Abstract
Attachment of gp120 to CD4 during HIV-1 entry triggers structural rearrangement in gp120 that enables binding to an appropriate coreceptor. Following coreceptor engagement, additional conformational changes occur in the envelope (Env), resulting in fusion of virion and cell membranes. Catalysts with redox-isomerase activity, such as protein disulfide isomerase (PDI), facilitate Env conversion from its inactive to its fusion-competent conformation. We report here that anti-PDI agents effectively block CXCR4 Env-mediated fusion and spread of virus infection. Exogenously added PDI, in turn, can rescue fusion from this blockade. We further find that PDI facilitates thiol/disulfide rearrangement in gp120 during conformational change, whereas inhibition of this redox shuffling prevents gp41 from assuming the fusogenic 6-helix bundle conformation. At the virus-cell contact site, gp120 induces assembly of PDI, CD4, and CXCR4 into a tetramolecular protein complex serving as a portal for viral entry. Our findings support the hypothesis that Env conformational change depends on a well-coordinated action of a tripartite system in which PDI works in concert with the receptor and the coreceptor to effectively lower the activation energy barrier required for Env conformational rearrangement.
Introduction
HIV-1 entry requires attachment of the gp120 subunit of the viral envelope (Env) glycoprotein to its primary receptor, CD4. This interaction induces a structural rearrangement in Env exposing conserved regions of the gp120 subunit, thereby enabling binding to an appropriate coreceptor, primarily the chemokine receptors CXCR4 or CCR5.1,2 Engagement of gp120 to the coreceptor triggers conformational changes in the gp41 subunit, leading to the formation of a 6-helix coiled-coil (trimer-of-hairpins) that involves exposure and subsequent insertion of the gp41 fusion peptide into the target cell membrane. This facilitates fusion of virion envelope with the host cell membrane and release of the viral capsid into the cytosol.3
Although the present model of HIV-1–mediated fusion and infection is thought to require only CD4 and a coreceptor, a reconstituted lipid bilayer system, which would incorporate only these components while excluding other cellular proteins, has not yet been developed to confirm this. Hence, it is conceivable that HIV-1 may rely on some additional catalyst to facilitate the conformational alterations in Env-driving membrane fusion. Using inhibitors of thiol (-SH)/disulfide (-S-S-) exchange, Ryser et al4 showed inhibition of HIV-1 replication and postulated that catalysts with redox-isomerase activity assist in Env refolding during entry. Three more recent papers5-7 provided additional support and novel insights supporting this hypothesis. The candidate-protein proposed to drive the -SH/-S-S- exchange is cell-surface localized protein disulfide isomerase (PDI). PDI is considered a redox-isomerase, based on its ability to catalyze the reduction, formation, and isomerization of disulfide bonds.8 Although an endoplasmic reticulum-resident protein, PDI is commonly detected at the cell surface.9 Given that PDI does not bear a transmembrane domain, its association with the plasma membrane occurs by way of noncovalent interactions with integral membrane proteins, lipids, or glycans.10
In addition to HIV-1, other animal viruses (baculovirus and vaccinia virus) have been shown to depend on the interchange of critical thiols and disulfides for successful entry. For example, thiol/disulfide interchange allows aggregation of the baculovirus fusion protein, gp64, into a fusogenic complex, which drives fusion of the viral envelope with the cell membrane.11 Vaccinia virus entry similarly depends on the reduction of critical disulfides in the core proteins, which disrupts viral structure and allows delivery of the viral core into the cytoplasm.12 The catalyst driving the thiol/disulfide exchange in these viruses has not been elucidated. However, it is reasonable to postulate the involvement of a mechanism that modulates the exchange of protons between a cell-surface associated donor (PDI) and viral proteins. In addition to viral fusion, other protein-mediated membrane fusion reactions involving -SH/-S-S- interchange have been documented. Examples of such -SH/-S-S–dependent fusions include exocytosis, fertilization, microsome fusion, and endocytosis.13
The present paper confirms the ability of PDI inhibitors to prevent Env-mediated fusion in an assay detecting the initial events leading to membrane fusion by way of the transfer of fluorescent dye from CD4+/CXCR4+ cells to their Env-expressing fusion partners. Our data also corroborate that PDI antagonists inhibit cell-to-cell transmission of HIV infection as measured by a decrease in reverse transcriptase activity in supernatants from treated, infected cells. They further support the notion that disulfide bonds in gp120 are reduced on attachment to CD4 on target cells and that such reduction occurs prior to, or concomitant with, the coreceptor binding.
In addition to corroborating earlier results, the present paper provides several novel, previously unreported, findings: (1) It demonstrates both by immunocolocalization and immunoprecipitation the presence of a tetramolecular cell surface-associated complex comprised of PDI, gp120, CD4, and CXCR4. (2) It reveals a new function of gp120 that appears to be involved in recruiting the other 3 components that contribute to the tetramolecular complex, as illustrated by the shift in immunocolocalization in the presence of gp120. (3) It shows that, although PDI is recruited into the tetramolecular protein complex at the cell surface, it is unlikely that this assembly takes place in GM1 lipid rafts. (4) This paper also provides evidence that treatment of target cells with the PDI inhibitor DTNB prevents gp41 from assuming the 6-helix bundle fusogenic conformation believed necessary to drive fusion. (5) This work extends published data further to show that only PDI associated with the fusion partner expressing CD4/CXCR4 contributes to fusion.
Materials and methods
Materials
Calcein am was purchased from Molecular Probes (Eugene, OR), Immobilon-P filters from Millipore (Bedford, MA), and bovine liver PDI from Calbiochem (La Jolla, CA). Enhanced chemifluorescence (ECF) substrate was obtained from Amersham Pharmacia Biotech (Buckinghamshire, United Kingdom). l-glutathione disulfide (GSSG), l-glutathione (GSH), and 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) were purchased from Sigma Chemical Co (St Louis, MO). Stromal cell-derived factor 1α (SDF-1α) was obtained from Antigenix America Inc (Huntington Station, NY). HIV-1 IIIB gp120 was produced as previously described14 by infection of BS-C-1 cells with the recombinant vaccinia virus vPE 50 at 5 PFU/cell.
Antibodies
Preparation of rabbit anti–HIV-1 gp41 serum directed against the N- and C-terminal gp41 heptad repeats was described elsewhere.15 The hybridoma producing the anti-gp41 monoclonal antibody (MAb), Chessie 8, was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) from Dr George Lewis.16 Rabbit anti-gp120 serum was generated by using recombinant gp120 IIIB as an immunizing antigen (K.S., unpublished observation, February 1992). The anti-CXCR4 MAb (4G10) was raised against a synthetic peptide corresponding to the N-terminus of CXCR4 (C.C.B. and Berger, unpublished observation, April 1996). Anti-actin polyclonal antibody (PAb) was purchased from Sigma Chemical Co. Phycoerythrin (PE)–labeled MAbs, Leu3A and 12G5, were obtained from BD Biosciences (San Jose, CA). Rat anti-CD4 MAb (clone YNB46.1.8) was purchased from Serotec (Raleigh, NC). Mouse anti-p56LCK and mouse antitransferrin receptor antibodies were obtained from Zymed Laboratories (San Francisco, CA). Rabbit anti-PDI PAb and mouse anti-PDI MAb were obtained from Stressgen Biotechnologies (San Diego, CA); alternative mouse anti-PDI MAb (clone RL-90) was purchased from Affinity Bioreagents (Golden, CO). The following secondary and control antibodies were used: antimouse antibody conjugated with Alexa Fluor 555 and antirat antibody conjugated with Alexa Fluor 647 (Molecular Probes), antirabbit antibody coupled with Cy2 and PE, as well as antimouse-PE (Jackson ImmunoResearch Laboratories, West Grove, PA), antirabbit and antimouse immunoglobulin G (IgG) antibodies conjugated with alkaline phosphatase (Pierce Chemical Co, Rockford, IL), isotype controls corresponding to the indicated MAb and irrelevant PAbs (R&D Systems, Minneapolis, MN; and BD Biosciences).
Cells
CD4+/CXCR4+ Sup T-1 cells (ATCC, Rockville, MD) and CD4+/CXCR4+/CCR5+ PM1 cells (M.A. Norcross, CDER, FDA) were grown in RPMI-1640 supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT). CHO-160 cells,17 constitutively expressing wild-type CXCR4-specific HXB2 Env, were cultured in Glasgow minimum essential medium (GMEM-S; BioSource International, Camarillo, CA) with 10% dialyzed FBS and 400 μM methionine sulfoximine (Sigma Chemical). HOS-CD4+/CXCR4– and HOS-CD4+/CXCR4+ cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from Dr Nathaniel Landau) were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS18,19 and HOS-CD4–/CXCR4– cells (ATCC) in Eagle minimal essential medium (EMEM) supplemented with 10% FBS.
Quantification of cell-cell fusion
Fusion was assessed by fluorescent dye (Calcein am) transfer from labeled CD4/CXCR4-expressing Sup T-1 or HOS-CD4+/CXCR4+ (target) cells to unlabeled envelope-expressing (Env+; host) CHO-160 cells. Sup T-1cells (7 × 106) were loaded with 0.02 μM Calcein am (in 100% dimethyl sulfoxide [DMSO]), incubated 40 minutes at 37°C, and excess dye was removed by 2 washes in phosphate-buffered saline (PBS). Next, labeled Sup T-1 cells were resuspended, added at a 5:1 ratio to unlabeled, adherent CHO-160 cells plated 16 hours earlier, and cocultured (1 hour, 37°C) to induce fusion. The role of redox shuffling in HIV-1 entry was studied by using the following membrane impermeable, thiol-reactive reagents as fusion inhibitors: (1) DTNB, (2) anti-PDI MAbs, and (3) bacitracin. Labeled target cells were treated with 0 to 5 mM DTNB or bacitracin (30 minutes, 37°C) or with MAb (15 minutes, 22°C). These agents did not interfere with the uptake of the fluorescent dye (data not shown). The contribution of PDI from Env-expressing membrane on fusion was assessed using CHO-160 cells treated with 1 or 5 mM DTNB (30 minutes, 37°C). After removing residual inhibitors, Sup T-1 cells were cocultured with Env+ cells for 1 hour at 37°C. Control cells were treated with PBS. Cells were treated with excess anti-CD3 MAb to control for potential steric hindrance by anti-PDI MAb. Fusion inhibitors did not affect the ability of labeled target cells to retain fluorescent dye (data not shown). To assess PDI's ability to rescue fusion after DTNB treatment, Sup T-1 cells were treated with 2.5 mM DTNB or PBS (control) (30 minutes, 37°C), washed twice in PBS, then 20 or 40 μg/mL PDI in 2 mM GSSG/20 mM GSH buffer was applied (90 minutes, 22°C). Cells that did not receive PDI were treated with GSSG/GSH buffer alone. The extent of fusion was quantified as the ratio of bound, dye-redistributed Sup T-1 cells to the total number of bound Sup T-1.
Virus infection of T cells
HIV-1LAV (0.005 RT cpm/cell) was adsorbed to 2 × 105pm-1 cells in serum-free medium (2 hours, 37°C). Virus replication was assessed in the presence of 1 mM DTNB, 25 μg/mL monoclonal, and 40 μg/mL polyclonal anti-PDI Abs. Normal mouse and rabbit IgG isotype controls were included at relevant concentrations. PDI inhibitors were added after the virus adsorption period. Cells were washed following virus adsorption and cultured in RPMI 1640 containing 10% FBS, 2 mM l-glutamine, and 0.01M HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid). Culture supernatant (62%) was removed every 2 days and replaced with fresh medium containing inhibitors where appropriate for the duration of the experiment. Harvested supernatants were assayed for reverse transcriptase (RT) activity to monitor the progression of infection.20 Curves shown reflect the best fits of sample means for duplicate wells that differed by not more than 15%.
Flow cytometry
To study the effects of DTNB on CD4 or CXCR4, Sup T-1 cells (1-2 × 106) treated with 2.5 mM DTNB or PBS (control) were incubated either with 0.5 μg gp120 IIIB/1 × 106 cells (30 minutes, 22°C) followed by anti-gp120 rabbit serum (15 minutes, 22°C) and PE-conjugated antirabbit secondary Ab, or with PE-labeled Leu3A (anti-CD4) or 12G5 (anti-CXCR4) Abs (15 minutes, 22°C). Cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytometer (BD Biosciences).
Measurement of intracellular Ca2+ mobilization
To assess the release of intracellular Ca2+, Sup T-1 cells were suspended in Hanks balanced salt solution and transferred (1 × 106 cells) to poly-l-lysine–coated coverslips for 30 minutes at 22°C. Cells were exposed to 2.5 μM Fura-2/am (Molecular Probes) and 2.5 mM DTNB or Hanks buffer (control) for 30 minutes at 37°C in the dark. Coverslips were mounted on the stage of an Axiovert 135 microscope (Zeiss, Göttingen, Germany) attached to the Attoflor Digital Fluorescence Microscopy System (Atto Instruments, Rockville, MD). Cells were exposed to alternating 340- and 380-nm light beams, and the intensity of light emission at 520 nm was measured. The ratio of light intensities, F340/F380, reflecting the changes in intracellular calcium concentration, [Ca2+]i, was followed in 60 to 70 single cells simultaneously. Release of intracellular Ca2+ was induced with 500 ng/mL SDF-1α 60 seconds after initiation of the measurement. A total of 227 treated and 169 untreated cells were analyzed.
SDS-PAGE and Western blot analysis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were performed as described previously.21 Briefly, proteins electrophoresed on 4% to 12% gradient Bis-Tris (tris(hydroxymethyl)aminomethane) polyacrylamide gels (Invitrogen Corporation, Carlsbad, CA) were transferred to Immobilon-P filters and reacted with primary antibodies, then blots were washed and incubated with alkaline phosphatase–conjugated goat antimouse or antirabbit IgG. After incubation with enhanced chemifluorescence substrate, dried blots were scanned on a Fuji Film Fluorescent Reader FLA-3000 at 473/520 nm excitation/emission wavelength with the Image Reader V 1.7 software package. Blots were quantified using Image Gauge V 3.45 (Fuji Film, Tokyo, Japan).
Labeling of gp120
Cell-surface thiols were labeled with 3-(N-maleimidylpropionyl) biocyatin (biotin maleimide; MPB; Molecular Probes). MPB (300 μM in RPMI) was added simultaneously with 10 μg gp120 IIIB/10 × 106 HOS-CD4–/CXCR4–, HOS-CD4+/CXCR4–, or HOS-CD4+/CXCR4+ cells and incubated for 30 minutes at 22°C on a rotator. To block PDI, 5 mM DTNB was applied to target cells prior to labeling. Unreacted MPB was quenched with 1 mM GSH. Labeled cells were washed twice and lysed in 20 mM sodium phosphate buffer (pH 7.5) containing 150 mM NaCl, 0.1% SDS, 1.5% Triton X-100 (TX-100), and protease inhibitor cocktail followed by incubation with the UltraLink Immobilized Streptavidin Plus (Pierce Chemical Co) (1 hour, 4°C). Immobilized streptavidin with bound proteins was washed 4 times in lysis buffer; biotin-labeled proteins were liberated from streptavidin by boiling in 4% lithium dodecyl sulfate (LDS) sample buffer, then analyzed for gp120 content by SDS-PAGE and immunoblotting with polyclonal anti-gp120 Ab.
Detection of 6-helix bundle conformation of gp41
CD4+/CXCR4+ cells were treated with 0 to 5 mM DTNB for 30 minutes at 37°C, washed, and cocultured with Env+ cells at a 3:1 ratio in the presence of anti–6 helix bundle PAb or rabbit PAb (control) for 1 hour at 37°C. Cells were washed twice, lysed (50 mM Tris-HCl, pH 8.0, 1.5% TX-100, 150 mM NaCl, and protease inhibitor cocktail), and incubated with protein G Sepharose beads (Pharmacia) for 1 hour at 4°C. Immobilized gp41 was washed 4 times in the lysis buffer and eluted from protein G beads by boiling in 2% LDS sample buffer, then analyzed for gp41 content by SDS-PAGE and immunoblotting with anti-gp41 MAb (Chessie 8). To control for potential loss of material, supernatants remaining after immunoprecipitation were tested for actin content by Western blotting using an anti-actin PAb.
Confocal microscopy
Analysis of PDI colocalization with CD4 and CXCR4 was carried out by using the following combinations of Abs: mouse anti-CXCR4 MAb (4G10) with antimouse Alexa Fluor 555–coupled secondary Ab, rat anti-CD4 MAb with antirat Alexa Fluor 647–coupled secondary Ab, and rabbit anti-PDI PAb with antirabbit Cy2–coupled secondary Ab. Sup T-1 cells were attached to poly-l-lysine–coated slides and treated in the presence or absence of gp120 IIIB in RPMI (0.5 μg/L × 106 cells) (30 minutes, 22°C). Cells were fixed in 2% paraformaldehyde and reacted with primary then secondary Abs. Cells were mounted in Fluoromount-G (Electron Microscopy Sciences, Fort Washington, PA), and images were acquired with a Zeiss LSM410 laser scanning confocal microscope equipped with a mixed-gas laser (488-, 568-, and 647-nm excitation lines) in sequential scans then analyzed with Zeiss LSM software (Thornwood, NY).
To test whether PDI is associated with GM1 rafts, Sup T-1 cells were treated with gp120 in RPMI (0.5 μg/1 × 106 cells) followed by anti-PDI rabbit PAb and cholera toxin B subunit coupled to Alexa Fluor 594 (45 minutes, 4°C). Further cross-linking with Cy2-coupled secondary antibodies was performed by incubation of cholera toxin–labeled cells for 10 minutes at 37°C.
Cells were then fixed, attached to poly-L-lysine slides, mounted, and visualized by confocal microscopy.
Co-immunoprecipitation
Anti-CXCR4 MAb, 4G10, was allowed to interact with Protein G Sepharose beads (Amersham Pharmacia Biotech AB, Uppsala, Sweden) for 1 hour followed by covalent cross-linking to Protein G beads with 20 mM dimethylpimelimidate for 30 minutes at 22°C. The reaction was stopped by incubation in 200 mM ethanolamine (pH 8) for 2 hours. HOS-CD4+/CXCR4+ cells (5 × 106) pretreated with 10 μg gp120 IIIB (40 minutes, 22°C) were washed and resuspended in lysis buffer (1% Brij97, 150 mM NaCl, 20 mM Tris [pH 8], 20 mM EDTA [ethylenediaminetetraacetic acid] and protease inhibitors; 4°C, 1 hour). The nuclear fraction was removed by centrifugation and 4G10/protein G beads were added to the supernatants for 14 hours at 4°C. The beads were washed extensively with lysis buffer, and proteins were eluted by boiling in 4% LDS sample buffer and then analyzed for gp120, CD4, and PDI content by SDS-PAGE and immunoblotting with the corresponding polyclonal rabbit sera.
Flotation gradient experiments
The protocol for analysis of membrane rafts has been published elsewhere.22 Briefly, to analyze PDI segregation into rafts, Sup T-1 cells (6 × 107) were treated with hypotonic buffer (10 mM Tris-HCl, pH 7.5, 4 mM EDTA and protease inhibitor cocktail) on ice. Postnuclear supernatant was lysed in cold TNE buffer containing 0.25% TX-100, 25 mM Tris-HCl, 150 mM NaCl, and 4 mM EDTA for 20 minutes on ice. Lysate was subsequently brought to 40% sucrose and overlaid with 30% followed by 5% sucrose in TNE buffer. Samples were subjected to flotation centrifugation at 181 298 g (46 000 rpm) for 16 hours at 4°C by using Beckman TLS-55 rotor. Ten fractions were recovered from the top of the gradient and analyzed by SDS-PAGE and Western blotting. Transferrin receptor (TfR) was used as a nonraft marker, whereas p56LCK (SrcK) was used as a raft-specific marker. Cellular cholesterol was depleted with 10 mM methyl-β-cyclodextrin (MβCD) for 10 minutes at 37°C prior to flotation centrifugation, where indicated. In some instances, cells were treated with gp120 IIIB (0.5 μg/1 × 106 cells) before lysis.
Results
Cell-cell fusion depends on expression of functional PDI on the target cell
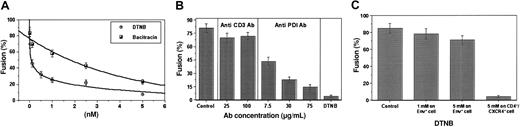
To determine whether PDI facilitates HIV-1 entry, CD4+/CXCR4+ cells (Sup T-1) were treated with the following: (1) DTNB, a nonspecific thiol blocker; (2) anti-PDI monoclonal antibodies (MAbs); and (3) bacitracin, a specific PDI inhibitor.9,23 Each PDI inhibitor significantly reduced the level of cell-cell fusion in a dose-dependent manner. The maximal doses of DTNB and bacitracin (5 mM) caused 91.1% and 72% inhibition of fusion, respectively (Figure 1A). Similarly, the highest dose of anti-PDI MAb (75 μg/mL) caused a significant reduction in fusion compared with an excess of anti-CD3 MAb used as a control for steric hindrance (14.5% versus 70% fusion; Figure 1B).
PDI inhibitors significantly decrease fusion between CD4+/CXCR4+ and Env+ cells. CD4+/CXCR4+ Sup T-1 (target) cells were treated with PDI inhibitors DTNB or bacitracin (A), as well as anti-PDI MAb or anti-CD3 control MAb (B) (30 minutes, 37°C), then washed and added to Env-expressing cells to assess fusion. (C) For reciprocal experiments to determine the effect of PDI inhibitors on the Env+ cells, CHO-160 cells were incubated under the conditions noted earlier in the presence and absence of DTNB (1 or 5 mM). Cells were then washed and cocultured with untreated Sup T-1 cells. Fusion of treated Sup T-1 and untreated CHO-160 cells was compared with fusion of untreated Sup T-1 and treated CHO-160 cells. Fusion was measured by fluorescent dye (Calcein am) redistribution from labeled Sup T-1 to unlabeled Env+ cells. Fusion was not observed in the control experiments when CD4+/CXCR4+ cells were mixed with Env– cells or in the presence of T-20/T-21 peptides, which are potent inhibitors of HIV-1 Env-mediated fusion3 (data not shown). Fusion (%) represents the ratio of bound, dye-redistributed Sup T-1 cells to the total number of bound Sup T-1. Each point (A) or bar (B) represents mean fusion ± SE for 200 cells counted.
PDI inhibitors significantly decrease fusion between CD4+/CXCR4+ and Env+ cells. CD4+/CXCR4+ Sup T-1 (target) cells were treated with PDI inhibitors DTNB or bacitracin (A), as well as anti-PDI MAb or anti-CD3 control MAb (B) (30 minutes, 37°C), then washed and added to Env-expressing cells to assess fusion. (C) For reciprocal experiments to determine the effect of PDI inhibitors on the Env+ cells, CHO-160 cells were incubated under the conditions noted earlier in the presence and absence of DTNB (1 or 5 mM). Cells were then washed and cocultured with untreated Sup T-1 cells. Fusion of treated Sup T-1 and untreated CHO-160 cells was compared with fusion of untreated Sup T-1 and treated CHO-160 cells. Fusion was measured by fluorescent dye (Calcein am) redistribution from labeled Sup T-1 to unlabeled Env+ cells. Fusion was not observed in the control experiments when CD4+/CXCR4+ cells were mixed with Env– cells or in the presence of T-20/T-21 peptides, which are potent inhibitors of HIV-1 Env-mediated fusion3 (data not shown). Fusion (%) represents the ratio of bound, dye-redistributed Sup T-1 cells to the total number of bound Sup T-1. Each point (A) or bar (B) represents mean fusion ± SE for 200 cells counted.
To examine whether PDI from CHO-160 cells24 contributes to fusion, Env+ cells were treated with DTNB before incubation with CD4+/CXCR4+ cells (Figure 1C). A minimal reduction in fusion occurred when Env+ cells were pretreated, whereas significant inhibition was observed when CD4+/CXCR4+ cells were pretreated with PDI inhibitor, indicating that fusion depends on the presence of functional PDI on the CD4/CXCR4-expressing target cell.
To determine whether the most efficacious PDI inhibitor, DTNB, alters gp120 binding to CD4 or CXCR4, we exposed CD4+/CXCR4+ cells to 2.5 mM DTNB, which caused 88% fusion inhibition, and measured binding of soluble gp120 IIIB (Figure 2A), Leu3A (CD4-specific MAb) (Figure 2B), and 12G5 (CXCR4-specific MAb) (Figure 2C) with the use of flow cytometry. DTNB failed to modify the binding under all conditions tested, supporting the concept of PDI involvement in a postbinding event. To further study the effect of DTNB on the target cells, we tested the ability of CXCR4 to induce release of intracellular Ca2+ after DTNB treatment. Treated and untreated CD4+/CXCR4+ cells responded similarly to CXCR4 ligand, SDF-1α, stimulation as evident by Ca2+ release in 71 of 227 (31%) treated versus 65 of 169 (38%) untreated cells (Figure 2D). Moreover, both cell groups had comparable kinetics in eliminating Ca2+ from the cytosol.
DTNB does not modify the Env binding site on the CD4 or CXCR4. (A) Sup T-1 cells treated with 2.5 mM DTNB or PBS (control) were incubated with 0.5 μg gp120/1 × 106 cells (30 minutes, 22°C) followed by anti-gp120 rabbit serum (15 minutes, 22°C) and PE-conjugated antirabbit secondary Ab or (B) with PE-labeled Leu3A (anti-CD4) MAb (15 minutes, 22°C). After staining, cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytofluorometer. Data shown are representative of 3 experiments. (C) Sup T-1cells treated with 2.5 mM DTNB or PBS (control) were incubated with 12G5 (anti-CXCR4) MAb. After staining, cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytofluorometer. (D) CXCR4 retains the ability to mobilize intracellular Ca2+ after DTNB treatment. Sup T-1 cells attached to poly-l-lysine–coated coverslips were loaded with 2.5 μM Fura-2/am and 2.5 mM DTNB or Hanks buffer (control). Fluorescence in cells was excited by alternating 340- and 380-nm light beams, and the emitted light intensity was measured at 520 nm. Data shown reflect a single-cell response in mobilizing intracellular Ca2+ induced with 500 ng/mL SDF-1α after following the baseline [Ca2+]i for 60 seconds and are representative of 3 experiments.
DTNB does not modify the Env binding site on the CD4 or CXCR4. (A) Sup T-1 cells treated with 2.5 mM DTNB or PBS (control) were incubated with 0.5 μg gp120/1 × 106 cells (30 minutes, 22°C) followed by anti-gp120 rabbit serum (15 minutes, 22°C) and PE-conjugated antirabbit secondary Ab or (B) with PE-labeled Leu3A (anti-CD4) MAb (15 minutes, 22°C). After staining, cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytofluorometer. Data shown are representative of 3 experiments. (C) Sup T-1cells treated with 2.5 mM DTNB or PBS (control) were incubated with 12G5 (anti-CXCR4) MAb. After staining, cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytofluorometer. (D) CXCR4 retains the ability to mobilize intracellular Ca2+ after DTNB treatment. Sup T-1 cells attached to poly-l-lysine–coated coverslips were loaded with 2.5 μM Fura-2/am and 2.5 mM DTNB or Hanks buffer (control). Fluorescence in cells was excited by alternating 340- and 380-nm light beams, and the emitted light intensity was measured at 520 nm. Data shown reflect a single-cell response in mobilizing intracellular Ca2+ induced with 500 ng/mL SDF-1α after following the baseline [Ca2+]i for 60 seconds and are representative of 3 experiments.
Exogenous PDI can rescue cell-cell fusion
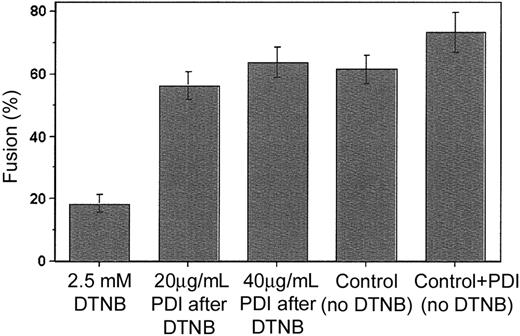
To confirm that PDI is a cofactor involved in viral entry, we designed a novel fusion-rescue experiment, whereby fusion was measured after the addition of exogenous PDI to cells previously treated with 2.5 mM DTNB. We found that PDI added at concentrations of 20 and 40 μg/mL following DTNB application and washout could restore Env-mediated fusion to levels comparable to those observed for the untreated controls (Figure 3). Together with the initial data showing fusion inhibition by specific PDI inhibitors (bacitracin and anti-PDI MAb), this finding provides strong evidence in support of PDI as a facilitator of HIV-1 entry.
Fusion can be rescued by the addition of exogenous PDI. Sup T-1 cells initially treated with 2.5 mM DTNB or PBS (control) were next incubated with 0 (control), 20 or 40 μg/mL PDI in 2 mM GSSG/20 mM GSH buffer (90 minutes, 22°C), then washed and cocultured with untreated Env-expressing cells (1 hour at 37°C). Bar no. 4 is the untreated control; bar no. 5 represents an additional control not exposed to DTNB but treated with 40 μg/mL PDI. The extent of fusion was measured following coincubation of Sup T-1 and Env+ cells as the ratio of bound, dye-redistributed Sup T-1 cells to the total number of bound Sup T-1. Each bar represents mean fusion ± SE for 200 cells counted.
Fusion can be rescued by the addition of exogenous PDI. Sup T-1 cells initially treated with 2.5 mM DTNB or PBS (control) were next incubated with 0 (control), 20 or 40 μg/mL PDI in 2 mM GSSG/20 mM GSH buffer (90 minutes, 22°C), then washed and cocultured with untreated Env-expressing cells (1 hour at 37°C). Bar no. 4 is the untreated control; bar no. 5 represents an additional control not exposed to DTNB but treated with 40 μg/mL PDI. The extent of fusion was measured following coincubation of Sup T-1 and Env+ cells as the ratio of bound, dye-redistributed Sup T-1 cells to the total number of bound Sup T-1. Each bar represents mean fusion ± SE for 200 cells counted.
Functional PDI is necessary for successful viral infection
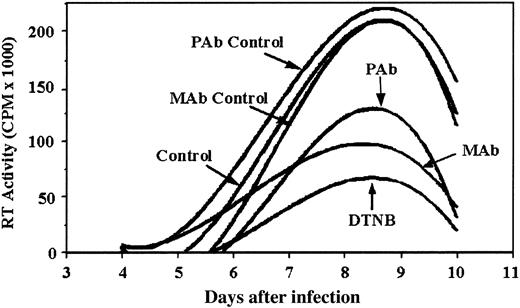
To confirm that PDI is essential for a spreading HIV-1 infection (ie, cell-to-cell transmission), we examined the effect of PDI inhibitors DTNB, anti-PDI MAb, and anti-PDI PAb on an acute infection of PM1 cells with CXCR4-tropic HIV-1LAV. Addition of PDI inhibitors after completion of virus adsorption and maintaining them throughout the course of infection reduced the spread of virus as shown by a significant reduction in the peak RT activity relative to the controls (Figure 4). DTNB maintained in cell culture at 1 mM had no effect on cell viability (data not shown; see Fenouillet et al5 ). These data support the concept that functional PDI is important for the spread of HIV-1 infection.
Functional PDI is necessary for successful HIV-1 infection. HIV-1LAV was incubated with pm-1 cells for a 2-hour adsorption period. Inhibitors (1 mM DTNB, 25 μg/mL monoclonal anti-PDI Abs, and 40 μg/mL polyclonal anti-PDI Abs) were added after the viral adsorption period and replenished with media replacement every 2 days thereafter (days 4, 6, and 8). Isotype controls for MAb and PAb were included at concentrations equivalent to anti-PDI Abs. Values shown correspond to RT activity measured in supernatants harvested during the course of infection on days 4, 6, 8, and 10 and reflect the best fits of sample means that differed by not more than 15%. Data shown are representative of 2 experiments.
Functional PDI is necessary for successful HIV-1 infection. HIV-1LAV was incubated with pm-1 cells for a 2-hour adsorption period. Inhibitors (1 mM DTNB, 25 μg/mL monoclonal anti-PDI Abs, and 40 μg/mL polyclonal anti-PDI Abs) were added after the viral adsorption period and replenished with media replacement every 2 days thereafter (days 4, 6, and 8). Isotype controls for MAb and PAb were included at concentrations equivalent to anti-PDI Abs. Values shown correspond to RT activity measured in supernatants harvested during the course of infection on days 4, 6, 8, and 10 and reflect the best fits of sample means that differed by not more than 15%. Data shown are representative of 2 experiments.
Gp120 undergoes disulfide rearrangement after CD4 engagement
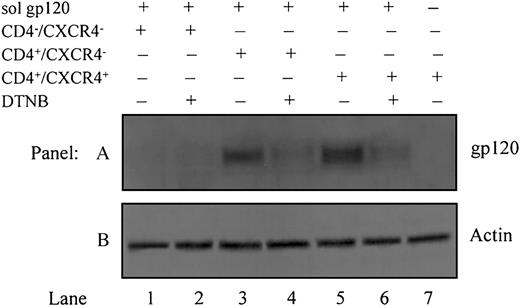
To determine whether gp120 undergoes disulfide rearrangement, releasing free thiols after engaging to CD4 and/or CXCR4, soluble recombinant gp120 IIIB was added to HOS-CD4–/CXCR4–, HOS-CD4+/CXCR4–, or HOS-CD4+/CXCR4+ cells concomitantly with the thiol group-marker, MPB. Assuming that PDI on the target cell catalyzes thiol/disulfide isomerization and enables gp120 to acquire its fusion-competent conformation, rearrangement of existing disulfide bonds in gp120 is required. Given that no free thiols are present in the gp120 ectodomain in its native conformation, incorporation of MPB indicates reduction of one or multiple disulfide bonds (Figure 5, lanes 3 and 5). DTNB (5 mM) blocked this effect, causing a significant decrease in MPB labeling (Figure 5, lanes 4 and 6). Labeling of gp120 thiols in HOS-CD4+/CXCR4– (devoid of only CXCR4, Figure 5, lane 3) is comparable to that in HOS-CD4+/CXCR4+ cells (Figure 5, lane 5). This indicates that CD4 is sufficient for allowing PDI to induce thiol/disulfide exchange in Env. Control HOS-CD4–/CXCR4– cells devoid of both CD4 and CXCR4 (Figure 5, lanes 1-2) or HOS-CD4+/CXCR4+ cells labeled in the absence of soluble gp120 (Figure 5, lane 7) showed no incorporation of MPB.
Gp120 undergoes disulfide-reduction prior to, or concomitant with, binding to CXCR4. All cells except for those in lane 7 were pretreated with soluble gp120 IIIB followed by labeling with biotin maleimide (MPB) to label free -SH groups. Lane 1, HOS-CD4–/CXCR4– cells (devoid of CD4 and CXCR4); lane 2, HOS-CD4–/CXCR4– cells pretreated with 5 mM DTNB to block PDI prior to MPB labeling; lane 3, HOS-CD4+/CXCR4– cells (devoid of only CXCR4); lane 4, HOS-CD4+/CXCR4–cells pretreated with 5 mM DTNB prior to MPB labeling; lane 5, HOS-CD4+/CXCR4+ cells (expressing both CD4 and CXCR4); lane 6, HOS-CD4+/CXCR4+ cells pretreated with 5 mM DTNB prior to MPB labeling; lane 7, HOS-CD4+/CXCR4+ cells that were not pretreated with gp120 (A). Residual supernatants after immunoprecipitation were analyzed for actin content with an anti-actin PAb (B). Data shown are representative of 2 experiments.
Gp120 undergoes disulfide-reduction prior to, or concomitant with, binding to CXCR4. All cells except for those in lane 7 were pretreated with soluble gp120 IIIB followed by labeling with biotin maleimide (MPB) to label free -SH groups. Lane 1, HOS-CD4–/CXCR4– cells (devoid of CD4 and CXCR4); lane 2, HOS-CD4–/CXCR4– cells pretreated with 5 mM DTNB to block PDI prior to MPB labeling; lane 3, HOS-CD4+/CXCR4– cells (devoid of only CXCR4); lane 4, HOS-CD4+/CXCR4–cells pretreated with 5 mM DTNB prior to MPB labeling; lane 5, HOS-CD4+/CXCR4+ cells (expressing both CD4 and CXCR4); lane 6, HOS-CD4+/CXCR4+ cells pretreated with 5 mM DTNB prior to MPB labeling; lane 7, HOS-CD4+/CXCR4+ cells that were not pretreated with gp120 (A). Residual supernatants after immunoprecipitation were analyzed for actin content with an anti-actin PAb (B). Data shown are representative of 2 experiments.
The band intensity in the anti-actin control (Figure 5B) indicates no loss of material during immunoprecipitation or electrophoresis. A fusion assay was carried out concurrently, showing that pretreating HOS-CD4+/CXCR4+ cells with 0.1, 1, or 5 mM DTNB inhibits fusion with Env+ CHO-160 cells in a dose-dependent manner (data not shown).
Inhibition of gp120 redox shuffling prevents formation of the gp41 6-helix bundle
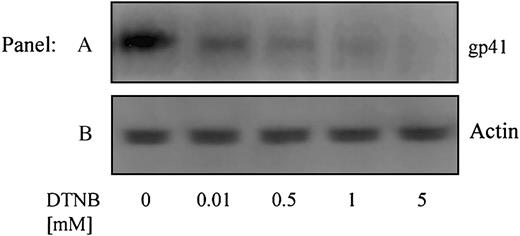
Using conformation-specific antibodies, we next attempted to determine how conformational changes leading to redox-isomerization in gp120 affect the activation (conformational modification) of gp41. An Ab specific for the 6-helix bundle conformation of gp41, that exists following Env-CXCR4-binding but prior to fusion,15 was used to immunoprecipitate gp41 from cells treated with DTNB (0-5 mM) (Figure 6A). The failure of this antibody to immunoprecipitate gp41 in the presence of DTNB, which blocks PDI active sites (vicinal thiols), indicates that PDI-induced redox shuffling in gp120 is necessary to enable formation of the fusogenic 6-helix bundle conformation of gp41. The intensity of the anti-actin controls (Figure 6B) indicates no loss of material during immunoprecipitation or electrophoresis.
PDI acts on Env prior to the 6-helix bundle conformation of gp41. Sup T-1 cells were treated with 0 to 5 mM DTNB. DTNB-treated cells were then cocultured with Env+ cells in the presence of anti–6 helix bundle PAb for 1 hour at 37°C. The cells were lysed, incubated with Protein G-coupled beads, then immobilized proteins were analyzed for gp41 content by SDS-PAGE and immunoblotting with anti-gp41 MAb (Chessie 8) (A). As a control, residual supernatants from immunoprecipitation were analyzed for actin content with an anti-actin PAb (B). Data shown are representative of 2 experiments.
PDI acts on Env prior to the 6-helix bundle conformation of gp41. Sup T-1 cells were treated with 0 to 5 mM DTNB. DTNB-treated cells were then cocultured with Env+ cells in the presence of anti–6 helix bundle PAb for 1 hour at 37°C. The cells were lysed, incubated with Protein G-coupled beads, then immobilized proteins were analyzed for gp41 content by SDS-PAGE and immunoblotting with anti-gp41 MAb (Chessie 8) (A). As a control, residual supernatants from immunoprecipitation were analyzed for actin content with an anti-actin PAb (B). Data shown are representative of 2 experiments.
PDI is associated with gp120/CD4/CXCR4 complex at the cell surface
To determine whether PDI colocalizes with CD4 and CXCR4 at the cell surface, we analyzed PDI distribution on the plasma membrane in the presence and absence of gp120 using scanning confocal microscopy. CD4 and CXCR4 exhibited significant colocalization as evident by the magenta-colored regions derived from the overlap of red CXCR4 and blue CD4 staining. PDI (green), which had a consistent patchy, discontinuous distribution, showed no overt colocalization with CD4 or CXCR4 in the absence of gp120, evident by the absence of white regions in the image (Figure 7A, left panel). In contrast, in the presence of gp120, PDI was distributed largely in the CD4/CXCR4 regions, indicating considerable colocalization with these 2 markers. Furthermore, this shows that, even though PDI is a nontransmembrane domain-bearing protein, it nevertheless segregates into very distinct membrane regions. Triple colocalization of PDI and receptors is indicated by the white, patchy regions on the cell resulting from green (PDI), red (CXCR4), and blue (CD4) overlap (Figure 7A, right panel).
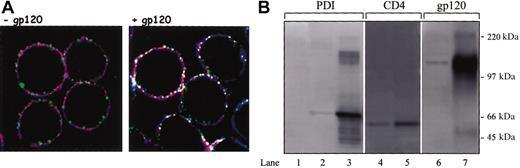
PDI colocalizes and is associated with CD4 and CXCR4 in the presence of gp120. (A) Sup T-1 cells were attached to poly-L-lysine slide and treated in the presence (right panel) or absence of soluble gp120 IIIB (left panel). Cells were then fixed in 2% paraformaldehyde and reacted with anti-CD4 rat MAb, anti-PDI rabbit PAb, and anti-CXCR4 mouse MAb. Subsequently, cells were incubated with secondary antirat conjugated with Alexa Fluor 647, antirabbit Cy2, and antimouse Alexa Fluor 555 Abs. Cells were mounted in Fluoromount-G and analyzed by confocal microscopy. PDI is shown as green, CXCR4 is shown as red, and CD4 is shown as blue; white indicates regions of triple, PDI/CD4/CXCR4 colocalization. The data shown are representative of the staining detected in more than 70% of analyzed cells. Original magnification, × 63. (B) HOS-CD4+/CXCR4+ cells were pretreated with gp120 IIIB followed by lysis and coimmunoprecipitation with the anti-CXCR4 MAb, 4G10. Lanes 1 to 3 were probed with anti-PDI PAb, lanes 4 and 5 with an anti-CD4 rabbit serum, and lanes 6 and 7 with an anti-gp120 rabbit serum. A negative control was prepared from HOS-CD4+/CXCR4+ cell lysate not exposed to 4G10 MAb (lane 1), lane 2 represents PDI coimmunoprecipitated from HOS-CD4+/CXCR4+ cells in the presence of anti-CXCR4 Ab, and lane 3 represents soluble PDI included as a control for band identity. Lane 4 represents CD4 coimmunoprecipitated from HOS-CD4+/CXCR4+ cells in the presence of anti-CXCR4 Ab. Lane 5 is a positive CD4 control prepared from HOS-CD4+/CXCR4+ cell lysate by adding the anti-CD4 MAb, Leu3A (3 μg/5 × 106 cells) and subsequent precipitation with protein G beads. Lane 6 indicates gp120 coimmunoprecipitation from HOS-CD4+/CXCR4+ cells in the presence of anti-CXCR4 Ab; lane 7 represents soluble gp120 included as a control for band identity.
PDI colocalizes and is associated with CD4 and CXCR4 in the presence of gp120. (A) Sup T-1 cells were attached to poly-L-lysine slide and treated in the presence (right panel) or absence of soluble gp120 IIIB (left panel). Cells were then fixed in 2% paraformaldehyde and reacted with anti-CD4 rat MAb, anti-PDI rabbit PAb, and anti-CXCR4 mouse MAb. Subsequently, cells were incubated with secondary antirat conjugated with Alexa Fluor 647, antirabbit Cy2, and antimouse Alexa Fluor 555 Abs. Cells were mounted in Fluoromount-G and analyzed by confocal microscopy. PDI is shown as green, CXCR4 is shown as red, and CD4 is shown as blue; white indicates regions of triple, PDI/CD4/CXCR4 colocalization. The data shown are representative of the staining detected in more than 70% of analyzed cells. Original magnification, × 63. (B) HOS-CD4+/CXCR4+ cells were pretreated with gp120 IIIB followed by lysis and coimmunoprecipitation with the anti-CXCR4 MAb, 4G10. Lanes 1 to 3 were probed with anti-PDI PAb, lanes 4 and 5 with an anti-CD4 rabbit serum, and lanes 6 and 7 with an anti-gp120 rabbit serum. A negative control was prepared from HOS-CD4+/CXCR4+ cell lysate not exposed to 4G10 MAb (lane 1), lane 2 represents PDI coimmunoprecipitated from HOS-CD4+/CXCR4+ cells in the presence of anti-CXCR4 Ab, and lane 3 represents soluble PDI included as a control for band identity. Lane 4 represents CD4 coimmunoprecipitated from HOS-CD4+/CXCR4+ cells in the presence of anti-CXCR4 Ab. Lane 5 is a positive CD4 control prepared from HOS-CD4+/CXCR4+ cell lysate by adding the anti-CD4 MAb, Leu3A (3 μg/5 × 106 cells) and subsequent precipitation with protein G beads. Lane 6 indicates gp120 coimmunoprecipitation from HOS-CD4+/CXCR4+ cells in the presence of anti-CXCR4 Ab; lane 7 represents soluble gp120 included as a control for band identity.
In addition, as more apparent in Figure 8A (right panel), gp120 appears to induce PDI aggregation on the cell surface reflected by the larger green areas relative to those observed for PDI in cells not treated with gp120 (left panel).
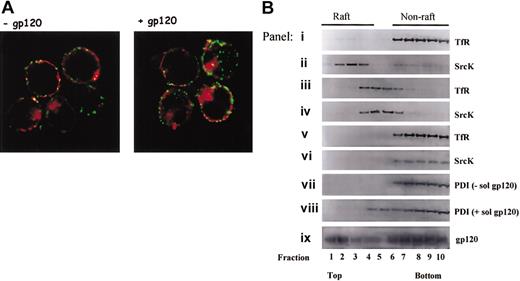
PDI fails to segregate in GM1 and TX-100–resistant rafts. (A) Sup T1 cells were treated in the presence or absence of gp120 then incubated with anti-PDI rabbit PAb and cholera toxin B subunit coupled to Alexa Fluor 594 for 45 minutes at 4°C. Raft aggregation was induced by incubation of cholera toxin-labeled cells in the presence of antirabbit Cy2-coupled Ab for 10 minutes at 37°C. Cells were then fixed, attached to poly-l-lysine slide, mounted, and analyzed by confocal microscopy. The data shown are representative of the staining detected in more than 90% analyzed cells. Original magnification, × 63. (B) Shown is glycerol gradient distribution of nonraft (TfR) and raft (SrcK) markers in cells lysed with 0.25% TX-100 (panels i-ii), cells lysed by hypotonic lysis without TX-100 (panels iii-iv), cells in which cholesterol was depleted with 10 mM MβCD followed by lysis in the presence of TX-100 (panels v-vi). Glycerol gradient distribution of PDI and gp120 in samples lysed with 0.25% TX-100 is shown in panels vii to ix. Cells in panel h were pretreated with soluble gp120 IIIB prior to detergent lysis. Data shown are representative of 4 experiments.
PDI fails to segregate in GM1 and TX-100–resistant rafts. (A) Sup T1 cells were treated in the presence or absence of gp120 then incubated with anti-PDI rabbit PAb and cholera toxin B subunit coupled to Alexa Fluor 594 for 45 minutes at 4°C. Raft aggregation was induced by incubation of cholera toxin-labeled cells in the presence of antirabbit Cy2-coupled Ab for 10 minutes at 37°C. Cells were then fixed, attached to poly-l-lysine slide, mounted, and analyzed by confocal microscopy. The data shown are representative of the staining detected in more than 90% analyzed cells. Original magnification, × 63. (B) Shown is glycerol gradient distribution of nonraft (TfR) and raft (SrcK) markers in cells lysed with 0.25% TX-100 (panels i-ii), cells lysed by hypotonic lysis without TX-100 (panels iii-iv), cells in which cholesterol was depleted with 10 mM MβCD followed by lysis in the presence of TX-100 (panels v-vi). Glycerol gradient distribution of PDI and gp120 in samples lysed with 0.25% TX-100 is shown in panels vii to ix. Cells in panel h were pretreated with soluble gp120 IIIB prior to detergent lysis. Data shown are representative of 4 experiments.
Previously, 2 reports provided evidence for the existence of trimolecular protein complexes comprised of gp120, CD4, and an appropriate coreceptor.25,26 Using a similar approach, we sought to determine whether PDI could be associated with such a trimolecular protein complex as strong supportive biochemical evidence for the colocalization data obtained by confocal microscopy. Using the CXCR4-specific MAb, 4G10, we were able to precipitate complexes containing PDI from CD4+/CXCR4+ cells in the presence of gp120 (Figure 7B). This observation has not been reported previously in any other study and provides biochemical evidence that PDI, gp120, CD4, and CXCR4 associate together. Indeed, this corroborates a model whereby PDI is interacting with the gp120/CD4/CXCR4 complex to form a transient tetramolecular protein complex at the cell surface, which serves as a portal for viral entry.
PDI fails to segregate in GM1 lipid rafts
Findings suggesting that CD4 associates with GM1 lipid rafts,27,28 coupled with the observations that disruption of cellular rafts inhibits viral entry and/or release,22,29,30 have raised awareness of the potential role of lipid rafts in HIV-1 biology. Therefore, we sought to study PDI association with lipid rafts. First, we evaluated PDI colocalization with the cholera toxin B subunit, which binds the well-known raft marker, GM1, using confocal microscopy (Figure 8A). We found that PDI does not colocalize with cholera toxin (as evident by the absence of yellow regions resulting from red-green overlap) in the presence or absence of gp120, which indicates a lack of PDI association with GM1 rafts. Second, we analyzed the association of PDI with rafts using equilibrium flotation gradient centrifugation, which relies on the insolubility of lipid rafts in TX-100 at 4°C (Figure 8B). Control panels a to f show the distribution of nonraft (TfR) and raft (SrcK) markers in the presence (panels i-ii) or absence (panels iii-iv) of TX-100 or under the conditions used for cholesterol depletion with β-methyl cyclodextrin (panels v-vi). A nonraft distribution of PDI is suggested for cells pretreated with gp120 (panel viii) or left untreated (panel vii). Panel ix shows that gp120 distributes in both raft and nonraft areas, even though PDI segregates in the nonraft regions in the presence of gp120 in the same sample (panel viii). Possible explanations for this are addressed in “Discussion.”
Discussion
Many enveloped viruses (influenza virus, baculovirus, Semliki Forest virus) rely on special conditions such as acidic pH, to drive the conformational change in the viral Env needed to mediate the membrane fusion process during viral entry.31 In contrast, other enveloped viruses, like HIV-1, bind to cell-surface receptors with high affinity and undergo a receptor-induced conformational change in Env to induce the membrane fusion process. Similar to low pH destabilization of a protein by protonation of titratable amino acid residues,32 the reduction of protein disulfide bridges11,33,34 and elevated temperature or urea35 can also destabilize the native protein structure at neutral pH and induce a conformational change. Supportive evidence by Carr et al35 and Carr and Kim36 that fusion proteins are trapped in the metastable state in their native structure and require activation energy before they can enter into a more stable configuration further substantiate this notion. Hence, the putative role of PDI in HIV-1 infection is to lower this activation energy to enable fusion proteins to undergo conformational change and acquire the more stable low-energy state.
An HIV-1 membrane fusion system in which only CD4 and a relevant coreceptor are imbedded in an artificial lipid bilayer has not yet been developed to determine whether other cofactors are required for the HIV-1 fusion process. Our data, however, as well as data from other laboratories,4-7 support the concept that a cell-surface–associated oxido-reductase is involved in HIV-1 entry. Our current findings indicate that thiol/disulfide rearrangement in gp120 catalyzed by PDI occurs post-CD4 engagement. Gallina et al6 similarly demonstrated that gp120 thiol/disulfide shuffling takes place after binding to the receptor. This, however, conflicts with the findings of Barbouche et al,7 suggesting that -SH/-S-S- exchange takes place after coreceptor engagement. The discrepancy between our findings and those of Barbouche et al7 may be due to the use of SDF-1α to block CXCR4 in these latter studies, rather than cells devoid of the coreceptor.
Our current study provides evidence that treatment of target cells with the PDI inhibitor, DTNB, prevents gp41 from assuming the 6-helix bundle conformation that drives fusion. This finding supports the concept that PDI-induced remodeling of gp120 is in some way transmitted to the gp41 subunit to activate its fusogenic properties and could further explain the inhibitory mechanism of anti-PDI agents.
In addition to PDI, other oxido-reductases having Cys-Xaa-Xaa-Cys (CXXC) motifs that modulate thiol/disulfide exchange may also play a role in HIV-1 entry. For example, thioredoxin was suggested to have a role in HIV-1 entry because of its reductive action on the target membrane in experiments using an arsenical compound known to block the vicinal thiols of the CXXC motifs of both PDI and thioredoxin.37 Inasmuch as both proteins share a high level of functional and structural similarity in their catalytic domains, it is conceivable that both are involved in this restructuring process dependent on the redox exchange.
We observed that PDI associated with Env-expressing membranes does not contribute to fusion. This may reflect an inability of PDI to freely dissociate from its host membrane, suggesting that it has a restricted range of motion. It further shows that the topology of PDI (its spatial orientation relative to gp120, CD4, or CXCR4) is critical for its ability to facilitate membrane fusion. This is reminiscent of the function of soluble NSF attachment protein receptor on the transport vehicle (v-SNARE) and its cognate t-SNARE at the target membrane, which must be in an antiparallel orientation to drive efficient membrane fusion during exocytosis.38
Importantly, we also observed that exogenous PDI can rescue cell-cell fusion that is inhibited by PDI antagonists. In this situation, the exogenous PDI could be exerting its effect by direct oxidoreductive activity on HIV Env. It is also possible that exogenous PDI is reducing 2-mercapto-5-nitrobenzoicacid (TNB) and liberating the catalytic thiol-reactive sites from previously blocked PDI.39 In either case, this strengthens the concept that fusion and inhibition of fusion are interchangeable, dynamic processes, which depend on -SH/-S-S- exchange at the cell surface.11
Given that CD4 associates with GM1 lipid rafts,27,28 failure to detect PDI in GM1 rafts appears to contradict our confocal microscopy (Figure 7A) and coimmunoprecipitation (Figure 7B) data, showing that PDI colocalizes and is associated with CD4 and CXCR4 in the presence of gp120. Despite the fact that other studies have challenged the role of lipid rafts as a place of viral entry,28,40 we recognize that results from our attempt to determine PDI localization and ascertain whether its action on gp120 takes place in these membrane domains is not sufficient to deny or support the significance of lipid rafts in HIV-1 entry. Still, several explanations may account for the observations in this study. First, it is possible that proteins weakly associated with the cell membrane, such as PDI, are difficult to detect in rafts using the TX-100 extraction conditions. In this situation, detergent may induce the dissociation of weakly bound proteins from the cell membrane. This was shown for glycosyl-phosphatidylinositol-anchored placental alkaline phosphatase (PLAP) whose association with the TX-100 insoluble membrane fraction significantly increased on Ab-induced lateral cross-linking stabilizing PLAP interactions with the membrane.41 Second, it is possible that PDI associates with rafts that are not TX-100 resistant. In this regard, Roper et al42 reported selective extraction of proteins from membrane rafts based on the type of the detergent used. Third, the failure to detect PDI in GM1 rafts by scanning confocal microscopy may indicate that PDI segregates in a different type of rafts (eg, GM3). There is increasing evidence to suggest that multiple cholesterol-based lipid rafts coexist on the cell surface.42,43 However, we cannot exclude the possibility that our results indicate PDI is indeed associated with nonraft regions on the membrane.
There are several possible scenarios regarding HIV-1 entry and the temporal and physical pattern of PDI association with the Env. First, assuming that PDI is associated with nonraft membrane domains, it is nevertheless possible that gp120 binding to CD4 occurs inside the GM1 lipid rafts. This interaction may initiate certain conformational changes in gp120, exposing previously buried disulfide bond-spanning gp120 regions. This, in turn, could facilitate the movement of Env-CD4 complexes out of GM1 rafts, perhaps to an area where PDI is located. Our data, as well those of Gallina et al,6 suggest that PDI action takes place after CD4 but prior to or concomitant with CXCR4 Env binding. Thus, PDI-induced gp120 conformational change may indeed occur in nonraft domains. Alternatively, if one assumes that PDI exists in rafts that are distinct from the GM1 type, bringing PDI into contact with gp120 and CD4 would require clustering of PDI rafts with GM1 rafts. In this scenario, PDI-induced gp120 conformational change and fusion could conceivably take place at the interface of 2 different types of rafts or in the PDI-specific rafts. Nevertheless, in either scenario the association of PDI with Env and its cellular receptors should take place, and indeed we provide biochemical evidence with the specific detection of a tetramolecular protein complex comprised of PDI, gp120, CD4, and CXCR4.
Given that current anti–HIV-1 therapies are often associated with severe side effects, identification of drugs with novel mechanisms of action is imperative. A common problem in designing anti–HIV-1 therapies comes from the fact that selective pressure induces generation of drug-resistant virus strains, which can effectively escape therapy.44 Escape mutants, however, are less likely to arise when the therapy targets sites that are critical for protein function or when multiple sites are being affected. Inhibitors of redox-active enzymes can prevent Env conformational change by blocking thiol/disulfide reorganization, thereby inhibiting membrane fusion and subsequent entry. Virus attempts to escape or circumvent these agents would require replacement or alterations of the cysteines and disulfide bonds involved in Env conformational change requiring significant structural modifications. Such changes would no doubt be difficult for the virus to acquire, making this process an attractive drug target.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-05-1390.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Drs Eric Freed and Akira Ono for helpful discussions regarding the analysis of raft proteins and advice on other aspects of this work. We thank Drs. Eric Freed, Barbara Rellahan, and Steven Kozlowski for critical review of this manuscript.

![Figure 2. DTNB does not modify the Env binding site on the CD4 or CXCR4. (A) Sup T-1 cells treated with 2.5 mM DTNB or PBS (control) were incubated with 0.5 μg gp120/1 × 106 cells (30 minutes, 22°C) followed by anti-gp120 rabbit serum (15 minutes, 22°C) and PE-conjugated antirabbit secondary Ab or (B) with PE-labeled Leu3A (anti-CD4) MAb (15 minutes, 22°C). After staining, cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytofluorometer. Data shown are representative of 3 experiments. (C) Sup T-1cells treated with 2.5 mM DTNB or PBS (control) were incubated with 12G5 (anti-CXCR4) MAb. After staining, cells were fixed in 3.7% formaldehyde and analyzed with FACScan cytofluorometer. (D) CXCR4 retains the ability to mobilize intracellular Ca2+ after DTNB treatment. Sup T-1 cells attached to poly-l-lysine–coated coverslips were loaded with 2.5 μM Fura-2/am and 2.5 mM DTNB or Hanks buffer (control). Fluorescence in cells was excited by alternating 340- and 380-nm light beams, and the emitted light intensity was measured at 520 nm. Data shown reflect a single-cell response in mobilizing intracellular Ca2+ induced with 500 ng/mL SDF-1α after following the baseline [Ca2+]i for 60 seconds and are representative of 3 experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/5/10.1182_blood-2003-05-1390/6/m_zh80050457370002.jpeg?Expires=1769111239&Signature=PDBBvKJvSwiEENM6jcP~GSCeB-GwDqS2P93PAB6BhfEdsPsxxyryRxARNjLHsXi2FVlnoyN5CC0PQNLTmd8b5Byx1xULEs2o4Tnc5CFCS6KLR0EBNgVpKSuyU6GQgoqh6AlKVXm7-T3kiRuk0q10PXRYQ3kEYCwMqj3YIpf794158CvJW-rXHVPiqCr6BH94MMZPvB1iYil~aYXPQ4dXB-brr543fFzmqiMDQ1oaUcUfDHisXkuVlqaF2-jCV3pBNhr258h0xBMk0K-uhw6rI7r53w4nRpzkIEzDWVKpXx40IlIrZ6kwbFP8uhQLVIiMav4FrlLUwWPruprxF0ujPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal