Abstract
SHP-1, an src homology 2 (SH2) domain containing protein tyrosine phosphatase, functions as a negative regulator of signaling downstream of cytokine receptors, receptor tyrosine kinases and receptor complexes of the immune system. Dephosphorylation of receptors and/or receptor-associated kinases has been described as the mechanism for the function of SHP-1. Here we demonstrate a novel mechanism by which SHP-1 down-regulates the Janus kinase–2 (Jak2)/signal transducer and activator of transcription-5 (Stat5) pathway downstream of the prolactin receptor (PRLR) and the erythropoietin receptor (EPOR) in a catalytic activity–independent manner. Structural/functional analysis of SHP-1 defined the C-terminal tyrosine residues (Y278, Y303, Y538, Y566) within growth factor receptor–bound protein 2 (Grb-2) binding motif to be responsible for delivering the inhibitory effects. Our results further indicate that these tyrosine residues, via recruitment of the adaptor protein Grb-2, are required for targeting the inhibitory protein suppressor of cytokine signaling–1 (SOCS-1) to Jak2 kinase. Finally, loss of SOCS-1 expression in SOCS-1–/– mouse embryonic fibroblast (MEF) cells led to attenuation in SHP-1 function to down-regulate PRL-induced Stat5 activation. All together, our results indicate that SHP-1 inhibits PRLR and EPOR signaling by recruitment and targeting of SOCS-1 to Jak2, highlighting a new mechanism of SHP-1 regulation of cytokine-receptor signaling.
Introduction
Cytokines such as prolactin (PRL) and erythropoietin (EPO) elicit a variety of biologic cell-specific responses by interacting with their respective receptors. The ability of these receptors to induce tyrosine phosphorylation plays a central role in generation and transduction of signaling pathways from the cell surface to the nucleus. Receptors for PRL and EPO belong to the class I cytokine receptor superfamily characterized by having no intrinsic kinase activity.1,2 The early events in signal generation by these 2 receptors are homodimerization of the receptor and activation of the Janus kinase–2 (Jak2) kinase.3,4 This leads to tyrosine phosphorylation of the receptor and to recruitment and, in some cases, activation of signaling molecules such as the transcription factor signal transducer and activator of transcription–5 (Stat5).5 Upon phosphorylation, Stat5 dimerizes and translocates to the nucleus, where it binds to its specific responsive elements and induces the transcription of target genes.6,7
Protein tyrosine phosphatases are important signaling enzymes, which along with protein tyrosine kinases control the initiation, propagation, and termination of cellular signaling.8 Src homology 2–containing protein tyrosine phosphatase–1 (SHP-1), also called protein tyrosine phosphatase 1C (PTP1-C), SHP, SH-PTP, and hematopoietic cell protein tyrosine phosphatase (HCP), is a cytoplasmic protein tyrosine phosphatase that contains 2 src homology 2 (SH2) domains, a single catalytic domain, and a C-terminal tail containing tyrosine residues.9-12 SHP-1 is expressed predominantly in hematopoietic cells. However, SHP-1 is also moderately expressed in a variety of nonhematopoietic cells, especially malignant epithelial cells.9,10,13,14 On the basis of the phenotype of SHP-1–deficient mice, SHP-1 is generally considered a negative regulator of signaling.15,16 Furthermore, biochemical and functional studies further established SHP-1 as a negative regulator of signaling downstream of a wide range of receptors including cytokine receptors, receptor tyrosine kinases (such as c-kit, colony-stimulating factor 1[CSF-1,] and epidermal growth factor [EGF] receptors) and receptor complexes of the immune system (such as B-cell receptor [BCR] and T-cell receptor [TCR]) (reviewed in Zhang et al17 ).
In cytokine-receptor signaling, abrogation in SHP-1 recruitment to EPOR led to hypersensitivity to EPO.18 In addition, loss of SHP-1 expression has been linked to hyperactivation of the Jak/Stat pathway downstream of interferon α (IFN-α)/IFN-β.19 As well, co-overexpression of SHP-1 with Jak2 in fibroblast cells led to diminished phosphorylation of Jak2 kinase.20 Together, these results indicate that SHP-1 is a negative regulator of cytokine-receptor signaling, however, the precise mechanisms of SHP-1 action are not well characterized.
Recent studies have demonstrated the importance of the C-terminal region of SHP-1 in the control of SHP-1 function. It has been suggested that the C-terminal tail of SHP-1 is involved in regulating the phosphatase activity and nuclear localization of the enzyme.21-24 More important, SHP-1 contains a number of C-terminal tyrosine residues that undergo phosphorylation upon mitogenic stimuli.25-28 These phosphotyrosine residues have been shown to serve an adaptor function by recruitment of growth factor receptor–bound protein 2 (Grb2).29 However, the biologic significance of SHP-1 phosphorylation and SHP-1/Grb2 complex formation specifically in modulating cytokine-receptor signaling is not yet elucidated.
In this report, we investigated the mechanism by which SHP-1 negatively regulates Stat5 activation downstream of prolactin receptor (PRLR). Our data provide evidence demonstrating that the negative function of SHP-1 in signaling can occur independently of its catalytic activity. Structure/activity studies showed that tyrosine residues at the C-terminal domain of SHP-1 deliver this inhibitory effect. We further determined that this is mediated by recruitment of Grb2 and its associated suppressor of cytokine signaling–1 (SOCS-1) protein and targeting SOCS-1 to Jak2. This process leads to the inactivation of Jak2, hence termination of PRL signaling. Together, our results imply that SHP-1 mediated SOCS-1/Jak2 association as a major mechanism by which SHP-1 regulates the Jak2/Stat5 pathway downstream of PRLR.
Materials and methods
Materials and antibodies
Human prolactin (hPRL) was obtained from Dr Parlaw (National Hormone and Peptide program, Harbor–University of California at Los Angeles [UCLA] Medical Center, Los Angeles, CA). Ovine PRL used for treatment of cells was obtained from Sigma (St Louis, MO). Monoclonal antibody to phosphotyrosine (4G10), monoclonal antibody to SHP-1, and polyclonal antibody to Jak2 were purchased from Upstate Biotechnology (Lake Placid, NY). Monoclonal antibody to phosphorylated Stat5 (Y694), polyclonal antibody to Stat5, and polyclonal antibody to SOCS-1 were obtained from Zymed Laboratories (South San Francisco, CA). Polyclonal antibody to the milk protein was a gift from Dr Nancy Hynes (Friedrich Miescher Institute, Basel, Switzerland). Monoclonal antibodies to Stat5 and Grb2 were purchased from BD Transduction Laboratories (Mississauga, ON, Canada). Polyclonal antibodies to Grb2 and SHP-1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase (HRP)–tagged monoclonal antibody to myc tag was purchased from Roche Diagnostics (Laval, QC, Canada). Monoclonal antibody to hemagglutinin (HA) was purchased from Berkeley Antibody (Berkeley, CA).
Plasmids and mutagenesis
Expression plasmid encoding the long form of PRLR was described previously.30 Expression plasmid encoding for EPOR was kindly provided by Dr Klingmuller (Max-Planck Institute for Biochemistry, Martinsried, Germany). Expression plasmids encoding SHP-1 wild-type (WT) and SHP-1C455S mutant form were obtained from Dr Axel Ullrich (Max-Planck Institute for Biochemistry). Expression plasmids encoding HA-tagged Grb2WT, Grb2R86K, Grb2W36, 193K were kindly provided by Dr Bruce J, Mayer (University of Connecticut Health Center, Farmington, CT). Expression plasmid encoding Myc-tagged SOCS-1 was a gift from Dr Akihiko Yoshimura (Life Sciences Institute, Kurume, Japan).
SHP-1δC (stop codon at amino acid 271 [aa271]); SHP-1Y278, 303, 538, 566F; and SHP-1CSY278, 303, 538, 566F were constructed with the use of SHP-1WT and SHP-1C455S cDNAs in PRK5 vector as templates (Figure 1A). Mutagenesis was performed according to the Kunkel method.31 The following primers were used for mutagenesis (all are 5′ to 3′; mutated codon is underlined): SHP-1δC, TTGCCCTTGTTTCATGGCCGCTGCC; SHP-1δKRK, CTCACTTCCTTCAGAGGGAACCC; Y278F, GAATGTTCTTGAAGCGGT TCTTGC; Y303F, GGCATTGATGAAGTCGGACCC; Y538F, GATGTTCCCGAACTC CGACTC; Y566F, CAGGTTCTCAAACA CATCCTC. The different mutant forms of SHP-1 were confirmed by sequencing (Figure 1A).
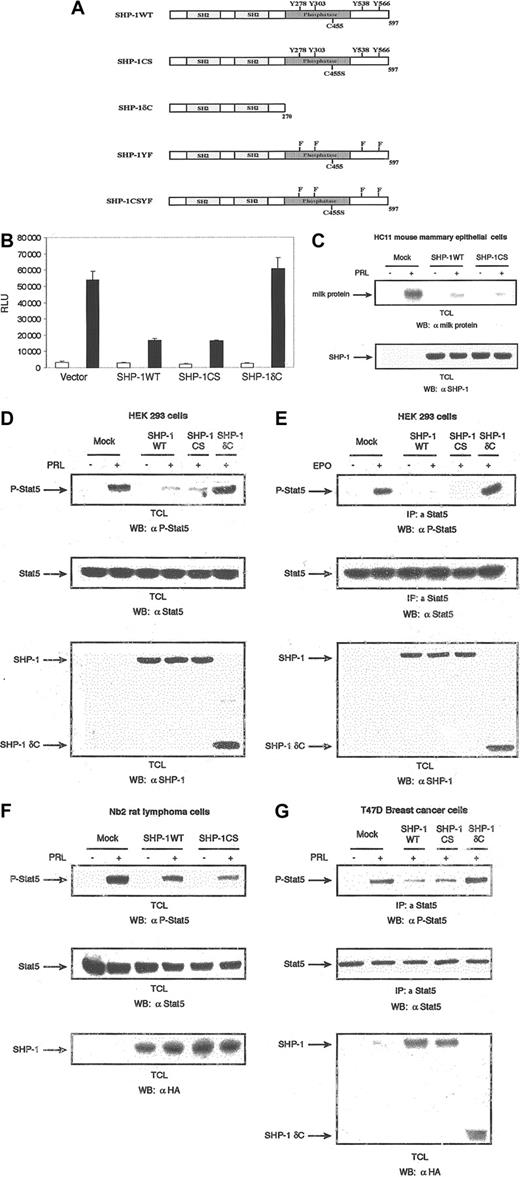
Effect of SHP-1 on Stat-5 activation. SHP-1 negatively regulates Stat5 activation independently of the phosphatase activity. (A) Schematic representation of SHP-1 wild type and its mutants. (B) The 293 cells were cotransfected with expression plasmids encoding PRLR, Stat5, β-casein gene promoter/luciferase reporter construct, and either vector, SHP-1WT, SHP-1CS, or SHP-1δC. Cells were starved and left unstimulated or stimulated with ovine PRL (oPRL) (1 μg/mL) overnight. Luciferase activity was assayed, and data were presented as relative light units. Each bar represents the mean ± standard deviation values of triplicate samples. The luciferase activity was normalized to the expression of β-galactosidase. □ indicates absence, and ▪, presence, of oPRL. (C) Differentiated HC11 cells were transfected with either vector alone or expression plasmids for the indicated forms of SHP-1. Cells were starved and stimulated with oPRL for 24 hours. Total cell lysates were immunoblotted with a polyclonal antibody to milk proteins (upper panel). The membrane was stripped and reblotted with polyclonal antibody to SHP-1 (lower panel). (D) The 293 cells were transiently cotransfected with expression vectors for PRLR, Stat5, and either vector or the indicated forms of SHP-1. Serum-starved cells were left untreated or treated with oPRL (1 μg/mL) for 10 minutes. Total cell lysates were analyzed by immunoblot with the use of specific antibodies directed against the phosphorylated form of Stat5 (upper panel). The membrane was stripped and reprobed with monoclonal antibody to Stat5 (middle panel), then with polyclonal antibody to SHP-1 (lower panel). (E) The 293 cells were transiently cotransfected with expression vectors for the EPOR, Stat5, and either vector or the indicated forms of SHP-1. Serum-starved cells were left untreated or treated with EPO (5 U/mL) for 10 minutes. Total cell lysates were immunoprecipitated with polyclonal antibody to Stat5 and immunoblotted with monoclonal antibody to phospho-Stat5 (upper panel). Total cell lysates from the same transfection were immunodetected with monoclonal antibody to Stat5 (middle panel), then with polyclonal antibody to SHP-1 (lower panel). (F) Nb2 rat pre–T lymphoma cells and Nb2 cells stably expressing HA-tagged forms of SHP-1WT and SHP-1CS were starved for 18 hours and then left untreated or treated with oPRL (1 μg/mL) for 10 minutes. Cell lysates were immunoblotted with a monoclonal antibody to phospho-Stat5 (upper panel), a monoclonal antibody to Stat5 (middle panel), and a monoclonal antibody to HA tag (lower panel). (G) T47D cells were transfected with either vector or expression plasmids for the indicated forms of HA-tagged SHP-1. Cells were serum starved for 18 hours and then stimulated with hPRL (1 μg/mL) for 10 minutes. Cells were lysed, and immunoprecipitations were performed with polyclonal antibody to Stat5, followed by immunoblotting with a monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reblotted with a monoclonal antibody to Stat5 (middle panel), then with a monoclonal antibody to HA tag (lower panel).
Effect of SHP-1 on Stat-5 activation. SHP-1 negatively regulates Stat5 activation independently of the phosphatase activity. (A) Schematic representation of SHP-1 wild type and its mutants. (B) The 293 cells were cotransfected with expression plasmids encoding PRLR, Stat5, β-casein gene promoter/luciferase reporter construct, and either vector, SHP-1WT, SHP-1CS, or SHP-1δC. Cells were starved and left unstimulated or stimulated with ovine PRL (oPRL) (1 μg/mL) overnight. Luciferase activity was assayed, and data were presented as relative light units. Each bar represents the mean ± standard deviation values of triplicate samples. The luciferase activity was normalized to the expression of β-galactosidase. □ indicates absence, and ▪, presence, of oPRL. (C) Differentiated HC11 cells were transfected with either vector alone or expression plasmids for the indicated forms of SHP-1. Cells were starved and stimulated with oPRL for 24 hours. Total cell lysates were immunoblotted with a polyclonal antibody to milk proteins (upper panel). The membrane was stripped and reblotted with polyclonal antibody to SHP-1 (lower panel). (D) The 293 cells were transiently cotransfected with expression vectors for PRLR, Stat5, and either vector or the indicated forms of SHP-1. Serum-starved cells were left untreated or treated with oPRL (1 μg/mL) for 10 minutes. Total cell lysates were analyzed by immunoblot with the use of specific antibodies directed against the phosphorylated form of Stat5 (upper panel). The membrane was stripped and reprobed with monoclonal antibody to Stat5 (middle panel), then with polyclonal antibody to SHP-1 (lower panel). (E) The 293 cells were transiently cotransfected with expression vectors for the EPOR, Stat5, and either vector or the indicated forms of SHP-1. Serum-starved cells were left untreated or treated with EPO (5 U/mL) for 10 minutes. Total cell lysates were immunoprecipitated with polyclonal antibody to Stat5 and immunoblotted with monoclonal antibody to phospho-Stat5 (upper panel). Total cell lysates from the same transfection were immunodetected with monoclonal antibody to Stat5 (middle panel), then with polyclonal antibody to SHP-1 (lower panel). (F) Nb2 rat pre–T lymphoma cells and Nb2 cells stably expressing HA-tagged forms of SHP-1WT and SHP-1CS were starved for 18 hours and then left untreated or treated with oPRL (1 μg/mL) for 10 minutes. Cell lysates were immunoblotted with a monoclonal antibody to phospho-Stat5 (upper panel), a monoclonal antibody to Stat5 (middle panel), and a monoclonal antibody to HA tag (lower panel). (G) T47D cells were transfected with either vector or expression plasmids for the indicated forms of HA-tagged SHP-1. Cells were serum starved for 18 hours and then stimulated with hPRL (1 μg/mL) for 10 minutes. Cells were lysed, and immunoprecipitations were performed with polyclonal antibody to Stat5, followed by immunoblotting with a monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reblotted with a monoclonal antibody to Stat5 (middle panel), then with a monoclonal antibody to HA tag (lower panel).
HA-tagged WT, CS, and δC forms of SHP-1 were generated by polymerase chain reaction with the use of the corresponding cDNAs in PRK5 as templates and the following oligonucleotides as primers: forward, 5′-CGCCATCGATATGCTGTCCCGTG-3′; and reverse, 5′-TTCGAATCTAGACAGTGATCCCAGGGC-3′. The polymerase chain reaction products were subcloned into the ClaI/XbaI sites of pOPRSVI/MCS vector containing HA-tag sequence between XhoI and ClaI. All constructs were sequenced prior to use.
Cell culture
The Nb2 prolactin-dependent rat pre–T lymphoma cells were grown in complete medium (RPMI media containing 10% fetal calf serum [Gibco, Burlington, ON, Canada], 10% horse serum [Sigma, St Louis, MO], 2 mM L-glutamine, 7.5% sodium bicarbonate, 5 μM 2 β-mercaptoethanol). Following overnight incubation in starvation medium (identical to complete medium but deprived of fetal bovine serum [FBS] and β-mercaptoethanol [β-ME]), cells were stimulated by ovine PRL (oPRL) (1 μg/mL) for the indicated times for individual experiments. Cells were pelleted in an Eppendorf tube and prepared for lysis. The 293, wild-type, and SOCS-1–/– mouse embryonic fibroblast (MEF) cells and T47D human breast cancer cells were grown in Dulbecco modified Eagle medium (DMEM) (4.5 g/L of glucose) containing 10% (vol/vol) fetal calf serum. Cells were serum starved overnight and then stimulated with prolactin (1 μg/mL) and prepared for lysis.
HC11, mouse mammary epithelial cells obtained from Dr Nancy Hynes (Friedrich Miescher Institute) and Dr Bernd Groner (Georg Speyer Haus, Frankfurt, Germany), were grown to confluency in RPMI 1640 medium containing 10% fetal calf serum (Life Technologies, Bethesda, MD), insulin (5 μg/mL), and epidermal growth factor (10 ng/mL). Cells were then induced by incubating them for 2 days in RPMI medium containing 10% fetal calf serum, insulin (5 μg/mL), and hydrocortisone (1 μM). Prior to experimentation, cells were starved in RPMI medium containing insulin (5 μg/mL), hydrocortisone (1 μM), and transferrin (10 μg/mL) and then stimulated with oPRL (1.5 μg/mL) for 24 hours. The human Jurkat leukemia T-cells were grown as suspension cultures in RPMI containing 10% fetal calf serum, 2 mM L-glutamine, 50 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.5). Cells were starved overnight and then stimulated with hPRL for 5 minutes.
Transient transfection
The human embryonic kidney 293 cells were grown in Dulbecco modified Eagle medium (DMEM) (4.5 g/L per liter of glucose) containing 10% (vol/vol) fetal calf serum. Approximately 5 × 105 cells were plated and then cotransfected by calcium phosphate precipitation with expression plasmids encoding PRLR, Jak2, Stat5, and indicated forms of SHP-1 or Grb2 as described for each experiment. After 18 hours of expression, the cells were serum starved overnight.
T47D cells (107 cells) were transfected by electroporation (Gene Pulser II; Bio-Rad, Hercules, CA) in 500 μL phosphate-buffered saline (240 V and 975 microfarads) with 10 μg expression plasmids encoding different forms of SHP-1 or Grb2. Following transfection, cells were plated in DMEM (10% fetal calf serum [FCS]) for 24 hours. The following day, cells were serum starved overnight before stimulation with PRL.
SOCS-1+/+ and SOCS-1–/– cells were transfected with the use of FuGENE 6 transfection reagent (Roche Diagnostics) in 35-mm plates according to the manufacturer's protocol.
Cell lysis, immunoprecipitation, and Western blotting
Cell pellets were lysed in RIPA lysis buffer (50 mM Tris-Hcl [tris(hydroxymethyl)aminomethane-Hcl], pH 7.4; 1% Nonidet P40 [NP-40]; 0.25% sodium deoxycholate; 150 mM NaCl; 1 mM EDTA [ethylenediaminetetraacetic acid]; 1 mM phenylmethylsulfonyl fluoride [PMSF]; 1 μg/mL each of aprotinin, leupeptin, and pepstatin; 1 mM sodium orthovanadate; and 1 mM NaF). The lysates were cleared by centrifugation at 13 000g for 10 minutes at 4°C to remove insoluble materials. Protein concentration was measured by means of the Bradford technique. Equal amounts of proteins obtained by whole-cell lysis were loaded and separated on an appropriate concentration of sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). For immunoprecipitations, equal amounts of proteins were incubated with the indicated antibodies in the presence of protein A–Sepharose beads for 3 hours at 4°C. Immunoprecipitates were washed 3 times with HNTG buffer (20 mM HEPES, pH 7.5; 150 mM NaCl; 0.1% Triton X-100; and 10% glycerol) and eluted in 20 μL sample buffer. Samples were then boiled for 10 minutes before loading on SDS-PAGE. Western blot analyses were performed with the indicated antibodies, and proteins were revealed with the use of chemiluminescence (Roche Diagnostics) according to manufacturer's instructions.
Luciferase assay
The assay was carried out as described previously.30 Briefly, 293 cells were transiently transfected with expression plasmids encoding PRLR, Stat5, and indicated forms of SHP-1 or Grb2 along with the β-casein gene promoter/luciferase construct and the expression plasmid for β-galactosidase with the use of the calcium phosphate method. After 24 hours, cells were incubated in the presence or absence of 1 μg/mL oPRL for 18 hours in starvation medium. Cells were lysed in lysis buffer (1% Triton X-100, 15 mM MgSO4, 4 mM EGTA [ethylene glycol tetraacetic acid], and 1 mM dithiothreitol [DTT]) and then centrifuged at 13 000g to clear the lysates. Luciferase activity was measured in relative light units and normalized to the expression of β-galactosidase. Results are presented as mean ± standard deviation of 3 separate experiments.
Far Western analysis
Far Western analysis was performed as described previously.32 Briefly, 293 cells were transfected with expression plasmids encoding PRLR, Jak2, and vector alone or indicated forms of SHP-1. Serum-starved cells were left untreated or treated with PRL for 5 minutes. Cell lysates were used for immunoprecipitations with polyclonal antibodies to SHP-1. Immunoprecipitates were separated on an 8% SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked in 10% nonfat dry milk in Tris-buffered saline–Tween (TBST) (50 mM Tris, pH 7.5; 200 mM NaCl; 0.05% Tween20) plus orthovanadate (1 mM) and EDTA (5 mM) overnight at 4°C. Glutathione S-transferase (GST)–Grb2 fusion protein was expressed in Escherichia coli and affinity purified by glutathione-Sepharose beads and then preincubated with 0.2 μg glutathione-conjugated horseradish peroxidase (GSH-HRP) (Sigma) for 30 minutes at room temperature. Binding reactions were performed by incubating the labeled probe with the blocked membranes (at a final concentration of 1 μg/mL) for 30 minutes at room temperature. Membranes were washed 3 times with TBST for 15 minutes. Bound proteins were revealed by chemiluminescence.
Results
SHP-1 negatively regulates cytokine-induced Stat5 activation independently of its catalytic activity
The Jak/Stat pathway has appeared as a central pathway in cytokine-receptor signaling including PRL receptor leading to transcriptional activation of target genes. The protein tyrosine phosphatase SHP-1 is known to be a critical negative regulator of cytokine signaling.18,19 While SHP-1 has been shown to block cytokine-induced tyrosine phosphorylation of Jak kinases, the mechanism by which SHP-1 regulates the Jak/Stat pathway has not been well characterized to date. Here we examined the role of SHP-1 in regulating Jak/Stat pathway downstream of PRLR leading to activation of the Stat5-responsive β-casein milk protein gene promoter. Human embryonic kidney (HEK) 293 cells were cotransfected with expression plasmids encoding the rat PRLR long form, Stat5, and either vector (as a control), SHP-1WT, or a catalytically inactive mutant of SHP-1 (SHP-1CS). As shown in Figure 1B, while PRL treatment led to induction of the β-casein gene promoter, overexpression of SHP-1WT significantly inhibited PRL signal, indicating that SHP-1 negatively regulates PRLR signaling. It was expected that overexpression of catalytically inactive SHP-1 would have no inhibitory effects on PRL ability to induce β-casein gene promoter activation. However, in samples overexpressing SHP-1CS, we detected inhibitory effects to the same extent as the effects exerted by SHP-1WT (Figure 1B). To further address the role of the catalytic domain of SHP-1 in blocking PRLR signaling, we examined the role of SHP1δC, a truncated mutant of SHP-1 containing only the 2 SH2 domains (Figure 1A), in regulating PRL signaling leading to gene promoter activation. Interestingly, in contrast to SHP-1CS, overexpression of SHP-1δC had no inhibitory effects on PRL-mediated signals (Figure 1B). This suggests that SHP-1 inhibits PRLR signaling by a mechanism independent of its catalytic activity but requires the C-terminal domain of SHP-1. To confirm the effects of SHP-1 in regulating PRL-mediated signals to milk protein gene activation, we examined the role of SHP-1 in PRL-induced milk protein gene expression in the mouse mammary epithelial HC11 cells. Differentiated cells were stimulated with PRL overnight following transient transfection with either SHP-1WT or SHP-1CS. Whole cellular lysates were subjected to immunoblotting with a polyclonal antibody to mouse milk proteins. As can be observed in Figure 1C, SHP-1WT as well as SHP-1CS inhibited prolactin-induced milk protein gene expression in mammary cells.
To further confirm our findings, we next examined the role of SHP-1 in modulating PRL-induced Stat5 tyrosine phosphorylation as an immediate downstream target of PRLR activation. Moreover, to extend our findings to other cytokine receptors using the Jak2/Stat5 pathway, we also examined the role of the different mutants of SHP-1 in EPOR-induced Stat5 activation. HEK 293 cells were cotransfected with expression plasmids encoding PRLR (Figure 1D) or EPOR (Figure 1E), along with expression vectors encoding Stat5 and the indicated forms of SHP-1. Whole-cell lysates as well as immunoprecipitations of Stat5 were subjected to immunodetection with a monoclonal antibody specific to phosphorylated tyrosine 694 of Stat5. As shown, both PRL- and EPO-mediated tyrosine phosphorylation of Stat5 were blocked in cells overexpressing wild-type or catalytically inactive form of SHP-1 but not in cells overexpressing the C-terminal deletion mutant form of SHP-1 (Figure 1D-E). Similar levels of Stat5 and SHP-1 expression were confirmed by reprobing the membranes with monoclonal antibody to Stat5 and with polyclonal antibody to SHP-1 (Figure 1D-E, middle and lower panels, respectively). Together, these results indicate that inhibition of PRL-mediated β-casein gene promoter activation and PRL/EPO–induced Stat5 tyrosine phosphorylation is independent of the catalytic activity of SHP-1 but requires its C-terminal domain.
As these results were unexpected, we chose to further confirm the function of the catalytic domain of SHP-1 in negative regulation of PRLR signaling to Stat5 activation by employing the PRL-responsive hematopoietic Nb2 cell model system. Using the PRL-dependent pre–T lymphoma Nb2 cells, we generated stably overexpressing HA-tagged SHP-1WT or HA-tagged SHP-1CS cell lines. Expression of the 2 SHP-1 genes was monitored by Western blot analysis, with the use of a monoclonal antibody to HA tag (Figure 1F, lower panel). Using these cell lines, we examined the role of SHP-1 catalytic activity in PRL-induced Stat5 tyrosine phosphorylation. As presented in Figure 1F, PRL stimulation led to tyrosine phosphorylation of Stat5; however, the amount of detectable phosphorylation was notably lower in SHP-1WT– and SHP-1CS–overexpressing Nb2 cells. These results confirm again that SHP-1 may inhibit PRL-induced Jak2/Stat5 pathway independently of its catalytic activity.
We next examined the role of SHP-1 in regulating PRLR-induced Stat5 activation in the T47D breast cancer cells as a nonhematopoietic model cell system. The T47D cells were transfected by electroporation with either a control vector or the expression plasmids encoding wild-type or different mutants of SHP-1 (Figure 1G). As can be observed, PRL treatment triggered phosphorylation of Stat5, and overexpression of either SHP-1WT or SHP-1CS inhibited PRL signals. Significantly, overexpression of the SHP-1δC mutant form was unable to block PRL-mediated tyrosine phosphorylation of Stat5. Together, the results indicate that in both hematopoietic and nonhematopoietic cells, SHP-1 regulation of PRLR signaling is mediated by noncatalytic activity–dependent mechanisms.
SHP-1's inhibitory effects on the Jak2/Stat5 pathway require C-terminal tyrosine residues
On the basis of the previous results, we next sought to define the functional domains at the C-terminal part of SHP-1 that contribute to the negative regulation of signaling. In mammalian cells, SHP-1 has been shown to undergo tyrosine phosphorylation and to associate with Grb2 in response to growth factor stimulation.25-29 However, the significance of SHP-1 tyrosine phosphorylation and its association with Grb2 in the regulation of cytokine-receptor signaling has not yet been characterized.
Previous studies have identified tyrosine residues 538 and 566 of SHP-1 as the major phosphorylation sites in vitro and in vivo, respectively.28,33,34 The sequences surrounding both Y538 (YGNI) and Y566 (YENL) conform to the consensus recognition motif for the SH2 domain of Grb2 (YXNX).35 There are, however, 2 other tyrosine residues, Y278 (YKNI) and Y303 (YINA) at the C-terminal part of SHP-1, that also fall within the Grb2-binding motif. To define the role of these tyrosine residues in SHP-1–negative regulation of PRLR signaling, we substituted phenylalanine for tyrosine at codons 278, 303, 538, and 566 in both SHP-1WT and SHP-1CS cDNAs and named them SHP-1YF and SHP-1CSYF, respectively (Figure 1A). We first confirmed that SHP-1 is tyrosine phosphorylated in response to PRL (Figure 2A) and associated with Grb2 in hematopietic as well as nonhematopoietic cells (Figure 2B). To test the role of the C-terminal tyrosine residues of SHP-1 in PRLR signaling leading to gene promoter activation, we initially examined the role of the previously described major phosphorylation sites of SHP-1 (Y538 and Y566) in regulating PRLR signaling. As can be seen in Figure 2C (left panel), individual mutations of Y538 and Y566 did not lead to recovery of PRLR signaling. However, simultaneous mutations of these 2 sites partially recovered PRLR signaling. This led us to consider the contribution of the other 2 tyrosine residues that are within the Grb2-binding motif, Y278 and Y303. Therefore, we examined the effects of SHP-1YF (Y278F, Y303F, Y538F, and Y566F) and SHP-1CSYF (Y278F, Y303F, Y538F, and Y566F) mutants on PRL-induced transcriptional activation. As shown in Figure 2C (right panel), overexpression of SHP-1WT inhibited PRL-mediated β-casein gene promoter activation. However, overexpression of SHP-1YF or SHP-1CSYF had no inhibitory effects on PRL-induced β-casein gene activation. These results indicate that tyrosine residues of SHP-1 within the Grb2-binding motif function to inhibit PRLR signaling.
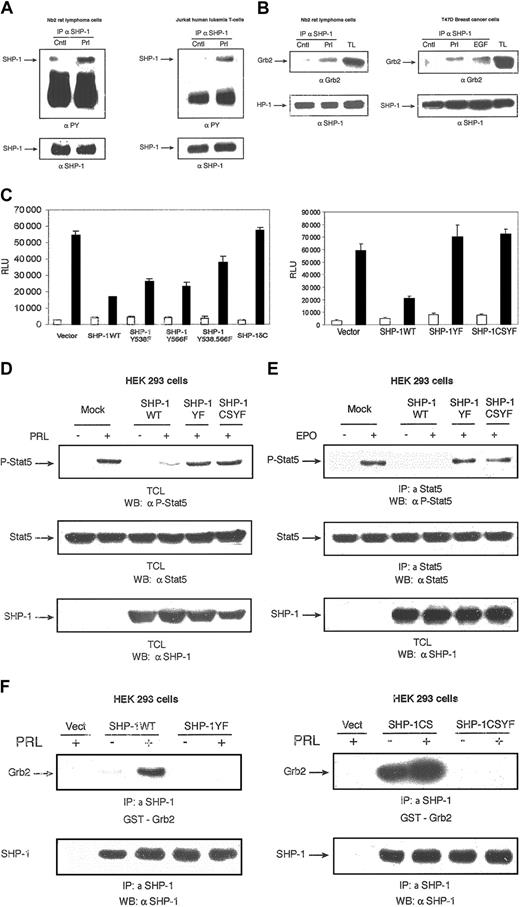
Effect of SHP-1 C-terminal tyrosine residues on PRLR signaling. SHP-1 C-terminal tyrosine residues deliver the inhibitory effect of SHP-1 in PRLR signaling. (A) Nb2 cells (left panel) and Jurkat cells (right panel) were starved overnight and then either left untreated or treated with PRL for 5 minutes. Cell lysates were used for immunoprecipitation with a polyclonal antibody to SHP-1 followed by immunoblotting with a monoclonal antibody to phosphotyrosine (4G10). The membranes were stripped and reblotted with a polyclonal antibody to SHP-1 (lower panels). (B) Nb2 cells were starved overnight and left unstimulated or stimulated with PRL for 5 minutes (left panel). T47D cells were starved overnight and left unstimulated or stimulated with hPRL (1 μg/mL) and EGF (20 ng/mL) (as a positive control) for 5 minutes (right panel). Cell extracts were immunoprecipitated with a polyclonal antibody to SHP-1 and subjected to Western blotting analysis with a monoclonal antibody to Grb2 (upper panels). The membranes were stripped and reblotted with a polyclonal antibody to SHP-1 (lower panels). TL indicates total cell extracts. (C) The 293 cells were cotransfected with expression plasmids for PRLR, Stat5, and the indicated forms of SHP-1 along with β-casein gene promoter/luciferase reporter construct. A luciferase assay was performed, and luciferase activity was normalized to the relative β-galactosidase values. Results represent means and standard deviations of 3 independent experiments. □ indicates absence, and ▪, presence, of oPRL. (D) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR, Stat5, and either vector or the indicated forms of SHP-1. Serum-starved cells were left unstimulated or stimulated with oPRL. Total cell lysates were probed with monoclonal antibody to phospho-Stat5 (upper panel), monoclonal antibody to Stat5 (middle panel), or polyclonal antibody to SHP-1 (lower panel). (E) Cell lysates from 293 cells overexpressing EPOR, Stat5, and either vector alone or the indicated forms of SHP-1 that were left unstimulated or stimulated with EPO (5 U/mL) were immunoprecipitated with polyclonal antibody to Stat5 and immunoblotted with monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reblotted with monoclonal antibody to Stat5 (middle panel) or polyclonal antibody to SHP-1 (lower panel). (F) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR and either vector alone or indicated forms of SHP-1 expression vectors. Serum-starved cells were left untreated or treated with PRL for 10 minutes. Cell lysates were immunoprecipitated with polyclonal antibody to SHP-1, and proteins were separated on SDS-PAGE and transferred to nitrocellulose membrane. Direct binding of GST-Grb2 fusion protein was assessed by far Western analysis (upper panels). The same membrane was stripped and reblotted with anti–SHP-1 (lower panels).
Effect of SHP-1 C-terminal tyrosine residues on PRLR signaling. SHP-1 C-terminal tyrosine residues deliver the inhibitory effect of SHP-1 in PRLR signaling. (A) Nb2 cells (left panel) and Jurkat cells (right panel) were starved overnight and then either left untreated or treated with PRL for 5 minutes. Cell lysates were used for immunoprecipitation with a polyclonal antibody to SHP-1 followed by immunoblotting with a monoclonal antibody to phosphotyrosine (4G10). The membranes were stripped and reblotted with a polyclonal antibody to SHP-1 (lower panels). (B) Nb2 cells were starved overnight and left unstimulated or stimulated with PRL for 5 minutes (left panel). T47D cells were starved overnight and left unstimulated or stimulated with hPRL (1 μg/mL) and EGF (20 ng/mL) (as a positive control) for 5 minutes (right panel). Cell extracts were immunoprecipitated with a polyclonal antibody to SHP-1 and subjected to Western blotting analysis with a monoclonal antibody to Grb2 (upper panels). The membranes were stripped and reblotted with a polyclonal antibody to SHP-1 (lower panels). TL indicates total cell extracts. (C) The 293 cells were cotransfected with expression plasmids for PRLR, Stat5, and the indicated forms of SHP-1 along with β-casein gene promoter/luciferase reporter construct. A luciferase assay was performed, and luciferase activity was normalized to the relative β-galactosidase values. Results represent means and standard deviations of 3 independent experiments. □ indicates absence, and ▪, presence, of oPRL. (D) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR, Stat5, and either vector or the indicated forms of SHP-1. Serum-starved cells were left unstimulated or stimulated with oPRL. Total cell lysates were probed with monoclonal antibody to phospho-Stat5 (upper panel), monoclonal antibody to Stat5 (middle panel), or polyclonal antibody to SHP-1 (lower panel). (E) Cell lysates from 293 cells overexpressing EPOR, Stat5, and either vector alone or the indicated forms of SHP-1 that were left unstimulated or stimulated with EPO (5 U/mL) were immunoprecipitated with polyclonal antibody to Stat5 and immunoblotted with monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reblotted with monoclonal antibody to Stat5 (middle panel) or polyclonal antibody to SHP-1 (lower panel). (F) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR and either vector alone or indicated forms of SHP-1 expression vectors. Serum-starved cells were left untreated or treated with PRL for 10 minutes. Cell lysates were immunoprecipitated with polyclonal antibody to SHP-1, and proteins were separated on SDS-PAGE and transferred to nitrocellulose membrane. Direct binding of GST-Grb2 fusion protein was assessed by far Western analysis (upper panels). The same membrane was stripped and reblotted with anti–SHP-1 (lower panels).
Next, we examined the regulation of PRL- and EPO-mediated Stat5 activation by the C-terminal tyrosine residues of SHP-1. As can be seen in Figure 2D-E, PRL as well as EPO stimulation of Stat5 activation was blocked in samples overexpressing SHP-1WT. However, PRL- and EPO-induced Stat5 activation was maintained in samples overexpressing SHP-1YF or SHP-1CSYF. Finally, to demonstrate that PRL-induced SHP-1/Grb2 complex formation is dependent on the tyrosine residues of SHP-1 within the Grb2-binding motif, 293 cells were cotransfected with expression plasmids encoding PRLR along with either SHP-1WT, SHP-1CS, SHP-1YF, or SHP-1CSYF. Following ligand stimulation, cell lysates were subjected to immunoprecipitation with a polyclonal antibody to SHP-1, and direct binding of Grb2 was assessed by far Western assay (Figure 2F). Notably, PRL-dependent Grb2/SHP-1WT interaction was observed. Significantly, more basal and PRL-induced interaction was detected in the presence of SHP-1CS. This is likely to be due to the absence of an autodephosphorylation mechanism of the enzyme. However, no interaction was found in samples overexpressing SHP-1YF or SHP-1CSYF. This result indicates that PRL-mediated SHP-1/Grb2 interaction requires Y278, Y303, Y538, and Y566 on SHP-1. Together, our results indicate that SHP-1–negative regulation of PRLR signaling is mediated through a phosphotyrosine-dependent mechanism and this mechanism is not limited to the PRLR system.
Grb2/SH3 domains mediate negative regulation of Stat5 activation induced by PRLR and EPOR
That Y278, Y303, Y538, and Y566 mediate SHP-1 negative regulatory effects on PRLR signaling and that these tyrosine residues are capable of direct binding to Grb2 strongly suggest that Grb2 is involved in mediating SHP-1 inhibitory effects in PRLR signaling. To verify this hypothesis, we examined the role of Grb2 in PRL-induced β-casein gene promoter activation as well as PRL- and EPO-induced Stat5 tyrosine phosphorylation. As shown in Figure 3A-C, overexpression of Grb2 led to inhibition of PRL- and EPO-induced signaling. Interestingly, the inactivation of either the SH2 domain (Grb2R86K) or the 2 SH3 regulatory domains of Grb2 (Grb2W36, 193K) had no inhibitory effects on PRL- or EPO-mediated signals. Finally experiments carried out using single SH3-domain inactive mutant forms of Grb2 (Grb2W36K) and (Grb2W193K) also indicated that inactivation of either the N-terminal (Grb2W36K) or the C-terminal (Grb2W193K) SH3-domain is critical for Grb2 negative regulatory effects on Stat5 activation (data not shown). Together, these results indicate that Grb2–SH3-domain associated proteins contribute to block Stat5 activation downstream of PRLR and EPOR.
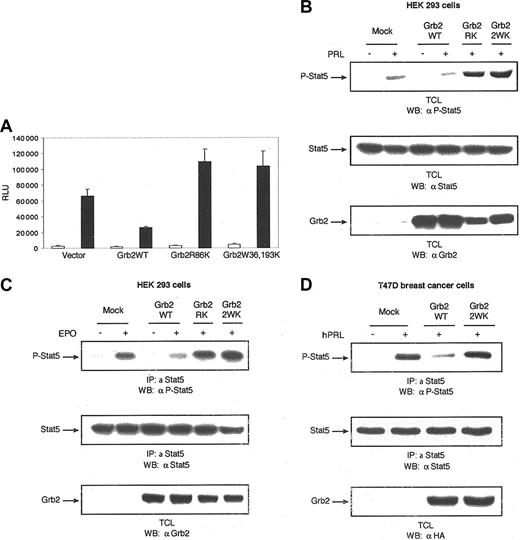
Effect of Grb2 on cytokine-induced Stat5 activation. Grb2 is a negative regulator of cytokine-induced Stat5 activation. (A) The 293 cells were cotransfected with expression plasmids for PRLR, Stat5, and the indicated forms of Grb2 along with β-casein gene promoter/luciferase reporter construct. A luciferase assay was performed, and luciferase activity was normalized to the relative β-galactosidase values. Results represent means and standard deviations of 3 independent experiments. □ indicates absence, and ▪, presence, of oPRL. (B) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR, Stat5, and either vector alone or the indicated forms of Grb2. Following 18 hours of serum starvation, cells were left unstimulated or stimulated with PRL for 10 minutes. Cell lysates were immunodetected by means of monoclonal antibodies to phospho-Stat5 (upper panel), Stat5 (middle panel), or Grb2 (lower panel). (C) The 293 cells cotransfected with expression plasmids encoding the EPOR, Stat5, and either vector alone or the indicated forms of Grb2. Following overnight starvation, cells were left unstimulated or stimulated with EPO (5 U/mL) for 10 minutes. Cell lysates were immunoprecipitated with polyclonal antibody to Stat5 and immunoblotted with monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reprobed with monoclonal antibodies to Stat5 (middle panel) or Grb2 (lower panel). (D) T47D cells were transfected with either vector alone or the indicated forms of HA-tagged Grb2 expression plasmids. Serum-starved cells were stimulated with hPRL (1 μg/mL) for 10 minutes and lysed. Immunoprecipitations were performed with polyclonal antibody to Stat5, followed by immunoblotting with monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reprobed with monoclonal antibodies to Stat5 (middle panel) or HA tag (lower panel).
Effect of Grb2 on cytokine-induced Stat5 activation. Grb2 is a negative regulator of cytokine-induced Stat5 activation. (A) The 293 cells were cotransfected with expression plasmids for PRLR, Stat5, and the indicated forms of Grb2 along with β-casein gene promoter/luciferase reporter construct. A luciferase assay was performed, and luciferase activity was normalized to the relative β-galactosidase values. Results represent means and standard deviations of 3 independent experiments. □ indicates absence, and ▪, presence, of oPRL. (B) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR, Stat5, and either vector alone or the indicated forms of Grb2. Following 18 hours of serum starvation, cells were left unstimulated or stimulated with PRL for 10 minutes. Cell lysates were immunodetected by means of monoclonal antibodies to phospho-Stat5 (upper panel), Stat5 (middle panel), or Grb2 (lower panel). (C) The 293 cells cotransfected with expression plasmids encoding the EPOR, Stat5, and either vector alone or the indicated forms of Grb2. Following overnight starvation, cells were left unstimulated or stimulated with EPO (5 U/mL) for 10 minutes. Cell lysates were immunoprecipitated with polyclonal antibody to Stat5 and immunoblotted with monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reprobed with monoclonal antibodies to Stat5 (middle panel) or Grb2 (lower panel). (D) T47D cells were transfected with either vector alone or the indicated forms of HA-tagged Grb2 expression plasmids. Serum-starved cells were stimulated with hPRL (1 μg/mL) for 10 minutes and lysed. Immunoprecipitations were performed with polyclonal antibody to Stat5, followed by immunoblotting with monoclonal antibody to phospho-Stat5 (upper panel). The membrane was stripped and reprobed with monoclonal antibodies to Stat5 (middle panel) or HA tag (lower panel).
To confirm these results in a PRLR physiologic cell system, PRL-mediated Stat5 tyrosine phosphorylation was investigated in T47D cells transiently overexpressing Grb2WT, Grb2W36, 193K. As shown in Figure 3D, PRL stimulation of T47D cells led to tyrosine phosphorylation of Stat5. Overexpression of Grb2WT impaired Stat5 phosphorylation in response to PRL stimulation; however, overexpression of the SH3 domains inactive mutant of Grb2 had no inhibitory effects on Stat5 tyrosine phosphorylation, confirming that the adaptor function of Grb2 to recruit proline-rich protein via its SH3 domains has inhibitory effects on the Jak2/Stat5 pathway.
SHP-1 C-terminal tyrosine residues, via recruitment of Grb2, target SOCS-1 to Jak2
Grb2 through its SH3 domains has been reported to interact with several proteins such as Sos, Grb2-associated binder 1 (Gab1), Gab2, c-Cbl, and Vav, as well as SOCS-1.36-40 Among these proteins, SOCS-1 is the only reported protein that needs both SH3 domains of Grb2 for optimal binding40 and is known as a potent inhibitor of the Jak/Stat pathway.41-43 To determine whether PRL induces Grb2/SOCS-1 interaction through the SH3 domains of Grb2, HEK 293 cells were cotransfected with expression plasmids encoding either Grb2WT, Grb2W36, 193K along with plasmids encoding PRLR and myc-tagged SOCS-1. Following PRL stimulation, proteins were immunoprecipitated with an antibody to Grb2 and subjected to Western blotting analysis with the use of monoclonal antibody to myc epitope. As can be seen in Figure 4A, SOCS-1 was found to coimmunoprecipitate with Grb2 following PRL stimulation. However, overexpression of the SH3 domains' inactive mutant of Grb2 led to inhibition in PRL-induced formation of Grb2/SOCS-1 complex. Since we have shown that PRL induces SHP-1/Grb2 interaction through the C-terminal tyrosine residues of SHP-1, we next examined whether PRL is able to induce SHP-1/SOCS-1 association and whether this process is regulated by SHP-1 C-terminal tyrosine residues within Grb2-binding motif. As can be seen in Figure 4B, PRL induces a clear association of SOCS-1 with both SHP-1WT and SHP-1CS but not with SHP-1YF or SHP-1CSYF. Together, these results indicate that PRL mediates association of SHP-1 with SOCS-1 through Grb2.
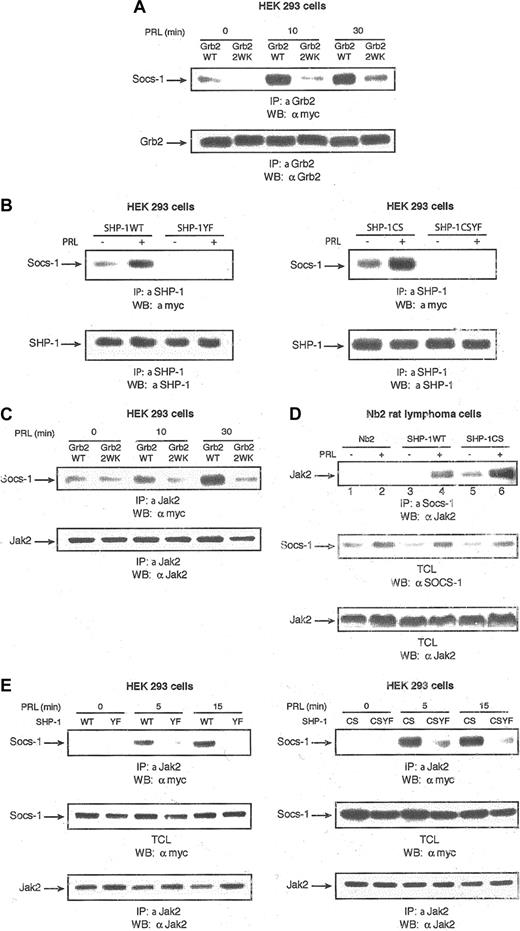
Grb2 and SHP-1 regulation of Jak2/SOCS-1 association. (A) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR, Stat5, and either vector alone or the indicated forms of Grb2. Following 18 hours of serum starvation, cells were stimulated with PRL for the times indicated. Cell lysates were used for immunoprecipitations with polyclonal antibody to Grb2 and immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panel). The same membrane was stripped and immunoblotted with monoclonal antibody to Grb2 (lower panel). (B) The 293 cells were cotransfected with expression vectors encoding PRLR, myc–SOCS-1, and the indicated forms of SHP-1. Serum-starved cells were left untreated or stimulated with PRL for 15 minutes. Cell lysates were immunoprecipitated with polyclonal antibody to SHP-1 and immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panels). The same membranes were stripped and reblotted with polyclonal antibody to SHP-1 (lower panels). (C) Cell lysates from 293 cells transfected with the same expression vectors as in panel A were stimulated with PRL for different times, immunoprecipitated with polyclonal antibody to Jak2, and then immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panel). The same membrane was stripped and immunoblotted with polycclonal antibody to Jak2 (lower panel). (D) Parental Nb2 cells as well as Nb2 cells stably overexpressing SHP-1WT and SHP-1CS were serum starved overnight and then left unstimulated or stimulated with PRL for 1 hour. Cell lysates were immunoprecipitated with polyclonal antibody to SOCS-1 and immunoblotted with polyclonal antibody to Jak2 (upper panel). Total cell lysates were immunoblotted with polyclonal antibodies to SOCS-1 (middle panel) and Jak2 (lower panel). (E) The 293 cells were transfected with the same expression vectors as panel B. Cells were starved overnight and stimulated with PRL for different times. Cell lysates were immunoprecipitated with polyclonal antibody to Jak2 and immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panels). The same membranes were stripped and reblotted with polyclonal antibody to Jak2 (lower panels). Total cell lysates from the same transfections were immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (middle panels).
Grb2 and SHP-1 regulation of Jak2/SOCS-1 association. (A) The 293 cells were transiently cotransfected with expression plasmids encoding PRLR, Stat5, and either vector alone or the indicated forms of Grb2. Following 18 hours of serum starvation, cells were stimulated with PRL for the times indicated. Cell lysates were used for immunoprecipitations with polyclonal antibody to Grb2 and immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panel). The same membrane was stripped and immunoblotted with monoclonal antibody to Grb2 (lower panel). (B) The 293 cells were cotransfected with expression vectors encoding PRLR, myc–SOCS-1, and the indicated forms of SHP-1. Serum-starved cells were left untreated or stimulated with PRL for 15 minutes. Cell lysates were immunoprecipitated with polyclonal antibody to SHP-1 and immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panels). The same membranes were stripped and reblotted with polyclonal antibody to SHP-1 (lower panels). (C) Cell lysates from 293 cells transfected with the same expression vectors as in panel A were stimulated with PRL for different times, immunoprecipitated with polyclonal antibody to Jak2, and then immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panel). The same membrane was stripped and immunoblotted with polycclonal antibody to Jak2 (lower panel). (D) Parental Nb2 cells as well as Nb2 cells stably overexpressing SHP-1WT and SHP-1CS were serum starved overnight and then left unstimulated or stimulated with PRL for 1 hour. Cell lysates were immunoprecipitated with polyclonal antibody to SOCS-1 and immunoblotted with polyclonal antibody to Jak2 (upper panel). Total cell lysates were immunoblotted with polyclonal antibodies to SOCS-1 (middle panel) and Jak2 (lower panel). (E) The 293 cells were transfected with the same expression vectors as panel B. Cells were starved overnight and stimulated with PRL for different times. Cell lysates were immunoprecipitated with polyclonal antibody to Jak2 and immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (upper panels). The same membranes were stripped and reblotted with polyclonal antibody to Jak2 (lower panels). Total cell lysates from the same transfections were immunoblotted with monoclonal antibody to myc tag detecting SOCS-1 (middle panels).
On the basis of these data, we sought to determine whether the C-terminal tyrosine residues of SHP-1 are able to block Jak2 kinase, thereby inhibiting Stat5 by recruitment of Grb2/SOCS-1 and targeting SOCS-1 to Jak2. To test this hypothesis, first we examined the ability of Grb2 to regulate PRL-induced Jak2/SOCS-1 association using Grb2 wild type and the SH3 domains' inactive mutant form of Grb2. Interestingly, following PRL stimulation more SOCS-1 was found to be associated with Jak2 in samples overexpressing Grb2WT but not Grb2W36, 193K (Figure 4C). This indicates that following PRL stimulation, Grb2 through its SH3 domains recruits SOCS-1 and targets SOCS-1 to Jak2, explaining the inhibitory effects of Grb2 in PRLR signaling.
Next we investigated whether SHP-1 is also able to regulate PRL-induced formation of the Jak2/SOCS-1 complex and the role of the C-terminal tyrosine residues of SHP-1 in this process. PRL-induced Jak2/SOCS-1 interactions were examined in the parental Nb2 cells and in Nb2 cells stably overexpressing SHP-1WT or SHP-1CS. Interestingly, we observed more Jak2/SOCS-1 interaction in cells overexpressing SHP-1WT compared with the parental Nb2 cells (Figure 4D; compare lines 4 and 2). This result indicates that SHP-1WT regulates PRL-induced Jak2/SOCS-1 association. Furthermore, we observed more PRL-induced Jak2/SOCS-1 interaction in Nb2 cells overexpressing SHP-1CS compared with cells overexpressing SHP-1WT (Figure 4D; compare lines 6 and 4). This is conceivably due to the enhanced recruitment of Grb2 to SHP-1CS, as discussed before. In agreement with previous studies indicating that SOCS-1 protein levels are induced in response to cytokine stimulation, it is interesting to note that in all 3 cell lines PRL treatment led to an increase in the levels of SOCS-1 (Figure 4D, middle panel). This result indicates that SHP-1 augments Jak2/SOCS-1 interactions downstream of PRLR.
We next determined the role of the tyrosine residues of SHP-1 within the Grb2-binding motif in regulating the PRL-induced Jak2/SOCS-1 interaction. As can be seen in Figure 4E, the PRL-induced Jak2/SOCS-1 association was completely abrogated in cells overexpressing either SHP-1YF or SHP-1CSYF compared with SHP-1WT and SHP-1CS, respectively, indicating the crucial function of SHP-1 C-terminal tyrosine residues in targeting SOCS-1 to Jak2. Together, our results indicate that SHP-1 regulates Jak2/SOCS-1 association downstream of PRLR, hence contributing to down-regulation of Stat5 activation.
Loss of SOCS-1 attenuates SHP-1 function to inhibit Stat5 activation downstream of PRLR
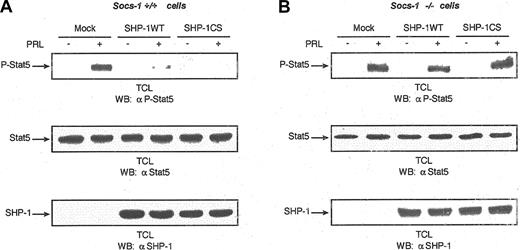
To determine the contribution of SOCS-1 to SHP-1–mediated inhibition of the Jak2/Stat5 pathway downstream of PRLR, we examined the effects of SHP-1WT and SHP-1CS in regulation of PRLR signaling using mouse embryonic fibroblast cells lacking the SOCS-1 gene (SOCS-1–/– MEFs). Similar to what was observed in 293, Nb2, and T47D cells, our results indicate that both SHP-1WT and SHP-1CS down-regulate Stat5 activation in SOCS-1+/+ cells (Figure 5A). However, Stat5 activation was refractory to the negative regulation by SHP-1WT or SHP-1CS in SOCS-1–/– cells (Figure 5B). The fact that the function of SHP-1CS to inhibit Stat5 activation was completely abolished in SOCS-1–/– cells indicates that SOCS-1 is the main player recruited by SHP-1 to block Stat5 activation downstream of PRLR. Furthermore, SHP-1WT significantly reduced Stat5 activation in SOCS-1+/+ cells. However, SOCS-1–/– cells were resistant to the function of SHP-1 in blocking Stat5 activation; in 6 separate experiments, more than 80% of the signal was maintained, demonstrating the importance of SOCS-1 in SHP-1–induced Jak2/Stat5 inhibition. Overall, our results demonstrate that the adaptor function of SHP-1 is critical and is required for its negative regulation of the Jak/Stat pathway.
Need for SOCS-1 in SHP-1–mediated negative regulation of Stat5 activation downstream of PRLR. (A) (B) Parental (SOCS-1+/+) (panel A) as well as SOCS-1–/– (panel B) MEF cells were cotransfected with expression plasmids encoding PRLR, Stat5, and either vector alone or the indicated forms of SHP-1 with the use of FuGENE 6 transfection reagent. Serum-starved cells were left untreated or treated with PRL for 15 minutes. Total cell lysates were imunoblotted with monoclonal antibody to phospho-Stat5 (upper panels). The same lysates were immunoblotted with monoclonal antibody to Stat5 (middle panels) and polyclonal antibody to SHP-1 (lower panels). These immunoblots were chosen as representative of 6 independent experiments.
Need for SOCS-1 in SHP-1–mediated negative regulation of Stat5 activation downstream of PRLR. (A) (B) Parental (SOCS-1+/+) (panel A) as well as SOCS-1–/– (panel B) MEF cells were cotransfected with expression plasmids encoding PRLR, Stat5, and either vector alone or the indicated forms of SHP-1 with the use of FuGENE 6 transfection reagent. Serum-starved cells were left untreated or treated with PRL for 15 minutes. Total cell lysates were imunoblotted with monoclonal antibody to phospho-Stat5 (upper panels). The same lysates were immunoblotted with monoclonal antibody to Stat5 (middle panels) and polyclonal antibody to SHP-1 (lower panels). These immunoblots were chosen as representative of 6 independent experiments.
Discussion
During the past few years, SHP-1 and SOCS-1 have been shown to represent 2 important nonredundant intracellular mechanisms for terminating cytokine signaling. The critical roles each of the 2 proteins plays in signal termination are clearly demonstrated in knockout mice of SHP-1 and SOCS-1. Both of these mouse model systems show immunologic abnormalities indicating loss of negative control mechanisms. As well, both SHP-1 and SOCS-1 have been implicated as tumor suppressor genes.44,45 The cooperation of these 2 proteins in the fine-tuning of signal termination shown by our study seems appropriate. Our data showed that both SHP-1WT and SHP-1CS led to the inhibition of Stat5 activation. This could be explained by the fact that catalytically inactive mutants of protein tyrosine phosphatases can “trap” substrates; however, the observation that SHP-1CSYF was unable to block cytokine-receptor signaling compared with SHP-1CS indicates that the negative regulation by SHP-1CS is due to the adaptor function of SHP-1. Furthermore, it is possible to reason that the loss of the inhibitory effects of SHP-1YF compared with SHP-1WT is due to the fact that phosphorylation of these tyrosine residues may directly modify the activity of the phosphatase domain, as has been proposed for SHP-2.13 This model can be ruled out since even in the absence of catalytic activity, mutation of the tyrosine residues in SHP-1CSYF is able to restore the signal compared with SHP-1CS. Finally, individual mutations of Y538 and Y566, major phosphorylation sites on SHP-1, did not lead to a significant recovery of PRLR signaling, whereas overexpression of SHP-1Y538, 566F was able to partially restore the signal (Figure 2C). Full recovery of PRL signaling was observed only when all 4 tyrosine residues within the Grb2-binding motif were mutated to phenylalanine. These results show that all 4 of the tyrosine residues contribute to the inhibitory effect of SHP-1.
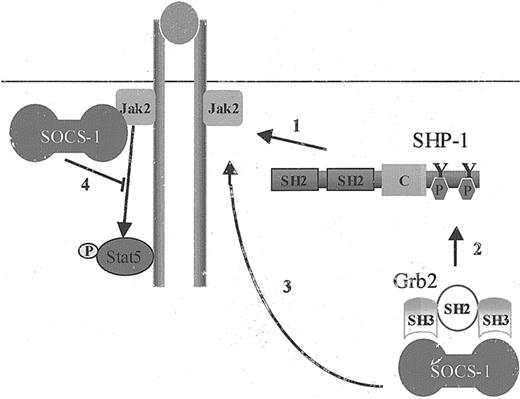
Our results demonstrate for the first time that SOCS-1 contributes to SHP-1 function in negative regulation of cytokine-receptor signaling, as the loss of SOCS-1 significantly compromises SHP-1's ability to down-regulate Stat5 activation. On the basis of these results, we propose that following ligand binding, SHP-1 is recruited to the receptor/Jak complex. SHP-1 recruitment leads to its phosphorylation on the C-terminal tyrosine residues, resulting in the recruitment of Grb2/SOCS-1 complex and targeting SOCS-1 to Jak2 kinase, consequently inhibiting cytokine-receptor signaling (Figure 6).
The adaptor role of SHP-1 in SHP-1–mediated negative regulation of cytokine-receptor signaling. The adaptor role of SHP-1 is critical in SHP-1–mediated negative regulation of cytokine-receptor signaling. The steps in this regulation are as follows: (1) SHP-1 protein tyrosine phosphatase is recruited to receptor/Jak2 complex upon receptor ligand binding. (2) This process results in SHP-1 tyrosine phosphorylation on its C-terminal tyrosine residues, leading to the recruitment of Grb2 and its associated protein SOCS-1. (3) SOCS-1 is targeted to Jak2, leading to (4) down-regulation of Stat5 activation.
The adaptor role of SHP-1 in SHP-1–mediated negative regulation of cytokine-receptor signaling. The adaptor role of SHP-1 is critical in SHP-1–mediated negative regulation of cytokine-receptor signaling. The steps in this regulation are as follows: (1) SHP-1 protein tyrosine phosphatase is recruited to receptor/Jak2 complex upon receptor ligand binding. (2) This process results in SHP-1 tyrosine phosphorylation on its C-terminal tyrosine residues, leading to the recruitment of Grb2 and its associated protein SOCS-1. (3) SOCS-1 is targeted to Jak2, leading to (4) down-regulation of Stat5 activation.
Together, our data present a novel mechanism by which SHP-1 negatively regulates the Jak/Stat pathway. We demonstrated that the C-terminal tyrosine residues of SHP-1 are mediators of this inhibitory effect by recruiting the inhibitory complex Grb2/SOCS-1 and targeting SOCS-1 to Jak2. We extended this mechanism of SHP-1 regulation of PRLR signaling to the EPOR signaling system, suggesting that this model could be a general regulatory mechanism downstream of cytokine receptors.
Prepublished online as Blood First Edition Paper, October 9, 2003; DOI 10.1182/blood-2003-07-2617.
Supported by grants MOP-13681 (S.A.) and MOP-53141 (J.J.L) from the Canadian Institutes of Health Research (CIHR). S.A. and J.J.L. are Research Scientists of the National Cancer Institute of Canada supported with funds provided by the Canadian Cancer Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Peter Gout for supplying the Nb2 cell line, Drs Nancy Hynes and Bernd Groner for supplying the HC11 cell line and anti–milk protein antibody, Dr Ursula Klingmuller for EPOR cDNA, and Drs Bruce Mayer and Motti Anafi for HA-tagged Grb2. We are grateful to Drs Axel Ullrich, Akihiko Yoshimura, and Paul Kelly for supplying materials used in this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal