).
Recombinant OPN inhibits AICD but not Fas-induced cell death. T cells were grown for 6 days in the presence (▪) and absence (□) of rOPN. Then, Fas-induced cell death was triggered using a soluble anti-Fas mAb (CH11), whereas AICD was induced by incubating cells in wells coated with an anti-CD3 mAb (OKT3). The rOPN (500 ng/mL) was added in the assays involving the rOPN-grown cells. Cell survival was assessed after 18 hours. Results are expressed as relative survival percentage (ie, the proportion of surviving cells relative to the corresponding control, untreated or treated with rOPN). Results are the means ± SD of 5 experiments each performed in duplicate. The P value was calculated using the Mann-Whitney U test. Similar results were obtained in 2 experiments in which apoptotic cells were detected by propidium iodide and annexin V staining (data not shown).
Discussion
This work shows that DALD patients produce high levels of OPN and suggests that OPN is involved in development of the autoimmune/lymphoproliferative pattern. The high OPN levels seem to be determined by a genetic component, since frequency of 3 haplotypes of the OPN gene were significantly altered in DALD patients with increased frequency of haplotype B and C and decreased frequency of haplotype A. Haplotype B or C carriers had an about 8-fold increased risk to develop DALD compared with homozygotes for haplotype A. Moreover, several relatives of DALD patients displayed high OPN levels and this phenotype was always cotransmitted with haplotype B or C.
These data suggest that haplotypes B and C are associated with production of high levels of OPN that, in turn, predispose patients to DALD. This possibility is supported by 2 sets of data. First, analysis of healthy subjects (both random subjects and DALD patients' relatives) showed that those carrying haplotypes B or C displayed higher OPN levels than homozygotes for haplotype A, which rules out the possibility that the high OPN levels were a consequence of the disease. Second, analysis of mRNAs for each haplotype in AC and AB heterozygotes showed that haplotype B and C are about 5-fold more expressed than haplotype A in each donor.
One possibility is that haplotypes B and C directly affect the transcription, maturation, or stability of the OPN mRNA. Alternatively, they can be in linkage disequilibrium with unidentified variations controlling OPN production. Increased stability could be directly ascribed to the changes characterizing haplotypes B and C. Both differ from haplotype A because of 2 synonymous variations in the coding region and one variation in the 3′ UTR. Haplotype C differs on account of a further variation in the 3′ UTR. The 3′ UTR variations seem to be particularly interesting, since polymorphisms in this region have been shown to stabilize the mRNA in other gene systems.26
OPN may favor development of DALD by either concurring in the apoptosis defect or influencing the immune effector response. The first model is suggested by the OPN inhibitory effect on AICD, which is a complex death response that involves up-regulation of both Fas and FasL, and other receptor systems such as those of TNF, TNF-related apoptosis-inducing ligand (TRAIL), and possibly IFN-γ.27-30 Since OPN did not inhibit cell death induced by anti-Fas mAb, it might exert its inhibitory effect by altering the function of the other receptor systems. This further apoptosis defect might exacerbate that due to the inherited alterations of the Fas pathway displayed by these patients. AICD, indeed, is involved in switching off the immune response and is defective in both DALD and ALPS (Fisher et al3 and A.C., unpublished observation, 2002).
The second model is suggested by the known effect of OPN on the immune effector response. In T cells, OPN potentiates proliferation, IFN-γ production, and CD40L expression, which in turn favor B-cell proliferation and antibody production.31 It is noteworthy that transgenic mice overexpressing OPN display accumulation of B1 lymphocytes, autoantibody production, and hypergammaglobulinemia, which recalls both the autoimmune/lymphoproliferative pattern and the direct correlation between OPN level and hypergammaglobulinemia in DALD patients.32 The high IL-10 levels displayed by both DALD and ALPS patients seem not to be in line with the pro-TH1 and anti-TH2 effect ascribed to OPN. However, a similar pattern, with high levels of both IL-10 and OPN, is also displayed by MLRlpr/lpr mice whose disease has been associated with a predominant TH1 response.21,33-35 The observation that an accelerated disease correlates with enhanced expression of IFN-γ versus IL-10 in MLRlpr/lpr substrains and that IL-10–deficient lpr/lpr mice develop an exacerbated disease suggests that IL-10 may be produced to counteract the TH1 response.35,36 However in mice, IL-10 has also been involved in some pathologic aspects of the disease such as production of antierythrocyte autoantibodies.37 In vitro studies in lpr/lpr mice suggested that, among lymphocytes, OPN is mainly produced by DN T cells, which are expanded in ALPS but not in DALD.38,39 However, OPN is ubiquitously expressed and increased levels in vivo could be ascribed to many cell types including epithelial cells that produce OPN constitutively.
Recent works associate OPN with “common” autoimmunities in both mice and humans. On the one hand, high levels of OPN have been detected in the kidney of patients with SLE,18,23 in the synovial fluid of patients with RA,26 in the brain of patients with MS,25 and in the spinal cord of rats with experimental autoimmune encephalomyelitis (EAE).25 Moreover, OPN-deficient mice are relatively resistant to progressive forms of EAE and have frequent remissions.40 Therefore, these studies showed that OPN may play a role in the pathogenesis of these diseases, but they did not assess the possibility that high production of OPN may be a genetic trait predisposing patients to autoimmunity. By contrast, the involvement of variant OPN haplotypes in the pathogenesis of SLE was suggested by Forton et al,40 who showed that patients display increased frequency of the 750T polymorphism with a relative risk (RR) equaling 1.63. In the Japanese population, Niino et al41 found that the +282 T>C polymorphism in exon VI was significantly associated with MS, and that the +750 C>T polymorphism in exon VII correlated with an early disease onset. However, no data were reported on the actual OPN level in these SLE and MS patients.
In conclusion, this work is the first report clearly showing a direct association between variations of the OPN gene, increased OPN production, and development of autoimmunity. We suggest that high OPN levels may act as a predisposing factor for DALD by acting in synergy with the Fas defect. Preliminary data show that it may be involved in classic ALPS, since we also found high levels in the sera of 4 of 4 patients with ALPS type Ia (data not shown). Defective Fas function and high OPN levels seem to be mostly independent factors since exogenous OPN did not affect cell death induced by anti-Fas mAb in vitro and high serum levels of OPN were found in healthy subjects displaying normal Fas function. In line with this model, Weber and Cantor22 showed that silencing of the OPN gene delays onset of some ALPS features in lpr/lpr mice. It must be underlined that concurrence of both factors does not necessarily induce development of DALD since some DALD patients' parents displayed both factors in the absence of signs of autoimmunity/lymphoproliferation. Therefore, other environmental or genetic factors may trigger the disease. The predisposing effect of OPN may depend on OPN's activity on lymphocyte proliferation, differentiation, and death. Therefore, OPN may play a role in both the lymphoproliferative and the autoimmune aspect of DALD. It is noteworthy that our previous data showed that DALD patients' families display increased frequency of common autoimmune diseases, such as type 1 diabetes mellitus, MS, RA, or SLE, in the absence of signs of lymphoproliferation.7 One possibility is that the increased OPN levels play a role in building up an autoimmune-prone background favoring development of these diseases. This possibility is supported by the increased levels of OPN detected in MS and SLE but must be validated by parallel analysis of OPN levels and the OPN gene in these diseases.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-05-1748.
Supported by Telethon grant no. E566 (Roma), Cofin Projects (Ministero dell'Istruzione dell'Università e della Ricerca [MIUR], Roma), Fondazione Italiana Sclerosi Multipla (FISM; Genova), AIDS Project (Istituto Superiore di Sanità [ISS], Roma), Associazione Italiana Ricerca sul Cancro (AIRC; Milano).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 2. Healthy subjects carrying the haplotype B or C display higher serum levels of OPN than homozygotes for haplotype A. Two groups of subjects were evaluated (ie, random healthy subjects [n = 50] and the healthy relatives belonging to the families shown in Figure 3 [n = 12]). The genotypes are indicated with different symbols as shown in the figure. The bold horizontal lines indicate the median values for each group; the dotted line, the 95th percentile of the controls. The P values were calculated using the Mann-Whitney U test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/4/10.1182_blood-2003-05-1748/6/m_zh80040456600002.jpeg?Expires=1769111291&Signature=MMlR5jl3AXKjgb7eK-px0Jw1KFCKwoSYvVrPsvb95dbRLKMA5kFd98xLH2rpYYygvdTIpwiwZFy7dBQcdwebNdfQjbPk-2raLQQZhWCzKQq2554V5I5Malr-MgEsxJMhRJ-YJwANwEfVaru3IqGnY9AXTwr~9sdOZyRJdvbeWhkTbfkjdP9btszY4uvpk7-heKQjYyrFShCa2ne05w9WI6uUDZ7do4aUEJGfoZtsmuYW9v9S3f0ZCL9A-x8Af9XHpmR7IGFg~knoHbrH8o99MN4vHHuvPdyBvIAXSs1rReUunm17miOEM73KH2eOlt5k3MNYN8AWt4XxcREcRSWY2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal