Abstract
Acquired somatic mutations in ATRX, an X-linked gene encoding a chromatin-associated protein, were recently identified in 4 patients with the rare subtype of myelodysplastic syndrome (MDS) associated with α thalassemia (ATMDS). Here we describe a series of novel point mutations in ATRX detected in archival DNA samples from marrow and/or blood of patients with ATMDS by use of denaturing high-performance liquid chromatography (DHPLC), a technique sensitive to low-level mosaicism. Two of the new mutations result in changes in amino acids altered in previously described pedigrees with germ line ATRX mutations (ATR-X syndrome), but the hematologic abnormalities were much more severe in the patients with ATMDS than in the corresponding constitutional cases. In one ATMDS case where DNA samples from several time points were available, the proportion of ATRX-mutant subclones correlated with changes in the amount of hemoglobin H. This study strengthens the link between acquired, somatic ATRX mutations and ATMDS, illustrates how molecular defects associated with MDS and other hematologic malignancies masked by somatic mosaicism may be detected by DHPLC, and shows that additional factors increase the severity of the hematologic phenotype of ATRX mutations in ATMDS.
Introduction
Mutations affecting the structure and/or synthesis of hemoglobin (HbA, α2β2) are most commonly observed in individuals from tropical and subtropical regions of the world.1-3 Sporadic, inherited mutations are occasionally found in individual families located outside of these areas.1 Perturbed patterns of hemoglobin production associated with abnormal erythropoiesis have also been detected as acquired abnormalities in patients with hematologic malignancy.4 For example, changes in the normal levels of fetal hemoglobin (HbF, α2γ2) and hemoglobin A2 (HbA2, α2δ2) may occur in acute leukemia and chronic myeloid disorders.5-7 In addition, there have been a number of reports describing α and β thalassemia as newly acquired traits in the context of hematologic malignancy.8-11 Acquired α thalassemia due to down-regulation of α globin synthesis is most easily recognized by demonstrating tetramers of excess β globin chains (β4, known as hemoglobin H [HbH]) in the peripheral blood, either by hemoglobin electrophoresis or supravital staining.12 When HbH is detected for the first time in an individual with a hematologic malignancy, the condition is often referred to as “acquired HbH disease.”9
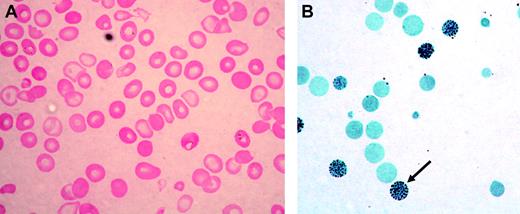
Acquired HbH disease has been most commonly reported in elderly northern European men who develop myelodysplastic syndrome (MDS) or, less often, other neoplastic myeloid disorders; this constellation is now referred to as α thalassemia–myelodysplastic syndrome (ATMDS; Mendelian Inheritance in Man [MIM] catalog no. 300448).9 The peripheral blood film in such patients is characterized by the presence of strikingly hypochromic, microcytic, and anisopoikilocytic red cells (Figure 1A), often intermingled with a variable proportion of apparently normal red cells, suggesting the coexistence of normal and abnormal hematopoietic clones. Stratification of red cell precursors in individual ATMDS cases has revealed a wide distribution of globin chain imbalance, with α/β globin chain synthesis ratios ranging from less than 0.1 to 1.0 (normal 0.9-1.2), indicating the presence of normal and abnormal clones contributing to erythropoiesis. Consistent with this, in some patients with ATMDS, the proportion of HbH waxes and wanes during the course of the illness and sometimes disappears entirely, especially during transformation to overt acute leukemia. This observation presumably reflects somatic mosaicism and clonal evolution in MDS.9
ATMDS peripheral blood findings. (A) Peripheral blood smear (hematoxylin-eosin; original magnification, × 400) from ATMDS patient no. 12 (Table 1) demonstrates a typical dimorphic red cell picture with severe hypochromia, microcytosis, and occasional poorly hemoglobinized “ghost cells.” (B) A hemoglobin H preparation (brilliant cresyl blue; original magnification, × 400) from patient no. 4 at time point 1 reveals many classic “golf ball cells” (arrow) with intracellular hemoglobin H inclusions.
ATMDS peripheral blood findings. (A) Peripheral blood smear (hematoxylin-eosin; original magnification, × 400) from ATMDS patient no. 12 (Table 1) demonstrates a typical dimorphic red cell picture with severe hypochromia, microcytosis, and occasional poorly hemoglobinized “ghost cells.” (B) A hemoglobin H preparation (brilliant cresyl blue; original magnification, × 400) from patient no. 4 at time point 1 reveals many classic “golf ball cells” (arrow) with intracellular hemoglobin H inclusions.
The common, inherited forms of α thalassemia result from deletions of one, 2, 3, or all 4 of the normal α globin genes (normal: αα/αα; thalassemic: –α/αα,–/αα,-α/-α,–/–α, or –/–) and less commonly from point mutations in the structural genes.13 In rare families, deletions of a remote critical regulatory region (HS-40) have been identified as a cause of α thalassemia.14 In addition, constitutional mutations in the gene encoding ATRX, an X-linked, chromatin-associated factor, have been shown to cause a rare condition (ATR-X syndrome, MIM no. 301040) where mild α thalassemia (usually < 10% HbH) is found in boys with severe mental retardation, facial dysmorphism, and urogenital abnormalities.15 In ATMDS, despite the presence of the thalassemic phenotype, extensive mapping and sequence analysis have revealed no mutations in the α globin cluster.9 Similarly, we have found no gross rearrangements in the ATRX gene in ATMDS (D.P.S., D.R.H., C.A.F., and R.J.G., unpublished data). However, it was recently shown that acquired, somatic point mutations in the ATRX gene (20 + 1G>A in the exon 1–intron 1 canonical splice donor site, and 236C>G; S79X) were present in myeloid but not lymphoid cells in 2 patients with ATMDS; ATRX mRNA splicing abnormalities were observed in 2 other patients with ATMDS in whom the causative genomic mutation remains unknown.16
The 2 ATMDS-associated ATRX mutations and 2 ATRX splicing abnormalities described to date have never been reported in ATR-X syndrome. One possible reason might be that the ATRX mutations associated with the much more severe hematopoietic impairment typical of ATMDS—where the number of HbH-containing erythrocytes usually ranges from 10% to 90% and the α/β globin chain synthesis ratio is often less than 0.1—are lethal during development when inherited. However, here we report 8 new ATMDS-associated acquired ATRX mutations and an additional ATRX base pair change of uncertain significance detected by denaturing high-performance liquid chromatography (DHPLC). This series includes one mutation identical to a previously described germ line ATRX mutation and another mutation that alters the same amino acid as a constitutional ATRX mutation. In addition, several of the newly characterized mutations result in the loss of highly conserved domains of the ATRX protein that are also lost in constitutional pedigrees. In each case, the ATMDS hematologic findings are more severely abnormal than in the previously described cases where the ATRX mutation was inherited rather than acquired. These findings expand the spectrum of acquired ATRX mutations and demonstrate that these are the primary mutations leading to α thalassemia in ATMDS. However, these observations also show that additional factors in the abnormal erythroid precursors of patients with ATMDS must contribute to the severe defect in α globin synthesis.
Patients, materials, and methods
DNA, polymerase chain reaction for DHPLC, and patients
All specimens were collected and analyzed with approval from patients (parents/legal guardians in the case of minor children) and with permission of the relevant institutional ethics committees. Hematologic data at the time of sample acquisition (samples were obtained between 1978 and 2002) from the 18 adult patients with chronic myeloid disorders analyzed to date are summarized in Table 1.
Hematology parameters and ATRX mutations detected to date in patients with ATMDS
Patient number (Oxford ATMDS Registry database no.) . | Patient age in years, sex, clinical diagnosis . | % HbH-containing cells by BCB . | % HbH by electrophoresis . | α/β globin synthesis ratio . | MCV, fL . | ATRX mutation cDNA; predicted protein abnormality if applicable (description) . |
|---|---|---|---|---|---|---|
| 1 (O51) | 60, M, MDS (RA) | 4 | 0 | NA | 69.8 | ND; -14G>A (5′UTR) polymorphism |
| 2 (O65) | 70, M, CMD NOS | 16 | 5.2 | NA | 62 | 20+1G>A (exon 1—intron 1 consensus splice donor site16 ) |
| 3 (O45) | 60, M, MDS (RA) | 35 | 8.7 | 0.11 | 67 | 236C>G, S79X16 |
| 4 (O27) | 76, M, CMD NOS | 50, < 1† | 25, < 1† | 0.09, 1.02† | 65, 78† | 576G>C; L192F |
| 5 (O46) | 70, M, MDS (RA) | 48 | 8.4 | 0.09 | 80.4 | 718T>G; C240G |
| 6 (O54) | 83, M, MDS (RAEB) | 75 | 37.6 | 0.09 | 69.3 | 794G>A; C265Y |
| 7 (O58) | 73, M, MDS (RA) | 16* | 0* | 0.58* | 82* | 2692G>C; D898H‡ |
| 8 (O53) | 76, M, MDS (RAEB) | 25 | NA | NA | 68.4 | Splicing abnormality: exclusion of exon 16, inclusion of segment from intron 16 (see Gibbons et al16 for details) |
| 9 (O61) | 67, M, MDS (RAEB) | 60* | 15* | 0.49* | 66* | Splicing abnormality: inclusion of segment from intron 18 (see Gibbons et al16 for details) |
| 10 (O29) | 86, M, MDS (?RARS) | 14* | 10* | 0.29* | 82* | 5567-3C>G (intron 23—exon 24 consensus splice acceptor site) |
| 11 (O50) | 79, M, MDS (RA) | 10* | 8* | NA | 82.6* | 6083G>C; R2028P |
| 12 (O42) | 71, M, MDS (RA) | 40 | 16.9 | 0.09 | 59.8 | 6884_6885insT; 2293Tfs2319X |
| 13 (O49) | 60, M, MDS (RA) | 25 | 13 | 0.07 | 69.4 | 7014_7017del; N2340fsX2341 |
| 14 (O26) | 61, M, MMM | 5* | 0* | 0.08* | 92* | 7049delT; E2351fsX2353 |
| 15 (O41) | 87, F, ET | 20 | NA | 0.62 | 65 | ND |
| 16 (O43) | 68, M, MDS (RA) | 6.6* | 0* | 0.60* | 88.6* | ND |
| 17 (O69) | 69, M, MDS (RA) | 0.5 | 0 | NA | 72 | ND |
| 18 (O68) | 68, M, MDS NOS | 0.1 | 0 | NA | 72.9 | ND |
Patient number (Oxford ATMDS Registry database no.) . | Patient age in years, sex, clinical diagnosis . | % HbH-containing cells by BCB . | % HbH by electrophoresis . | α/β globin synthesis ratio . | MCV, fL . | ATRX mutation cDNA; predicted protein abnormality if applicable (description) . |
|---|---|---|---|---|---|---|
| 1 (O51) | 60, M, MDS (RA) | 4 | 0 | NA | 69.8 | ND; -14G>A (5′UTR) polymorphism |
| 2 (O65) | 70, M, CMD NOS | 16 | 5.2 | NA | 62 | 20+1G>A (exon 1—intron 1 consensus splice donor site16 ) |
| 3 (O45) | 60, M, MDS (RA) | 35 | 8.7 | 0.11 | 67 | 236C>G, S79X16 |
| 4 (O27) | 76, M, CMD NOS | 50, < 1† | 25, < 1† | 0.09, 1.02† | 65, 78† | 576G>C; L192F |
| 5 (O46) | 70, M, MDS (RA) | 48 | 8.4 | 0.09 | 80.4 | 718T>G; C240G |
| 6 (O54) | 83, M, MDS (RAEB) | 75 | 37.6 | 0.09 | 69.3 | 794G>A; C265Y |
| 7 (O58) | 73, M, MDS (RA) | 16* | 0* | 0.58* | 82* | 2692G>C; D898H‡ |
| 8 (O53) | 76, M, MDS (RAEB) | 25 | NA | NA | 68.4 | Splicing abnormality: exclusion of exon 16, inclusion of segment from intron 16 (see Gibbons et al16 for details) |
| 9 (O61) | 67, M, MDS (RAEB) | 60* | 15* | 0.49* | 66* | Splicing abnormality: inclusion of segment from intron 18 (see Gibbons et al16 for details) |
| 10 (O29) | 86, M, MDS (?RARS) | 14* | 10* | 0.29* | 82* | 5567-3C>G (intron 23—exon 24 consensus splice acceptor site) |
| 11 (O50) | 79, M, MDS (RA) | 10* | 8* | NA | 82.6* | 6083G>C; R2028P |
| 12 (O42) | 71, M, MDS (RA) | 40 | 16.9 | 0.09 | 59.8 | 6884_6885insT; 2293Tfs2319X |
| 13 (O49) | 60, M, MDS (RA) | 25 | 13 | 0.07 | 69.4 | 7014_7017del; N2340fsX2341 |
| 14 (O26) | 61, M, MMM | 5* | 0* | 0.08* | 92* | 7049delT; E2351fsX2353 |
| 15 (O41) | 87, F, ET | 20 | NA | 0.62 | 65 | ND |
| 16 (O43) | 68, M, MDS (RA) | 6.6* | 0* | 0.60* | 88.6* | ND |
| 17 (O69) | 69, M, MDS (RA) | 0.5 | 0 | NA | 72 | ND |
| 18 (O68) | 68, M, MDS NOS | 0.1 | 0 | NA | 72.9 | ND |
Patients no. 2, 3, 8, and 9 were described in Gibbons et al16 ; all other ATRX mutations are newly described here. The precise gDNA mutation responsible for the splicing abnormality detected in cDNA from patients no. 8 and no. 9 is currently unknown.
HbH indicates hemoglobin H; MDS, myelodysplastic syndrome; BCB, brilliant cresyl blue supravital stain; MCV, mean corpuscular volume (normal, 81-99 fL); NA, data not available or assay not performed; ND, no pathologic mutation detected; cDNA, complementary deoxyribonucleic acid; ET, essential thrombocythemia; MMM, myelofibrosis with myeloid metaplasia; CMD NOS, chronic myeloid disorder not otherwise specific; MDS NOS, myelodysplastic syndrome not otherwise specified; RA, refractory anemia; RAEB, refractory anemia with excess blasts; RARS, refractory anemia with ringed sideroblasts (French-American-British Co-operative Group Classification was used to describe MDS subtypes, as most predated the recent reclassification by the World Health Organization); and UTR, untranslated region. ? indicates diagnostic uncertainty.
Patient had been transfused with packed red cells prior to obtaining sample, and HbH level, MCV, and globin synthesis ratio may reflect admixture of patient and donor cells.
Samples were obtained at 2 time points for patient no. 4 (see “Sensitivity of DHPLC for detection of mutations in the ATRX gene”).
Base pair change of uncertain significance, possible polymorphism (see “Comparison of acquired and germ line ATRX mutations”).
Genomic DNA was extracted from heparin-anticoagulated peripheral blood and/or bone marrow via either a phenol-chloroform method or a resin-based DNA extraction kit (Nucleon BACC2; Nucleon Biosciences, Coatbridge, United Kingdom). DNA amplification was performed by polymerase chain reaction (PCR); reagents included GeneAmp PCR Buffer II (Applied Biosystems, Foster City, CA), MgCl2 (concentration 1.5 mM to 3 mM depending on the specific amplicon; Applied Biosystems), dNTPs (200 μM; Roche, Mannheim, Germany), forward and reverse primers (list available on request; final concentration 0.8 μM; Invitrogen, Paisley, United Kingdom), 100 ng template DNA, and a 5:1 ratio of AmpliTaq Gold DNA polymerase (total 1 U; Applied Biosystems) to Pwo DNA polymerase (total 0.2 U; Roche). Pwo polymerase was included to provide 3′-5′ exonuclease activity and thereby increase replication fidelity and improve DHPLC performance characteristics. Amplicons were designed to cover the entire 5′ untranslated region, protein coding region, canonical splice donor and splice acceptor sites, and the 3′ polyadenylation consensus signal of ATRX. Total reaction volume was 50 μL. Reactions were carried out on a PTC-200 Peltier Thermocycler (MJ Research, Waltham, MA). Amplification conditions for each amplicon are available on request.
DHPLC
PCR-amplified samples were transferred directly into a skirted 96-well plate (Thermo Life Sciences, Basingstoke, United Kingdom) and warmed to 95°C, then cooled to room temperature over approximately 1 hour to promote heteroduplex formation. A wild-type DNA control was included for each amplicon. DNA from patients with known or suspected ATRX constitutional mutations (ie, pure mutant DNA from patients with ATR-X syndrome) was mixed with equal amounts of wild-type DNA before heteroduplexing, as was DNA from suspected new cases of ATR-X syndrome. However, DNA from patients with ATMDS was not mixed, in view of the tissue DNA admixture in unfractionated blood and marrow characteristic of chronic myeloid disorders. For DHPLC analysis, we used the WAVE 3500HT DNA Fragment Analysis System (Transgenomic, Omaha, NE). Briefly, 5 μL of each DNA sample was injected into a high-throughput DNASep column and eluted over 4 minutes through a 260-nM photodetector, with concentrations of buffer A (ie, 0.1 M triethylammonium acetate [TEAA]; Transgenomic) and buffer B (ie, 0.1 M TEAA and 25% acetonitrile in ultrapure water) adjusted automatically as calculated by the Navigator software package (Transgenomic). All samples were run at a minimum of 2 different oven temperatures, including at least one temperature for which the Navigator software predicted each segment of the exonic component of the amplicon would be under partially denaturing conditions.
Subcloning and sequencing
Subcloning was performed using the pGEM-T Easy Vector System (Promega; Madison, WI) and XL1-Blue competent cells (Stratagene, La Jolla, CA) or DH5α cells (Invitrogen). Because the Pwo polymerase results in amplicons with blunt ends, A-tailing was performed to facilitate insertion into this plasmid vector, which has single 3′-T overhangs at either end of the insertion site. For A-tailing, the PCR product was first purified using either PEG-NaOAc and 95% ethanol or a QIAquick PCR Purification Kit (Qiagen, Crawley, United Kingdom), then 2 μL purified fragment was added to 1 μL 10 X Taq DNA polymerase reaction buffer with MgCl2 (Roche), dATP (200 μM; Roche), Taq DNA polymerase (5 U; Roche), and 6 μLdH2O. This mixture was incubated at 70°C for 30 minutes, and 2 μLof the reaction product was used in the ligation reaction. The ligation product was transformed into competent cells according to a heat-shock protocol recommended by the manufacturer. After transformation, cells were grown overnight at 37°C in 5% CO2 on duplicate LB/Ampicillin/IPTG/X-Gal (Sigma and Bio101, Carlsbad, CA) plates; white colonies were harvested and grown in 3 mL SOC medium at 37°C in 5% CO2 for 18 hours. Plasmid DNA was extracted using QIAprep Spin Miniprep Kit (Qiagen). The presence of inserts of appropriate length in the extracted plasmids was confirmed by digesting at 37°C for 1 hour using EcoRI restriction endonuclease (Roche) in SuRE/Cut Buffer (Roche); digests were electrophoresed through a 1% agarose gel stained with ethidium bromide and visualized with a phosphoimager.
Sequencing of subclones bearing inserts of the proper length was performed using the ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) and the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The generated sequence data were analyzed using the Sequencher v4.1.2 software package (Gene Codes, Ann Arbor, MI). Mutations and polymorphisms are described here using the nomenclature and numbering scheme suggested by den Dunnen and Antonarakis.17 Although all the ATMDS mutations in this study were detected at the genomic DNA (gDNA) level, they are described in terms of cDNA for the sake of clarity because of the lack of a consensus numbering system for ATRX gDNA.
Hemoglobin H and globin chain analysis
Fresh peripheral blood was incubated for 4 hours with 1% brilliant cresyl blue (Merck, Poole, United Kingdom) in 0.9% NaCl, smeared on a glass slide and examined for intraerythrocytic hemoglobin H inclusions.
Hemoglobin subtype analysis was performed on an automated high-performance liquid chromatography unit (Variant Hemoglobin Testing System; Bio-Rad, Hercules, CA) using a protocol recommended by the manufacturer.
Measurement of globin chain synthesis was performed according to the method of Weatherall et al.18,19 Briefly, an enriched reticulocyte preparation derived from fresh heparinized marrow was resuspended in 2.5% fetal calf serum and RPMI 1640 without leucine (Gibco BRL, Grand Island, NY), incubated for 1 hour at 37°C with 50 μCi (1.85 MBq) [3H]leucine (Amersham) and ferrous ammonium sulfate (Sigma), then cooled and washed to eliminate unincorporated radiolabeled amino acid. Cells were then lysed with distilled water. Globin chains were precipitated with 2% HCl in acetone with trace amounts of β-mercaptoethanol (Sigma) and separated by Whatman CM-23 cellulose column chromatography using a urea-phosphate buffer gradient at pH 6.4 in the presence of dithiothreitol (Sigma). Absorbance of collected eluted fractions was measured at 280 nM on a spectrophotometer, and aliquot radioactivity was quantified on a scintillation counter.
Results
Following the identification of ATRX mutations or splicing abnormalities in 4 patients with ATMDS as described,16 further investigation was hampered by the rarity of the syndrome, the mixed cellularity of archival samples, and the admixture of neoplastic clonal and residual normal polyclonal progenitors that is characteristic of MDS. To overcome the latter 2 limitations, we developed a screening protocol to search for mutations in the relatively large (10.1 kb mRNA) ATRX gene using DHPLC.
Sensitivity of DHPLC for detection of mutations in the ATRX gene
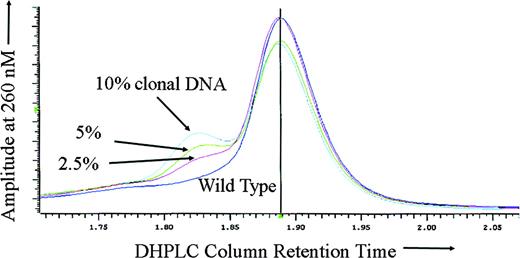
To test the sensitivity of DHPLC for ATRX mutation detection in the setting of somatic mosaicism, DNA derived from the purified peripheral blood neutrophils of a previously described16 patient with ATMDS with an ATRX mutation in the canonical splice donor site at the exon 1–intron 1 boundary (20 + 1G>A) was mixed with wild-type DNA in varying proportions. As expected, with either 100% mutant DNA (derived from the patient's purified peripheral blood granulocytes) or 100% wild-type DNA alone, no heteroduplex formation was detected, and DHPLC analysis of the amplicon containing exon 1 revealed only a single narrow homoduplex peak. However, for all of the mixtures tested (containing 80%, 60%, 40%, 20%, 10%, 5%, and 2.5% mutant granulocyte DNA) either a distinct second heteroduplex peak or an abnormal “shoulder” on the homoduplex peak was apparent (Figure 2).
Sensitivity of DHPLC for mosaic mutation detection. DHPLC tracing of the following mixtures of abnormal (purified neutrophil) DNA from ATMDS patient no. 2 with a 20 + 1G>A ATRX mutation and normal, wild-type ATRX DNA: 10% mutation-enriched DNA (from the patient's purified peripheral blood granulocytes) with 90% wild-type (cyan), 5% mutant DNA with 95% wild-type (green), 2.5% mutant DNA with 97.5% wild-type (red), and 100% wild-type DNA (purple). The left peak or “shoulder” represents DNA heteroduplexes, the larger central peak represents homoduplexes. Even 2.5% abnormal DNA is clearly differentiated from the pure wild-type DNA. If the peripheral blood granulocytes from the patient with ATMDS included the progeny of any residual normal clones (ie, if the granulocyte DNA was not 100% ATRX mutant), the actual mutant DNA would be even less than 2.5% and the sensitivity correspondingly greater.
Sensitivity of DHPLC for mosaic mutation detection. DHPLC tracing of the following mixtures of abnormal (purified neutrophil) DNA from ATMDS patient no. 2 with a 20 + 1G>A ATRX mutation and normal, wild-type ATRX DNA: 10% mutation-enriched DNA (from the patient's purified peripheral blood granulocytes) with 90% wild-type (cyan), 5% mutant DNA with 95% wild-type (green), 2.5% mutant DNA with 97.5% wild-type (red), and 100% wild-type DNA (purple). The left peak or “shoulder” represents DNA heteroduplexes, the larger central peak represents homoduplexes. Even 2.5% abnormal DNA is clearly differentiated from the pure wild-type DNA. If the peripheral blood granulocytes from the patient with ATMDS included the progeny of any residual normal clones (ie, if the granulocyte DNA was not 100% ATRX mutant), the actual mutant DNA would be even less than 2.5% and the sensitivity correspondingly greater.
To assess the ability of DHPLC to detect a wide range of pathogenic ATRX mutations, we screened archival DNA samples from boys with ATR-X syndrome that included 49 unique constitutional ATRX mutations involving 17 of the 35 exons (list available on request). DHPLC clearly resolved 48 of the 49 mutations (98%), including examples of substitutions, insertions, and deletions. The only mutation that the WAVE system missed was a 580G>A in exon 8; the available archival DNA from this pedigree was partially degraded and amplified poorly, and the DHPLC system was able to detect several other single base-pair changes in this region, including 576G>C and 599G>C. Subsequently, screening of new blood samples from undiagnosed boys with clinical features of constitutional ATR-X syndrome using our DHPLC conditions revealed 5 new germ line ATRX mutations (data not shown) including 3 in exons without previously described mutations, expanding the number of validated amplicons to 20.
Characterization of ATRX mutations in patients with ATMDS
Since the DHPLC method appeared sensitive and able to detect a wide range of mutations, we next analyzed unfractionated archival DNA samples from the 14 patients with myeloid disorders and hypochromic red cell indices summarized in Table 1. This screen detected the 9 novel ATRX mutations listed in Table 1 as well as a new polymorphism (-14G/A).
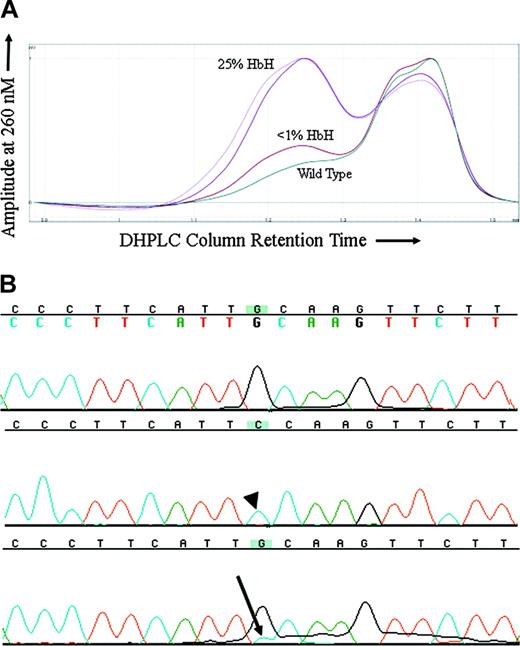
Because MDS is a clonal disorder and previous findings had suggested that the hematologic features of ATMDS might vary in a manner reflecting this clonality, it was of interest to compare the markers of α thalassemia with the proportion of the abnormal clone. For ATMDS patient no. 4, archival DNA samples from 2 different time points when the patient had very different amounts of HbH were available for analysis. In all the other patients, either the amount of HbH remained stable over time or an archival DNA sample was only available from one time point. DHPLC analysis of an amplicon containing ATRX exon 8 from patient no. 4 showed a second peak suggestive of a heteroduplex (Figure 3A). This peak was not seen in the wild-type control or any other ATMDS samples screened at the same time. The size of the peak was greater in a sample of bone marrow obtained when patient no. 4 had 25% HbH by electrophoresis and 50% HbH-containing cells by supravital staining compared with a subsequent marrow sample when he had less than 1% HbH-containing cells and undetectable HbH on electrophoresis (Figure 3A). Subcloning and sequencing of this amplicon revealed 8 of 23 (35%) of the clones with a 576G>C; L192F substitution (Figure 3B) when patient no. 4 had 25% HbH by electrophoresis, and only 3 of 40 (7.5%) clones when he had less than 1% HbH, demonstrating a correlation between the proportion of ATRX mutation–containing clones and the severity of the thalassemic phenotype. The L192F mutation alters an amino acid in a loop-helix-loop motif in one of the critical zinc-finger DNA-binding domains of the ATRX protein.20
576G>C ATRX mutation in a patient with ATMDS. (A) DHPLC tracing of the exon 8 amplicon from patient no. 4. The lighter and darker purple tracings represent unfractionated peripheral blood and bone marrow from a sample obtained when the patient had 50% erythrocytes with HbH inclusions by brilliant cresyl blue (BCB) staining and 25% HbH on electrophoresis. The brick red tracing represents unfractionated marrow from a time point 6 months later when the amount of HbH by electrophoresis had fallen to less than 1% and only rare HbH inclusions were detectable on supravital staining. The dark green tracing is a wild-type control. (B) Big Dye–generated sequence of 2 subclones (top 2 lines) and unfractionated marrow DNA (bottom line) from patient no. 4, exon 8 (coding strand 5′ to 3′, running left to right). The top line is sequence of a subclone with normal DNA (ie, 576G), the middle line is from one of the subclones demonstrating the 576G>C mutation (arrowhead), and the bottom line represents DNA from unfractionated marrow where a contribution from cytidine (blue, with arrow) can be seen as a minor peak under the primary fluorochrome representing guanidine (black).
576G>C ATRX mutation in a patient with ATMDS. (A) DHPLC tracing of the exon 8 amplicon from patient no. 4. The lighter and darker purple tracings represent unfractionated peripheral blood and bone marrow from a sample obtained when the patient had 50% erythrocytes with HbH inclusions by brilliant cresyl blue (BCB) staining and 25% HbH on electrophoresis. The brick red tracing represents unfractionated marrow from a time point 6 months later when the amount of HbH by electrophoresis had fallen to less than 1% and only rare HbH inclusions were detectable on supravital staining. The dark green tracing is a wild-type control. (B) Big Dye–generated sequence of 2 subclones (top 2 lines) and unfractionated marrow DNA (bottom line) from patient no. 4, exon 8 (coding strand 5′ to 3′, running left to right). The top line is sequence of a subclone with normal DNA (ie, 576G), the middle line is from one of the subclones demonstrating the 576G>C mutation (arrowhead), and the bottom line represents DNA from unfractionated marrow where a contribution from cytidine (blue, with arrow) can be seen as a minor peak under the primary fluorochrome representing guanidine (black).
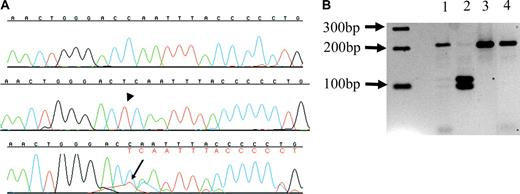
In 2 additional patients with ATMDS from whom only one DNA sample was available, the proportion of subclones with an ATRX mutation also roughly correlated with the amount of HbH. Patient no. 11 had 10% HbH-containing cells and 8% HbH by electrophoresis, and after subcloning, 10% of clones were found to harbor a 6083G>C; R2028P mutation. Patient no. 12 had 40% HbH-containing cells and 16.9% HbH by electrophoresis; 30% of clones contained the insertion 6884_6885insT; 2293Tfs2319X (Figure 4A). The latter mutation creates a new HinfI restriction site, and HinfI (New England Biolabs) digestion confirmed the presence of the mutation in both a mutant subclone and unfractionated marrow-derived DNA from patient no. 12 (Figure 4B).
6884_6885 insT ATRX mutation in a patient with ATMDS. (A) Sequences from patient no. 12, exon 33 (5′ to 3′, running left to right). The top line is a sequence from a normal subclone, the middle line represents one of the 10% of subclones bearing the thymidine insertion 6884_6885insT (arrowhead) that results in a frame shift, and the lower line is unfractionated marrow DNA, where the inserted thymidine (red, with arrow) can be seen as a secondary peak under the 3′ cytidine (blue); this is followed by a series of secondary peaks (denoted in red letters under primary base calls) as a result of the frame shift. (B) Hinf I digest from patient no. 12. The predicted normal exon 33 amplicon digestion fragments are 218-bp and 21-bp long. The T insertion at 6884_6885insT expands the amplicon from 239 bp to 240 bp and creates a new restriction site, leading to 3 digestion fragments of lengths 115 bp, 104 bp, and 21 bp. A 2-log DNA ladder (New England Biolabs) is at far left. Lane 2 is a digest of a subclone containing 6884_6885insT, and has the predicted 115-bp and 104-bp fragments. Lane 3 is a digest of a subclone from patient no. 12 where sequencing did not show the mutation, and only the 218-bp fragment can be seen, similar to lane 4, a wild-type control with only the 218-bp fragment. Lane 1 is a digest of amplified unfractionated marrow DNA from patient no. 12, and contains both the normal 218-bp fragment as well as faint bands corresponding to the 115-bp and 104-bp fragments from the minor population of mutant DNA.
6884_6885 insT ATRX mutation in a patient with ATMDS. (A) Sequences from patient no. 12, exon 33 (5′ to 3′, running left to right). The top line is a sequence from a normal subclone, the middle line represents one of the 10% of subclones bearing the thymidine insertion 6884_6885insT (arrowhead) that results in a frame shift, and the lower line is unfractionated marrow DNA, where the inserted thymidine (red, with arrow) can be seen as a secondary peak under the 3′ cytidine (blue); this is followed by a series of secondary peaks (denoted in red letters under primary base calls) as a result of the frame shift. (B) Hinf I digest from patient no. 12. The predicted normal exon 33 amplicon digestion fragments are 218-bp and 21-bp long. The T insertion at 6884_6885insT expands the amplicon from 239 bp to 240 bp and creates a new restriction site, leading to 3 digestion fragments of lengths 115 bp, 104 bp, and 21 bp. A 2-log DNA ladder (New England Biolabs) is at far left. Lane 2 is a digest of a subclone containing 6884_6885insT, and has the predicted 115-bp and 104-bp fragments. Lane 3 is a digest of a subclone from patient no. 12 where sequencing did not show the mutation, and only the 218-bp fragment can be seen, similar to lane 4, a wild-type control with only the 218-bp fragment. Lane 1 is a digest of amplified unfractionated marrow DNA from patient no. 12, and contains both the normal 218-bp fragment as well as faint bands corresponding to the 115-bp and 104-bp fragments from the minor population of mutant DNA.
Comparison of acquired and germ line ATRX mutations
A 576G>C; L192F mutation identical to that observed in ATMDS patient no. 4 was previously detected in a boy with the constitutional ATR-X syndrome who had 0.1% HbH-containing erythrocytes. While this strongly supports the pathologic significance of the acquired mutation in the patient with ATMDS, it is not clear why the patient with ATMDS had a much more severe hematologic phenotype than the patient with ATR-X.
An acquired ATRX mutation resulting in an amino acid change similar to that previously found in an ATR-X pedigree was found in ATMDS patient no. 5. This patient with ATMDS had a 718T>G; C240G mutation and 48% HbH-containing red cells, while the ATR-X pedigree proband had 719G>T; C240F and only 6% HbH-containing cells. Both mutations result in a loss of the same cysteine residue in the highly conserved plant homeodomain (PHD) finger, a zinc-finger motif that has been identified in many proteins, some of which appear to be involved in chromatin-mediated transcriptional regulation.21 The consensus PHD motif and adjacent cysteine-rich domain (protein residues 220 to 268) is the most common site of germ line ATRX mutations associated with ATR-X syndrome (Figure 5A).20 Loss of C240 may result in similar loss of a disulfide bond and unfolding of the PHD motif with both germ line and acquired mutations, but again the severity of the red cell abnormality was much greater in the patient with ATMDS. A 794G>A; C265Y mutation was found in ATMDS patient no. 6; although this also results in a loss of a cysteine residue in the PHD finger and may lead to protein unfolding, no directly comparable germ line ATRX mutations have yet been described.
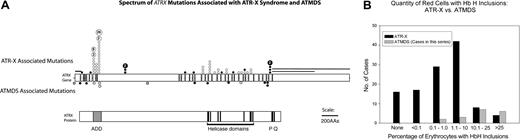
Acquired versus constitutional ATRX mutations. (A) Comparison of the spectrum of ATRX mutations described in boys with ATR-X syndrome with those found in ATMDS. The ATRX gene (top, with introns not to scale) is aligned with ATRX protein (bottom) to allow comparison of mutation site with functionally important protein domains such as the ADD (ie, ATRX, DNMT3, DNMT3L) zinc-finger domain (ADD includes a C2-C2 type of zinc finger and the closely located PHD motif), helicase domains, P-box, and Q-box. ATR-X–associated mutations are annotated above the schematic representation of the ATRX gene at the mutation loci, while mutations associated with ATMDS are denoted below. • represents mutations predicted to cause protein truncation (ie, frame shifts and nonsense mutations) and null mutations, whereas ○ represents amino acid changes, including in-frame insertions and deletions. Unfilled boxes denote the newly described -14G>A polymorphism and 2692G>C; D898H base-pair change of uncertain significance as described in the text. (B) Comparison of the fraction of erythrocytes containing HbH inclusions in 118 patients with ATR-X syndrome with the fraction detected in the 18 patients with ATMDS analyzed in this study. Transfused and untransfused patients with ATMDS are tallied together; the possibility of dilution by donor erythrocytes contributing to artifactually decreased HbH-containing cells in the former group should be recognized.
Acquired versus constitutional ATRX mutations. (A) Comparison of the spectrum of ATRX mutations described in boys with ATR-X syndrome with those found in ATMDS. The ATRX gene (top, with introns not to scale) is aligned with ATRX protein (bottom) to allow comparison of mutation site with functionally important protein domains such as the ADD (ie, ATRX, DNMT3, DNMT3L) zinc-finger domain (ADD includes a C2-C2 type of zinc finger and the closely located PHD motif), helicase domains, P-box, and Q-box. ATR-X–associated mutations are annotated above the schematic representation of the ATRX gene at the mutation loci, while mutations associated with ATMDS are denoted below. • represents mutations predicted to cause protein truncation (ie, frame shifts and nonsense mutations) and null mutations, whereas ○ represents amino acid changes, including in-frame insertions and deletions. Unfilled boxes denote the newly described -14G>A polymorphism and 2692G>C; D898H base-pair change of uncertain significance as described in the text. (B) Comparison of the fraction of erythrocytes containing HbH inclusions in 118 patients with ATR-X syndrome with the fraction detected in the 18 patients with ATMDS analyzed in this study. Transfused and untransfused patients with ATMDS are tallied together; the possibility of dilution by donor erythrocytes contributing to artifactually decreased HbH-containing cells in the former group should be recognized.
Three of the new ATRX mutations (6884_6885insT; 2293Tfs2319X, 7014_7017del; N2340fsX2341, and 7049delT; E2351fsX2353 in patient nos. 12-14, respectively) result in a frame-shift and generation of a premature stop codon near the C-terminus of ATRX. This is predicted to result in the loss of 2 highly conserved ATRX domains: the P-box (protein residues 2385-2401), an element conserved among SNF2-like family members involved in transcriptional regulation, and the Q-box, a stretch of glutamine residues (protein 2415-2425) representing a potential protein interaction domain.22 Several germ line ATRX mutations resulting in the loss of these domains have been described (Figure 5A).23 In contrast to patients with ATMDS, all of the affected individuals in those ATR-X pedigrees have only rare HbH-containing cells.
ATMDS patient no. 10 had a 5567-3C>G ATRX mutation in the consensus splice acceptor site 5′ to exon 24 (in intron 23), and patient no. 11 had a 6083G>C; R2028P mutation in exon 27—both located in the region between the third and fourth of the 7 highly conserved helicase domains of ATRX. Archival mRNA was not available from patient no. 10 (deceased) with 5567-3C>G in order to confirm the presumed splicing abnormality. Although mutations in the region of the ATRX helicase domains are relatively common in germ line ATR-X patients (Figure 5), the associated hematologic phenotypes in these pedigrees are quite variable, and the 2 mutations detected in the patients with ATMDS have not been described to date in ATR-X pedigrees.
The functional consequence (if any) of the remaining 2 newly described ATRX base-pair changes, in patients no. 1 and no. 7, respectively, is uncertain. Unlike the other 8 patients with ATMDS with novel ATRX mutations described here, in these cases no mosaicism was apparent on sequencing and/or subcloning. In addition, these single base-pair ATRX changes were also present in Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines derived from the patients. The -14G>A change in patient no. 1 is located in a conserved area of the 5′ untranslated region of the ATRX gene, but we were able to obtain gDNA from this deceased patient's daughter (with her informed consent) and found that she is heterozygous for -14G and -14A, demonstrating that this base-pair change is a germ line polymorphism. In patient no. 7, the 2692G>C; D898H mutation is in the middle of exon 10, 3′ to the PHD motif; this region of ATRX is not highly conserved in the mouse.22 Unfortunately, patient no. 7 is also dead, and no other genetic material is availabe to help resolve the mutation versus polymorphism issue (eg, DNA from buccal smears or fibroblast cell lines derived from the patient or from a surviving daughter). Neither -14G>A nor the 2692G>C; D898H variant have been detected in previous studies of pedigrees with ATR-X syndrome, patients with ATMDS, or healthy controls.
Patients in whom no ATRX mutation was detected
No pathologic mutations in ATRX were found in patients no. 15 through no. 18 or patient no. 1 with chronic myeloid disorders, HbH inclusions, and hypochromic, microcytic red cells. These patients were atypical of ATMDS in several respects. In patients no. 1 and no. 16 through no. 18, the amount of HbH was so small that it was not detectable by electrophoresis. Patients no. 17 and no. 18 had only very rare (0.01% and 0.3%) HbH-containing erythrocytes on supravital staining. Patient no. 15 was the only female patient with ATMDS studied; approximately 90% of patients with ATMDS described have been men.9
Discussion
The data presented here add further support to recent observations16 showing that point mutations in the ATRX gene at Xq13.3 are strongly associated with the rare ATMDS subtype and are responsible for the associated α thalassemia seen in such cases. However, these data also raise several new questions regarding the mechanism by which α globin expression is down-regulated in this condition.
The normal role of ATRX protein in the cell is not known, but it is a member of the SWI/SNF group of proteins that are thought to influence a wide range of nuclear processes (eg transcription, replication, recombination) via effects on chromatin.23,24 ATRX is found in the nucleus where it localizes to pericentromeric heterochromatin during interphase and mitosis.25,26 Consistent with this, ATRX has been shown to interact with the major heterochromatin-associated protein HP1.25 A proportion of ATRX is also found in promyelocytic leukemia (PML) bodies,27 which contain a variety of nuclear factors that influence apoptosis, regulate gene expression, and control progression through the cell cycle. These bodies are characteristically disrupted in AML subtype M3.28 ATRX has also recently been shown to form a stable complex with Daxx, a protein that is found in PML bodies and has been implicated in the regulation of apoptosis.27
Perhaps the best guide to the role played by ATRX in the cell comes from analysis of the consequences of germ line ATRX mutations in the inherited ATR-X syndrome. Such mutations are associated with widespread changes in the patterns of DNA methylation in the genome together with alterations in gene expression.24 At present, the best documented of these changes in gene expression is that which occurs at the α globin cluster, where ATRX mutations are consistently associated with down-regulation of gene expression, resulting in mild α thalassemia. As shown here and in our previous report,16 acquired ATRX mutations in the ATMDS syndrome may be associated with a relatively severe degree of α thalassemia. At present it is not yet clear how ATRX contributes to normal α globin gene expression, or at what stage(s) of development and red cell differentiation ATRX is involved.
The findings presented here raise a new issue in our understanding of the pathophysiology of ATMDS. The dramatically reduced α/β globin synthesis ratios, the consequent hematologic changes, and the relatively high levels of HbH in ATMDS cases with acquired ATRX mutations demonstrate that the associated α thalassemia is, in general, more severe in ATMDS than in ATR-X syndrome. Individuals with ATMDS have been described in whom up to 90% of the peripheral blood erythrocytes contain HbH inclusions,9 whereas patients with constitutional ATRX mutations have much lower levels of HbH (90% have 10% HbH inclusions or less, while 53% have ≤ 1% HbH inclusions) and some have no HbH inclusions (Figure 5B).29 One explanation may be that the spectrum of acquired somatic ATRX mutations in ATMDS may include severe, null mutations that are not seen in patients with the inherited ATR-X syndrome because such mutations may be lethal early in development.16,30 However, in this study we found a patient with severe hypochromic microcytic anemia and up to 50% of red cells containing HbH inclusions who has acquired the same mutation (576G>C; L192F) previously identified in a patient with ATR-X syndrome who had minimal red cell changes and only very rare HbH inclusions. We also found an ATRX mutation (718T>G; C240G) in another patient with ATMDS whose mutation affects the same amino acid as a congenital ATR-X case, as well as a series of ATRX mutations that may have similar functional consequences as previously described constitutional cases. In each instance the ATMDS hematologic phenotype was more severe than in the corresponding ATR-X cases. Although it seems almost certain that the ATRX mutation is responsible for the α thalassemia in both groups, the degree to which α globin expression is down-regulated must depend on other interacting molecular and/or cellular factors. This might have been expected, since we have previously noted that in ATR-X syndrome, the number of HbH inclusions may vary over several orders of magnitude in different pedigrees with the same ATRX mutation, and there is also some variation within affected families. It will be important to understand which factors are modifying the effects of mutations in ATRX.
To date, all 12 of the patients with ATMDS in whom we have identified clearly pathogenic ATRX mutations or ATRX splicing abnormalities have the typical features of this rare syndrome with hypochromic, microcytic anisopoikilocytic red cells associated with substantial amounts of HbH and significantly reduced α/β globin chain synthesis ratios. The only ATMDS patients with an α/β globin chain synthesis ratio more than 0.2 in whom we found an ATRX mutation were those who had received red cell transfusions prior to study, which likely affected their red cell indices and globin chain synthesis ratio.
The red cell indices of patients with ATMDS in general are quite distinct from the majority of patients with MDS, who typically have normocytic or macrocytic anemia.31 We have observed other patients with MDS and similar hypochromic microcytic red cell morphology but with only rare or no HbH inclusions; to date, no mutations in the ATRX gene have been detected in such individuals. It is not clear whether this is a result of insensitivity of our current detection techniques to very small subpopulations of cells with an ATRX mutation, or whether a different molecular mechanism might be responsible.
It seems unlikely that ATRX mutations play a key role in the development of MDS. Patients with the inherited ATR-X syndrome do not show any evidence of genome instability and have no increased incidence of MDS or hematologic malignancy.29 Furthermore, if ATRX mutations were involved in the origin or evolution of MDS, α thalassemia should be encountered more frequently in such patients. It seems likely that mutations in ATRX have come to attention in MDS only because the resulting α thalassemia has an easily detectable and dramatic red cell phenotype.
Before we studied this series of patients with ATMDS with DHPLC, the ATRX gene had been sequenced in a subset of the patients without detection of additional mutations. Several possibilities might account for difficulty in finding ATRX mutations in ATMDS. Mutations in MDS are often lineage restricted.32 This also appears to be true in ATMDS, where the mutations described above (with the exception of the one that may represent a polymorphism) were found in DNA isolated from myeloid cells but not DNA from lymphoblastoid cell lines derived from the patients, where such cell lines were available. Although malignant stem cell transformation has been described in MDS, the usual cell of origin is thought to be a committed myeloid progenitor and lymphocytes are often not involved in the neoplastic clone.33-35 In addition, ATRX analysis could be confounded by somatic mosaicism in ATMDS. These issues are pertinent to genetic and pathophysiologic investigations in all types of MDS, and therefore devising a strategy to identify ATRX mutations in unfractionated samples of DNA from patients with ATMDS provides a good model for developing general approaches for mutation analysis in MDS and related hematologic malignancies.
Direct sequencing can detect point mutations in mixtures of genomic DNA containing 20% to 30% mutant DNA.36 Denaturing gradient gel electrophoresis (DGGE) is more sensitive than sequencing (DGGE can detect < 5% mutant DNA) but is cumbersome and only a few samples can be analyzed at a time.37 Recently, DHPLC has proven to be a powerful, sensitive, high-throughput technique for mutation detection38 that may be particularly useful in the setting of germ line and somatic mosaicism.39-41 DHPLC is therefore an attractive technique for mutational analysis of neoplastic conditions such as MDS, including the ATMDS subtype, where the abnormal cells are not easy to separate from normal hemopoietic cells. This is also relevant to archival DNA obtained from unfractionated blood or marrow specimens.
Somatic mosaicism has likely contributed to the difficulty many investigators have faced in describing specific molecular lesions associated with various MDS phenotypes and cytogenetic patterns— compare, for example, the many specific mutations that have been recognized in acute leukemia42 with the relatively small number fully characterized in MDS.43 The present study demonstrates that DHPLC can be a powerful tool for detecting mutations in the setting of mosaicism in MDS. Therefore, in addition to confirming the importance of ATRX in ATMDS, our findings have important implications for identifying other genes that may be mutated in MDS.
Prepublished online as Blood First Edition Paper, October 30, 2003; DOI 10.1182/blood-2003-09-3360.
Supported by the Medical Research Council in the United Kingdom (R.J.G., C.A.F., D.R.H.). D.P.S. is a Mayo Foundation Research Scholar supported by the Mayo Foundation, Rochester, MN.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal