Abstract
Drosophila enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) homology 1 (EVH1) domain proteins regulate signal transduction at the neuronal and immunologic synapse. Despite shared cell biologic machinery at these synapses, the regulation of client proteins that transmit synaptic activity to the nucleus is likely to be different. Homer-3, a member of the EVH1 family, is expressed in the thymus, suggesting a role for this protein in T-cell signal transduction. Upon T-cell receptor (TCR) engagement, Homer-3 was recruited to the contact area of Jurkat cells to anti-CD3 and CD28 antibody–coated beads prior to actin accumulation and was subsequently translocated into the nucleus. Overexpression of Homer-3 reduced transcriptional activation via the serum response element (SRE) in response to anti-CD3 antibody, phorbol ester, or dominant active Ha-Ras. Consistent with these results, knockdown of Homer-3 increased SRE activation. Homer-3 coprecipitated with CCAAT/enhancer binding protein β (C/EBPβ), one of the transcription factors that binds to the SRE and has a consensus motif binding to EVH1 domain. Moreover, Homer-3 and its EVH1 domain fragment reduced transcriptional activation of C/EBPβ. These findings suggest that Homer-3 may be involved in the regulation of SRE activation in T cells via interaction between its EVH1 domain and C/EBPβ.
Introduction
The importance of compartmentalization in lymphocyte activation is only beginning to be appreciated. Early signaling events following T-cell receptor (TCR) ligation require lipid rafts and polymerization of the actin cytoskeleton, which serve to assemble signaling complexes.1-5 Drosophila enabled/vasodilator-stimulated phosphoprotein (Ena/VASP) homology 1 (EVH1) domain–containing proteins can be divided into 4 subgroups: the Ena/VASP, Wiskott-Aldrich syndrome protein (WASP), Spred, and Homer families.6 The EVH1 domain was first identified as a homologous region of about 110 amino acids present in the N-terminal region of the Ena/VASP family. Members of the Ena/VASP family are key adaptor molecules in regulating actin filament assembly.7-9 WASP family proteins regulate actin dynamics through binding and activation of the actin-related protein 2/3 (Arp2/3) complex, which nucleates new actin filaments.10-12 Spreds are Sprouty-related suppressors of Ras signaling. Spreds constitutively associate with Ras and suppress phosphorylation of Raf.13
The Homer family was first identified as an immediate early gene expressed in rat brain after electroconvulsive seizures or electrical stimulation that leads to long-term potentiation of hippocampal glutamatergic synapses.14,15 Members of the Homer family bind to a variety of proteins, including group I metabolic glutamate receptors (mGluR), inositol triphosphate receptors, ryanodine receptors, and Shank (Src homology 3 domain and ankyrin repeat-containing).14,16-18 All of these interactions have been shown to be mediated by the EVH1 domain of the Homer molecule. The EVH1 domains of Homer family members recognize a PPxxFr motif,16,19 whereas the EVH1 domains of Ena/VASP family interact with an FPPPP motif.20,21 Crystal structure analyses have defined the N-terminal domain of the Homer family as a class II EVH1 domain.19 Except for short isoforms resulting from alternative splicing, all the long forms of Homers have a coiled-coil domain (CCD) in the C-terminal region, which is required for the formation of Homer dimers and tetramers.22 In addition, the CCD comprises 1 or 2 typical leucine zipper motifs, which can promote further multimerization.17,23,24 As a result of component domains, members of the Homer family may function as scaffolds, coupling receptors, and adaptor proteins in synapse assembly and signaling. In neurons Homers can bring together membrane mGluR and endoplastic reticulum–associated intracellular inositol triphosphate receptors.25
The EVH1 domain proteins Ena-VASP–like (Evl)/mammalian enabled (Mena) and WASP/WASP family verprolin-homologous protein (Wave) are found in neuronal cells and T lymphocytes and are implicated in intracellular synapse formation in both cell types.26-28 The functions of the Homer family have been studied mainly in neurons. Homer-1 is localized to excitatory synapses and modulates calcium signaling, axon guidance, and dendritic spine remodeling.29-31 Expression of short Homer-1a, which lacks the CCD, is rapidly induced in the hippocampus following electroconvulsive seizure and promotes the disassembly of the signal complex.14,15,32 Homer-2 (Cupidin) is also enriched in the postsynaptic density fraction and interacts with actin cytoskeleton, mGluR, and activated Rho family guanosine 5′-triphosphatase (GTPase), Cdc42.23 A recent study has demonstrated that Homer-2 modulates G protein–coupled receptors by regulating regulators of G protein signaling (RGS) proteins and phospholipase C β (PLCβ) GTPase-activating protein (GAP) activities.33
The differential expression of Homer family members initiated an interest in determining the functional implications of Homer family members in immune cell signaling. Homer-3 is the unique Homer isoform expressed in the thymus.22 Gray et al29 have demonstrated that Homer-3 interacts with dynamin-3 within the neuronal synapse and modulates dendritic spine function, whereas its functions in the hematopoietic system have not yet been investigated. In the present study, we describe a novel role for Homer-3 following T-cell activation. Homer-3 is recruited to the immunologic synapse prior to actin accumulation. Subsequently, Homer-3 is translocated into the nucleus in an EVH1 domain–dependent manner and regulates serum response element (SRE) activation in T cells. Inhibition of SRE activation by Homer-3 may depend on the interaction between its EVH1 domain and CCAAT/enhancer binding protein β (C/EBPβ). These observations implicate Homer-3 in the negative regulation of SRE activation in T cells.
Materials and methods
Cells and reagents
Jurkat E6.1 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in Iscoves modified Dulbecco medium including 10% heat-inactivated fetal bovine serum, 50 μM 2-mercaptoethanol, and 20 μg/mL gentamycin. Mouse CD4+ T cells were isolated from the spleens of Friend leukocyte virus B–sensitive (FVB) mice by magnetic bead purification (Dynal Biotech, Oslo, Norway). Purity was confirmed by flow cytometry analysis. Goat anti–Homer-3 antibody, which recognizes the C-terminus of Homer-3 (blocking peptide for anti–Homer-3 antibody, mouse antiactin antibody, and mouse C/EBPβ antibody), were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-Ezrin antibody was purchased from Cell Signaling Technology (Beverly, MA). Rabbit anti-WASP antibody and mouse anti-Vav1 antibody were purchased from Upstate Biotechnology (Lake Placid, NY). Mouse anti-Myc antibody was purchased from Covance (Princeton, NJ). Mouse anti-CD3ζ antibody was purchased from Zymed (South San Francisco, CA).
Expression constructs
A plasmid including Homer-3 cDNA was kindly provided by Dr Paul Worley (Johns Hopkins Medical School, Baltimore, MD).22 Simple modular architecture research tool (SMART)34 predicted EVH1 domain from amino acid residues 1-110 of human Homer-3 and coiled-coil domains from residues 192-243 and 282-338 (the sequence number is based on human Homer-3A11).35 According to the prediction, the fragment corresponding to residues 1-110 was generated by polymerase chain reaction (PCR) from the plasmid using primers 5′-cgcggggaattcttatgtccacagccagggag-3′ and 5′-gcgcccgtcgacgtcttcacttcctggaactt-3′ and named EVH. The fragment corresponding to residues 192-361 was generated using primers 5′-cgcggggtcgacgctgcaggacagcaacaaca-3′ and 5′-gcgcccagatctatcagggcgcagcctcagcc-3′ and named CCD. The full-length fragment was also generated using primers 5′-cgcggggaattcttatgtccacagccagggag-3′ and 5′-gcgcccagatctatcagggcgcagcctcagcc-3′ and named Full. These fragments were subcloned into pCMV-Myc (Clontech, Palo Alto, CA). The accuracy of these constructs was confirmed by sequencing and molecular size analysis of overexpressed proteins. Green fluorescence protein (GFP)–Ha-RasG14V construct was kindly provided by Dr Joseph Avruch (Massachusetts General Hospital).36 GFP-WASP construct was provided by Dr Marco Lopez (Brigham and Women's Hospital, Boston, MA).37 A 250–base pair (bp) fragment of the k10 promoter cloned upstream of luciferase gene and C/EBP constructs were provided by Dr Joel Habener (Massachusetts General Hospital) and Dr Marco Lopez.38 The SRE luciferase reporter, which contains 3 tandem repeats of the murine c-fos SRE inserted upstream of luciferase gene, was provided by Dr Michael White (University of Texas Southwestern Medical Center, Dallas).39
Conjugate formation assays
Jurkat cells were stimulated with antibody-coated beads as described previously.40 Briefly, the cells were mixed with Dynabeads CD3/CD28 or CD19 (Dynal Biotech) at a ratio of 1:2. After centrifugation for 3 minutes at 100g, the cell-bead mixture was incubated for an additional 2 to 10 minutes at 37°C. Conjugates were then resuspended, plated onto poly-l-lysine–coated glass slides, and fixed for 10 minutes at room temperature in 3.5% paraformaldehyde and 0.1% Tween 20 in phosphate-buffered saline (PBS).
Spreading assays
Cells were stimulated with antihuman CD3 antibody (OKT3) or antimouse CD3e antibody (Pharmingen, San Diego, CA) on glass slides as described previously.41 Briefly, the glass slides (Fisher Scientific, Hanover Park, IL) were treated with a 0.01% wt/vol poly-l-lysine solution (Sigma-Aldrich, St Louis, MO) for 30 minutes at room temperature. After 4 washes with distilled water, they were coated with 10 μg/mL anti-CD3 antibody for 2 hours at 37°C in a moist chamber. After 4 washes with PBS, the cells were plated onto the coated glass slides. After incubation at 37°C for 2 to 10 minutes, the cells were fixed for 10 minutes at room temperature with 3.5% paraformaldehyde and 0.1% Tween 20 in PBS.
PMA stimulation in solution
Jurkat cells were resuspended in culture medium including 50 ng/mL phorbol myristate acetate (PMA) or vehicle, incubated at 37°C for 10 minutes, plated onto poly-l-lysine–coated glass slides, and then fixed.
Immunocytochemical staining
Fixed cells were washed with PBS, quenched with 0.1 M NH4Cl for 5 minutes at room temperature, and washed with PBS. After blocking with 1% bovine serum albumin (BSA) in PBS, the cells were labeled with primary antibodies followed by 3 washes for 5 minutes each. After labeling with secondary antibodies conjugated with Alexa Fluor (Molecular Probes, Eugene, OR), the slides were mounted with Aqua Poly/Mount (Polysciences, Warrington, PA). The samples were viewed under a fluorescence microscope (Olympus, Tokyo, Japan) or confocal laser scanning microscope (Leica Microsystems, Chicago, IL).
Nuclear extraction and immunoblotting
Jurkat cells (1.5 × 107) were stimulated at 37°C with OKT3 (5 μg/mL) for 0, 10, 60, and 240 minutes. The nuclear extract was obtained according to the protocol of Schreiber et al.42 Briefly, the cells were washed with cold PBS, resuspended in 1 mL hypotonic buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], pH7.9; 10 mM KCl; 0.1 mM EDTA [ethylenediaminetetraacetic acid]; 0.1 mM EGTA [ethyleneglycotetraacetic acid]; 1 mM dithiothreitol [DTT]; 1 mM sodium vanadate; 10 μg/mL leupeptin; 10 μg/mL aprotinin) for 15 minutes, mixed vigorously with 62.5 μL of 10% nonidet P-40 (NP-40), and then incubated for 15 minutes at 4°C. After centrifugation at 10 000g for 1 minute, the pellet was washed twice with hypotonic buffer. The pellet was resuspended in hypertonic buffer (20 mM HEPES, pH 7.9; 400 mM NaCl; 1 mM EDTA; 1 mM EGTA; 1 mM DTT; 1 mM sodium vanadate; 10 μg/mL leupeptin; 10 μg/mL aprotinin) for 30 minutes. After centrifugation at 10 000g for 15 minutes, the supernatant was used as the nuclear extract. The nuclear extract (15 μg total proteins) was resolved on a 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel and blotted with anti-p65 antibody (Upstate Biotechnology) or anti–Homer-3 antibody.
Construction of small interfering RNA to Homer-3
Two sets of oligonucleotides were generated in the DNA Core Facility of Massachusetts General Hospital (Boston, MA) as described43 : no. 1 targeting nucleotides 218-236 of coding sequence of Homer-3, 5′-gatccccttcccagaagttcgggcattcaagagatgcccgaacttctgggaagtttttggaaa-3′ and 5′-tcgatttccaaaaacttcccagaagttcgggcatctcttgaatgcccgaacttctgggaagggg-3′; no. 2 targeting nucleotides 138-156, 5′-gatcccctgtgtaccgcatcatcagcttcaagagagctgatgatgcggtacacatttttggaaa-3′ and 5′-tcgatttccaaaaatgtgtaccgcatcatcagctctcttgaagctgatgatgcggtacacaggg-3′. Each set of olignulceotides was phosphorylated, annealed, and subcloned into pSuppress (a generous gift from Dr Daniel Billadeau; Mayo Clinic, New York, NY) to express small interfering RNA (SiRNA) downstream of the RNA polymerase III–dependent H1 promoter as described.43 The accuracy of these constructs was confirmed by sequencing.
Luciferase assay
The induction and determination of luciferase activation were performed as described previously.44 Briefly, 5 × 106 Jurkat cells were resuspended in 250 μL culture medium and electroporated (250 V, 950 μF) with luciferase reporter and expression constructs. Upon electroporation, the cells were diluted in 2.5 mL culture medium and incubated for 24 or 20 hours followed by a further 4 hours' incubation with 5 μg/mL OKT3 or 50 ng/mL PMA. The cells were lysed in cell culture lysis reagent (Promega, Madison, WI). Luciferase activity was determined with Luciferase Assay System (Promega) according to the manufacturer's instructions. The electroporation efficiency was normalized by quantifying GFP expression with flow cytometry for each assay. The staining with annexin V and 7-aminoactinomycin (BD Biosciences, San Jose, CA) confirmed similar viability of the cells after electroporation (data not shown).
Immunoprecipitation of C/EBPβ
Jurkat cells (1 × 107) were electroporated with 10 μg Myc construct expressing full-length Homer-3. After electroporation, 2 × 107 cells were resuspended in 10 mL culture medium and incubated for 24 hours. After 10 minutes' stimulation with PMA (50 ng/mL), the nuclear extract was obtained as described in “Nuclear extraction and immunoblotting.” For immunoprecipitation, 300 μL nuclear extract was diluted with 500 μL distilled water including 1 mM DTT, 1 mM sodium vanadate, 10 μg/mL leupeptin, 10 μg/mL aprotinin, and then incubated with 2 μg rabbit normal IgG or anti-C/EBPβ (a generous gift from Dr Joel Habener) overnight at 4°C. The immune complex was collected with 20 μL of protein A–agarose beads.
Pull-down assay using C/EBP consensus oligonucleotide agarose conjugates
Nuclear extract was obtained from Jurkat cells electroporated with Myc construct either empty or full-length Homer-3 as described in “Nuclear extraction and immunoblotting.” The nuclear extract was incubated with C/EBP consensus oligonucleotide (TGC AGA TTG CGC AAT CTG CA) agarose conjugates (Santa Cruz Biotechnology) or its mutant oligonucleotide (TGC AGA GAC TAG TCT CTG CA) agarose conjugates (Santa Cruz Biotechnology), and the complex was used for Western blotting according to the manufacturer's instructions.
Results
The intracellular distribution of Homer-3 in T cells after T-cell receptor engagement
Previous study using Western blot analysis has demonstrated that Homer-3 is expressed in T cells.22 We have confirmed this observation (data not shown). To establish antibody specificity, we performed immunohistochemical staining of Homer-3 in various tissues. Light microscopic examinations revealed that Homer-3 was detected in Purkinje neurons in the cerebellum (data not shown) as previously described.22
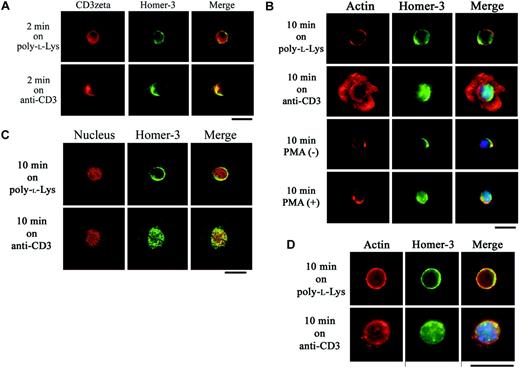
To observe the intracellular distribution of endogenous Homer-3 in T cells after polarization of TCR, Jurkat cells were activated with anti-CD3 and anti-CD28 antibody–coated beads. This system has properties of an immunologic synapse based on synergy of TCR and integrin signals.40,45 As previously demonstrated, Ezrin and WASP were recruited to the cell-bead contact area along with actin accumulation27,28,40 (Figure 1A). However, sequential observation revealed Homer-3 recruitment to the immunologic synapse prior to filamentous actin redistribution (Figure 1A). Surprisingly, on prolonged stimulation, Homer-3 was translocated into the nucleus following actin accumulation at the synaptic interface (Figure 1A). Homer-3 redistribution was not observed when anti-CD19 antibody–coated beads were incubated with Jurkat cells (Figure 1A).
Ezrin, WASP, and Homer-3 localization in Jurkat cells stimulated with anti-CD3/CD28 antibody–coated beads. (A) Antibody-coated bead T-cell conjugates demonstrated redistribution of endogenous Homer-3 following T-cell receptor activation. F-actin accumulation (phalloidin-Alexa Fluor 594, red) in the contact area was induced by stimulation with anti-CD3/CD28 antibody–coated beads for 10 minutes, whereas no increase in actin accumulation was present at 2 minutes in Jurkat cells. Ezrin (green) and WASP (green) were recruited to the contact area along with actin accumulation after 10 minute stimulations. In contrast, Homer-3 (green) was recruited to the contact area after 2 minutes' stimulation, where increased actin accumulation was not observed. After actin accumulation in the contact area, Homer-3 was translocated into the nucleus (DAPI [4,6 diamidino-2-phenylindole], blue). Representative images showed that neither recruitment to the contact area nor nuclear translocation of Homer-3 was observed in the cells stimulated with anti-CD19 antibody–coated beads. (B) Jurkat cells were electroporated with Myc construct expressing coiled-coil domain (CCD) or EVH1 domain fragment of Homer-3. Twenty-four hours later, the cells were stimulated with anti-CD3/CD28 antibody-coated beads for 2 minutes and then stained with anti-Myc antibody. The CCD fragment of Homer-3 was recruited to the contact area, whereas the EVH1 domain fragment was not. Representative images were presented from experiments performed at least 3 times. Original magnification × 1000.
Ezrin, WASP, and Homer-3 localization in Jurkat cells stimulated with anti-CD3/CD28 antibody–coated beads. (A) Antibody-coated bead T-cell conjugates demonstrated redistribution of endogenous Homer-3 following T-cell receptor activation. F-actin accumulation (phalloidin-Alexa Fluor 594, red) in the contact area was induced by stimulation with anti-CD3/CD28 antibody–coated beads for 10 minutes, whereas no increase in actin accumulation was present at 2 minutes in Jurkat cells. Ezrin (green) and WASP (green) were recruited to the contact area along with actin accumulation after 10 minute stimulations. In contrast, Homer-3 (green) was recruited to the contact area after 2 minutes' stimulation, where increased actin accumulation was not observed. After actin accumulation in the contact area, Homer-3 was translocated into the nucleus (DAPI [4,6 diamidino-2-phenylindole], blue). Representative images showed that neither recruitment to the contact area nor nuclear translocation of Homer-3 was observed in the cells stimulated with anti-CD19 antibody–coated beads. (B) Jurkat cells were electroporated with Myc construct expressing coiled-coil domain (CCD) or EVH1 domain fragment of Homer-3. Twenty-four hours later, the cells were stimulated with anti-CD3/CD28 antibody-coated beads for 2 minutes and then stained with anti-Myc antibody. The CCD fragment of Homer-3 was recruited to the contact area, whereas the EVH1 domain fragment was not. Representative images were presented from experiments performed at least 3 times. Original magnification × 1000.
In order to identify the domain of Homer-3 responsible for the distribution to the immunologic synapse, we first asked whether overexpressed Homer-3 behaved similarly in Jurkat cells. Overexpression of Homer-3 in Jurkat cells did not interfere with the actin cytoskeleton architecture (data not shown). Overexpressed Homer-3 preceded actin accumulation at the immunologic synapse, and this effect was mediated by the C-terminal CCD but not by the N-terminal EVH1 domain (Figure 1B).
To confirm the coclustering with TCR and the nuclear translocation of Homer-3 in response to TCR stimulation, we turned to a system that readily permits visualization of the coclustering and the nuclear translocation following TCR engagement. On glass slides coated with poly-l-lysine only, endogenous Homer-3 was detected in the cytoplasmic region without significant colocalization with CD3ζ (Figure 2A). After 2 minutes' stimulation on glass slides coated with antihuman CD3 antibody (OKT3), Homer-3 coclustered with CD3ζ (Figure 2A). On the other hand, after 10 minutes' stimulation on OKT3-coated glass slides, Homer-3 was translocated into the nucleus (Figure 2A). The nuclear translocation was also observed in Jurkat cells activated in solution with PMA (Figure 2B). Confocal laser scanning microscopy also confirmed the nuclear translocation of Homer-3 following TCR stimulation (Figure 2C).
Coclustering with CD3ζ and nuclear translocation of Homer-3 in response to TCR stimulation in T cells. (A) Jurkat cells were incubated for 2 minutes at 37°C on glass slides coated with poly-l-lysine only or poly-l-lysine and anti-CD3 antibody (OKT3). After fixation, the cells were stained with anti-CD3ζ antibody and anti–Homer-3 antibody followed by secondary antibodies-Alexa Fluor 594 (red) or 488 (green). After incubation on the glass slides coated with OKT3, Homer-3 (green) coclustered with CD3ζ (red). (B) Jurkat cells were incubated for 10 minutes at 37°C on glass slides coated with poly-l-lysine only or poly-l-lysine and OKT3. After fixation, the cells were stained with anti–Homer-3 antibody followed by secondary antibody-Alexa Fluor 488 (green), phalloidin-Alexa Fluor 594 (red), and DAPI (blue) for nuclear staining. Homer-3 (green) was distributed in the cytoplasmic region after incubation on the glass slides coated with poly-l-lysine only. After incubation on the glass slides coated with OKT3, F-actin (red) was rearranged into circumferential rings and Homer-3 was translocated into the nucleus (blue). The nuclear translocation of Homer-3 was also observed in Jurkat cells stimulated with PMA in solution for 10 minutes. (C) Jurkat cells were incubated for 10 minutes as in panel B. After fixation, the cells were stained with anti–Homer-3 antibody followed by secondary antibody-Alexa Fluor 488 (green) and propidium iodide (red) for nuclear staining. The cells were observed with confocal laser scanning microscopy, which confirmed the nuclear translocation of Homer-3 following TCR stimulation. (D) Primary CD4+ T cells were isolated from mouse spleen and stimulated on antimouse CD3 antibody–coated glass slides. While Homer-3 was distributed in the cytoplasmic region of the cells incubated on the glass slides coated with poly-l-lysine only, the translocation into the nucleus was observed after stimulation with anti-CD3 antibody. Representative images were presented from experiments performed at least 3 times. Bars, 10 μm.
Coclustering with CD3ζ and nuclear translocation of Homer-3 in response to TCR stimulation in T cells. (A) Jurkat cells were incubated for 2 minutes at 37°C on glass slides coated with poly-l-lysine only or poly-l-lysine and anti-CD3 antibody (OKT3). After fixation, the cells were stained with anti-CD3ζ antibody and anti–Homer-3 antibody followed by secondary antibodies-Alexa Fluor 594 (red) or 488 (green). After incubation on the glass slides coated with OKT3, Homer-3 (green) coclustered with CD3ζ (red). (B) Jurkat cells were incubated for 10 minutes at 37°C on glass slides coated with poly-l-lysine only or poly-l-lysine and OKT3. After fixation, the cells were stained with anti–Homer-3 antibody followed by secondary antibody-Alexa Fluor 488 (green), phalloidin-Alexa Fluor 594 (red), and DAPI (blue) for nuclear staining. Homer-3 (green) was distributed in the cytoplasmic region after incubation on the glass slides coated with poly-l-lysine only. After incubation on the glass slides coated with OKT3, F-actin (red) was rearranged into circumferential rings and Homer-3 was translocated into the nucleus (blue). The nuclear translocation of Homer-3 was also observed in Jurkat cells stimulated with PMA in solution for 10 minutes. (C) Jurkat cells were incubated for 10 minutes as in panel B. After fixation, the cells were stained with anti–Homer-3 antibody followed by secondary antibody-Alexa Fluor 488 (green) and propidium iodide (red) for nuclear staining. The cells were observed with confocal laser scanning microscopy, which confirmed the nuclear translocation of Homer-3 following TCR stimulation. (D) Primary CD4+ T cells were isolated from mouse spleen and stimulated on antimouse CD3 antibody–coated glass slides. While Homer-3 was distributed in the cytoplasmic region of the cells incubated on the glass slides coated with poly-l-lysine only, the translocation into the nucleus was observed after stimulation with anti-CD3 antibody. Representative images were presented from experiments performed at least 3 times. Bars, 10 μm.
To observe the distribution of Homer-3 in primary T cells, CD4+ T cells were isolated from mouse spleen. Although the spreading on antimouse CD3 antibody–coated glass slides was not as extensive as what we observed with Jurkat cells, immunostaining for endogenous Homer-3 confirmed nuclear translocation (Figure 2D). The specific staining of Homer-3 was abolished after incubation of anti–Homer-3 antibody with blocking peptide (data not shown).
To identify the domain responsible for Homer-3 nuclear localization, Jurkat cells were electroporated with Myc-tagged constructs expressing fragments of Homer-3. Twenty-four hours later, viable cells were stimulated on glass slides coated with OKT3. While both isolated EVH1 domain and CCD fragments were detected in the cytoplasmic region on the glass slides coated with poly-l-lysine only, the EVH1 domain fragment was detected in the nucleus after 10 minutes' incubation on OKT3 (Figure 3). In contrast, the CCD fragment was localized in the cytoplasm and formed aggregates (Figure 3). These subcellular localization studies for the first time suggest a novel role for Homer-3 in T-cell signaling.
Nuclear translocation of EVH1 domain fragment in Jurkat cells stimulated with anti-CD3 antibody. Jurkat cells were electroporated with Myc construct expressing EVH1 domain or coiled-coil domain (CCD) fragment of Homer-3. Twenty-four hours later, the cells were incubated for 10 minutes at 37°Con glass slides coated with poly-l-lysine only or poly-l-lysine and OKT3 and then stained with anti-Myc antibody. Both fragments were detected in the cytoplasmic region after incubation on poly-l-lysine–coated glass slides. After stimulation with OKT3, the EVH1 domain fragment was detected in the nucleus, whereas the CCD fragment formed cytoplasmic aggregates. Representative images were presented from experiments performed at least 3 times. Bars, 10 μm.
Nuclear translocation of EVH1 domain fragment in Jurkat cells stimulated with anti-CD3 antibody. Jurkat cells were electroporated with Myc construct expressing EVH1 domain or coiled-coil domain (CCD) fragment of Homer-3. Twenty-four hours later, the cells were incubated for 10 minutes at 37°Con glass slides coated with poly-l-lysine only or poly-l-lysine and OKT3 and then stained with anti-Myc antibody. Both fragments were detected in the cytoplasmic region after incubation on poly-l-lysine–coated glass slides. After stimulation with OKT3, the EVH1 domain fragment was detected in the nucleus, whereas the CCD fragment formed cytoplasmic aggregates. Representative images were presented from experiments performed at least 3 times. Bars, 10 μm.
To quantify the nuclear translocation of Homer-3 following TCR stimulation, the nuclear fractions were isolated from Jurkat cells and probed for nuclear proteins by Western blot analysis. Consistent with published reports, p65 accumulated in the nucleus following TCR stimulation (Figure 4). As shown in Figure 4, the amount of Homer-3 was also increased in the nucleus 10 minutes after TCR stimulation and then reduced gradually (Figure 4).
Accumulation of Homer-3 in the nucleus following TCR stimulation. Jurkat cells were stimulated with OKT3 (5 μg/mL) for 0, 10, 60, or 240 minutes at 37°C, and then nuclear extract was obtained as described in “Materials and methods.” The nuclear extract (15 μg of total proteins per well) was resolved on a 7.5% SDS-PAGE gel and blotted with anti-p65 antibody or anti–Homer-3 antibody. As already known, p65 accumulated in the nucleus after TCR stimulation. The amount of Homer-3 was also increased in the nucleus at 10 minutes after TCR stimulation and then decreased gradually. * indicates nonspecific bands with anti-p65 antibody.
Accumulation of Homer-3 in the nucleus following TCR stimulation. Jurkat cells were stimulated with OKT3 (5 μg/mL) for 0, 10, 60, or 240 minutes at 37°C, and then nuclear extract was obtained as described in “Materials and methods.” The nuclear extract (15 μg of total proteins per well) was resolved on a 7.5% SDS-PAGE gel and blotted with anti-p65 antibody or anti–Homer-3 antibody. As already known, p65 accumulated in the nucleus after TCR stimulation. The amount of Homer-3 was also increased in the nucleus at 10 minutes after TCR stimulation and then decreased gradually. * indicates nonspecific bands with anti-p65 antibody.
Homer-3 regulates SRE activation in Jurkat cells
EVH1 domain proteins have been shown to be important in regulating early signal transduction events, modulating membrane actin dynamics, and actin-based motility in various models.6,25 In neuronal synaptic signaling, the Homer family of proteins have been noted to function as immediate early genes.14,22 Several reports have shown that within minutes of TCR ligation, a network of signaling molecules enhances the transcription of immediate early genes, including c-fos.46-49 Transcription of c-fos depends on the SRE in its promoter.48 Since subcellular distribution of Homer-3 is dynamically regulated during the early phase of T-cell activation and SRE-dependent reporter assay is a well-characterized system to monitor immediate early gene activation, we examined its potential involvement in SRE-mediated transcriptional activation.
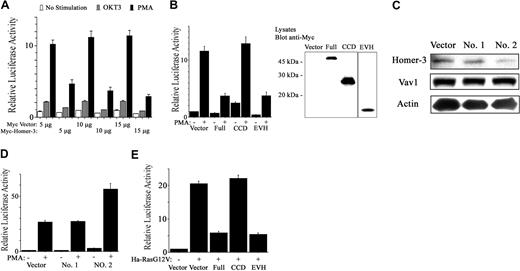
TCR activation or PMA was used to induce SRE-luciferase expression in the presence or absence of overexpressed forms of Homer-3 (Figure 5A). Overexpression of full-length Homer-3 reduced SRE activation induced by OKT3 and especially by PMA (Figure 5A). To identify the domain responsible for the reduction of SRE activation, Jurkat cells were electroporated with expression vectors containing fragments of Homer-3. Overexpression of the EVH1 domain fragment reduced SRE activation induced by PMA, although this fragment was less abundantly expressed than the other fragments as measured by Western blotting (Figure 5B). Reduction of SRE activation was not due to increased cell death after electroporation, since each experimental population showed no increase in annexin V and 7-AAD staining (data not shown).
Homer-3 regulates SRE activation in Jurkat cells. (A) Jurkat cells were electroporated with SRE luciferase reporter (1 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing full-length Homer-3 (5, 10, or 15 μg). Twenty hours later the cells were stimulated with OKT3 (5 μg/mL) or PMA (50 ng/mL) for an additional 4 hours. Luciferase activity was determined as described in “Materials and methods.” Overexpression of Homer-3 reduced SRE activation induced by OKT3 and especially by PMA. Three independent experiments were performed with duplicate samples. Histograms represent the mean ± SE (A-B, D-E). (B) Jurkat cells were electroporated with SRE luciferase reporter (1 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing each fragment of Homer-3 (15 μg). Twenty hours later the cells were stimulated with PMA (50 ng/mL) for an additional 4 hours. Luciferase activity was determined as described in “Materials and methods.” The expression of each fragment was confirmed with Western blotting. Overexpression of the EVH1 fragment reduced SRE activation induced by PMA, although the expression level of EVH1 fragment was much less than that of the other fragments. (C) Jurkat cells were electroporated with constructs expressing SiRNA directed against Homer-3 (no. 1, nucleotides 218-236 of coding sequence; no. 2, nucleotides 138-156; 10 μg). After 72 hours' incubation, the cells were lysed and blotted for endogenous Homer-3 expression. Only SiRNA construct of Homer-3 no. 2 suppressed Homer-3 expression, whereas expression level of Vav1 and actin was not affected. (D) To assess SRE activation under knockdown of Homer-3, Jurkat cells were electroporated with SRE luciferase reporter (2 μg), GFP construct (0.2 μg), and SiRNA constructs (10 μg). After 68 hours' incubation, the cells were stimulated with PMA (50 ng/mL) for an additional 4 hours. Luciferase activity was determined as described in “Materials and methods.” SRE activation was elevated only after electroporation of SiRNA construct of Homer-3 no. 2. (E) Jurkat cells were electroporated with SRE luciferase reporter (1 μg), GFP-Ha-RasG12V, or empty construct (2.5 μg) and Myc construct either empty or expressing each fragment of Homer-3 (15 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of full-length or EVH1 domain fragments of Homer-3 reduced SRE activation induced by Ha-RasG12V. Immunoblotting confirmed expression of GFP-Ha-RasG14V and each fragment of Homer-3 (data not shown).
Homer-3 regulates SRE activation in Jurkat cells. (A) Jurkat cells were electroporated with SRE luciferase reporter (1 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing full-length Homer-3 (5, 10, or 15 μg). Twenty hours later the cells were stimulated with OKT3 (5 μg/mL) or PMA (50 ng/mL) for an additional 4 hours. Luciferase activity was determined as described in “Materials and methods.” Overexpression of Homer-3 reduced SRE activation induced by OKT3 and especially by PMA. Three independent experiments were performed with duplicate samples. Histograms represent the mean ± SE (A-B, D-E). (B) Jurkat cells were electroporated with SRE luciferase reporter (1 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing each fragment of Homer-3 (15 μg). Twenty hours later the cells were stimulated with PMA (50 ng/mL) for an additional 4 hours. Luciferase activity was determined as described in “Materials and methods.” The expression of each fragment was confirmed with Western blotting. Overexpression of the EVH1 fragment reduced SRE activation induced by PMA, although the expression level of EVH1 fragment was much less than that of the other fragments. (C) Jurkat cells were electroporated with constructs expressing SiRNA directed against Homer-3 (no. 1, nucleotides 218-236 of coding sequence; no. 2, nucleotides 138-156; 10 μg). After 72 hours' incubation, the cells were lysed and blotted for endogenous Homer-3 expression. Only SiRNA construct of Homer-3 no. 2 suppressed Homer-3 expression, whereas expression level of Vav1 and actin was not affected. (D) To assess SRE activation under knockdown of Homer-3, Jurkat cells were electroporated with SRE luciferase reporter (2 μg), GFP construct (0.2 μg), and SiRNA constructs (10 μg). After 68 hours' incubation, the cells were stimulated with PMA (50 ng/mL) for an additional 4 hours. Luciferase activity was determined as described in “Materials and methods.” SRE activation was elevated only after electroporation of SiRNA construct of Homer-3 no. 2. (E) Jurkat cells were electroporated with SRE luciferase reporter (1 μg), GFP-Ha-RasG12V, or empty construct (2.5 μg) and Myc construct either empty or expressing each fragment of Homer-3 (15 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of full-length or EVH1 domain fragments of Homer-3 reduced SRE activation induced by Ha-RasG12V. Immunoblotting confirmed expression of GFP-Ha-RasG14V and each fragment of Homer-3 (data not shown).
To further examine the involvement of Homer-3 in SRE activation, SRE-induced luciferase activity was determined following SiRNA-mediated knockdown of endogenous Homer-3 expression. These experiments relied upon transfection of the RNA polymerase III–dependent H1 promoter constructs, in which the H1 promoter drives transcription of a homologous hairpin RNA, as described.43 Two constructs targeting different sequences of the Homer-3 mRNA were tested. One of the 2, SiRNA construct no. 2, substantially decreased endogenous Homer-3 protein expression within 3 days after electroporation (Figure 5C). In the SRE activation assay, electroporation of the SiRNA construct no. 2 resulted in increased SRE activation relative to the knockdown-defective construct no. 1 (Figure 5D). Taken together, these observations suggest that Homer-3 is involved in down-regulation of SRE activation induced by PMA stimulation in T cells.
It has been previously shown that phorbol esters activate the Ras pathway, leading to SRE activation.50-52 Furthermore, PMA-induced activation of the SRE was partially inhibited by dominant-negative c-Raf, extracellular signal-related kinase 1 (ERK1), or ERK2, and also by dominant-negative mitogen-activated kinase kinase 1 (MEKK1), but was not inhibited by dominant-negative RhoA.53 Thus, we investigated the involvement of Homer-3 in Ras-induced SRE activation. Constitutively active Ha-RasG12V increased SRE activation in Jurkat cells (Figure 5E). This activation decreased in the presence of overexpressed Homer-3 (Figure 5E). The EVH1 domain fragment also reduced Ras-induced SRE activation (Figure 5E).
Homer-3 binds to C/EBPβ and suppresses its activity
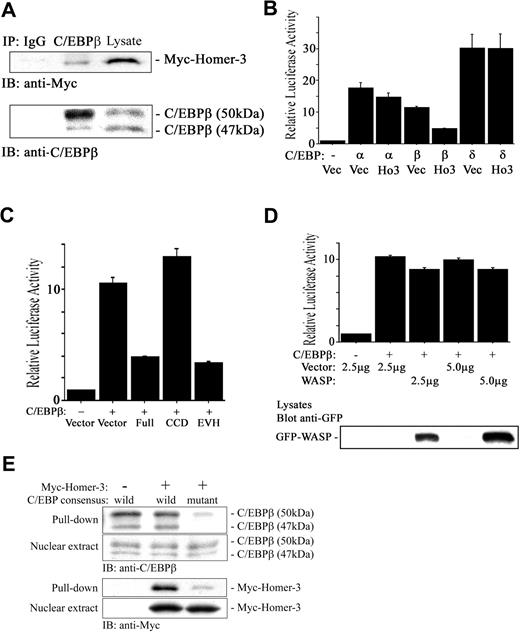
The SRE interacts with several transcription factors including the serum response factor (SRF), Ets-like gene-1 (Elk-1), and C/EBPβ.54-57 SRF and C/EBPβ interact in vivo through the DNA binding domain of SRF and the C-terminus of C/EBPβ.52 Therefore, like Elk-1, C/EBPβ may be recruited to the SRE not only by binding to the DNA, which is not a high-affinity site, but also by protein-protein interaction with SRF. The association of SRF and C/EBPβ is dramatically stimulated by activated Ras.52 Given that our database analysis identified C/EBPβ as having a consensus motif binding to Homer family EVH1 domain (PPxxFr; r is lysine in human, and arginine in mouse and rat C/EBPβ) in the N-terminal region, which functions as a transcriptional activator58 (Figure 6 underlined), and that overexpression of Homer-3 did not interfere with phospholyration of Elk-1 induced by PMA (data not shown), we hypothesized that Homer-3 binds to C/EBPβ and inhibits its transcriptional activation. Myc-tagged Homer-3 coprecipitated with endogenous C/EBPβ, suggesting an interaction between the 2 proteins (Figure 7A, middle lane). To observe the effect of Homer-3 overexpression on C/EBPβ activation, we determined Keratin 10 gene (k10)–promoter–dependent luciferase activity, which is specifically induced by members of the C/EBP family.38 Keratin 10 is expressed early during the differentiation of keratinocytes and is essential for epidermal barrier function.59 A previous report has demonstrated that the k10 promoter contains 3 functional C/EBP binding sites and that C/EBPα and β transactivate a k10 promoter–luciferase reporter.38 Overexpression of Homer-3 reduced C/EBPβ activation most effectively among C/EBP isoforms (Figure 7B). Overexpression of the Homer-3 EVH1 domain fragment also reduced the activity induced by C/EBPβ (Figure 7C), strongly suggesting another important functional role for the EVH1 domain in transcriptional activation. On the other hand, WASP, a well-known EVH domain–containing protein, did not attenuate the activation induced by C/EBPβ (Figure 7D).
Alignment of human C/EBP family. C/EBPβ has a consensus motif binding to Homer family EVH1 domain (PPXXFr, bold and underlined; r is lysine in human, and arginine in mouse and rat C/EBPβ). * indicates amino acid residue preserved among isoforms.
Alignment of human C/EBP family. C/EBPβ has a consensus motif binding to Homer family EVH1 domain (PPXXFr, bold and underlined; r is lysine in human, and arginine in mouse and rat C/EBPβ). * indicates amino acid residue preserved among isoforms.
Homer-3 binds to C/EBPβ and suppresses its transcriptional activity. (A) Jurkat cells were electroporated with Myc construct expressing full-length Homer-3. Twenty-four hours later, the cells were stimulated with PMA (50 ng/mL) for 10 minutes, and then nuclear extract was obtained as described in “Materials and methods.” The nuclear extract was incubated with 2 μg rabbit IgG (left lane) or anti-C/EBPβ antibody (middle lane) overnight, and the immune complex was collected with protein A–agarose beads. The immune complex and nuclear extract (right lane) were resolved on a 7.5% SDS-PAGE gel and blotted with anti-Myc antibody. Immunoblotting with mouse anti-C/EBPβ antibody confirmed that rabbit anti-C/EBPβ antibody precipitated full-length C/EBPβ (50 kDa), which contains the consensus motif binding to the EVH1 domain. (B) Jurkat cells were electroporated with k10 promoter–dependent luciferase reporter (2 μg), each isoform of C/EBP or empty construct (5 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing full-length Homer-3 (2.5 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of Homer-3 reduced k10 promoter–dependent luciferase activity induced with C/EBPβ. Three independent experiments were performed with duplicate samples. Histograms present the mean ± SE (B-D). (C) Jurkat cells were electroporated with k10 promoter–dependent luciferase reporter (2 μg), C/EBPβ, or empty construct (5 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing each fragment of Homer-3 (2.5 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of full-length or EVH1 domain fragments of Homer-3 reduced k10 promoter–dependent luciferase activity induced by C/EBPβ. (D) Jurkat cells were electroporated with k10 promoter–dependent luciferase reporter (2 μg), C/EBPβ, or empty construct (5 μg), and GFP-WASP or empty construct (2.5 or 5 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of WASP did not affect C/EBPβ-induced luciferase activity compared with that of Homer-3. WASP expression was confirmed with Western blotting. (E) Pull-down assay using C/EBP consensus oligonucleotide agarose conjugates. Jurkat cells were electroporated with Myc construct either empty (left lane) or expressing full-length Homer-3 (middle and right lanes). Twenty-four hours later, the cells were stimulated with PMA (50 ng/mL) for 10 minutes and then nuclear extract was obtained as described in “Materials and methods.” The nuclear extract was incubated with wild-type (left and middle lane) or mutant (right lane) C/EBP consensus oligonucleotide agarose conjugates. The complex was used for Western blotting.
Homer-3 binds to C/EBPβ and suppresses its transcriptional activity. (A) Jurkat cells were electroporated with Myc construct expressing full-length Homer-3. Twenty-four hours later, the cells were stimulated with PMA (50 ng/mL) for 10 minutes, and then nuclear extract was obtained as described in “Materials and methods.” The nuclear extract was incubated with 2 μg rabbit IgG (left lane) or anti-C/EBPβ antibody (middle lane) overnight, and the immune complex was collected with protein A–agarose beads. The immune complex and nuclear extract (right lane) were resolved on a 7.5% SDS-PAGE gel and blotted with anti-Myc antibody. Immunoblotting with mouse anti-C/EBPβ antibody confirmed that rabbit anti-C/EBPβ antibody precipitated full-length C/EBPβ (50 kDa), which contains the consensus motif binding to the EVH1 domain. (B) Jurkat cells were electroporated with k10 promoter–dependent luciferase reporter (2 μg), each isoform of C/EBP or empty construct (5 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing full-length Homer-3 (2.5 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of Homer-3 reduced k10 promoter–dependent luciferase activity induced with C/EBPβ. Three independent experiments were performed with duplicate samples. Histograms present the mean ± SE (B-D). (C) Jurkat cells were electroporated with k10 promoter–dependent luciferase reporter (2 μg), C/EBPβ, or empty construct (5 μg), GFP construct (0.2 μg), and Myc construct either empty or expressing each fragment of Homer-3 (2.5 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of full-length or EVH1 domain fragments of Homer-3 reduced k10 promoter–dependent luciferase activity induced by C/EBPβ. (D) Jurkat cells were electroporated with k10 promoter–dependent luciferase reporter (2 μg), C/EBPβ, or empty construct (5 μg), and GFP-WASP or empty construct (2.5 or 5 μg). Twenty-four hours later, luciferase activity was determined as described in “Materials and methods.” Overexpression of WASP did not affect C/EBPβ-induced luciferase activity compared with that of Homer-3. WASP expression was confirmed with Western blotting. (E) Pull-down assay using C/EBP consensus oligonucleotide agarose conjugates. Jurkat cells were electroporated with Myc construct either empty (left lane) or expressing full-length Homer-3 (middle and right lanes). Twenty-four hours later, the cells were stimulated with PMA (50 ng/mL) for 10 minutes and then nuclear extract was obtained as described in “Materials and methods.” The nuclear extract was incubated with wild-type (left and middle lane) or mutant (right lane) C/EBP consensus oligonucleotide agarose conjugates. The complex was used for Western blotting.
Interaction of Homer-3 with C/EBPβ does not impair binding of C/EBPβ to the consensus sequence
To see if interaction of Homer-3 with C/EBPβ impairs binding of C/EBPβ to the consensus sequence, pull-down assay was performed after overexpression of Homer-3. C/EBP consensus oligonucleotide agarose conjugates pulled down endogenous C/EBPβ (Figure 7E, left lane), and its precipitation was not impaired even after overexpression of Homer-3 (Figure 7E, middle lane), suggesting that the interaction of Homer-3 with C/EBPβ did not impair binding of C/EBPβ to the consensus oligonucleotide. Furthermore, overexpressed Homer-3 was also pulled down (Figure 7E, middle lane), indicating that Homer-3 bound to the C/EBP consensus oligonucleotide (TGC AGA TTG CGC AAT CTG CA) with C/EBPβ. The mutation of the C/EBP consensus oligonucleotide (TGC AGA GAC TAG TCT CTG CA) significantly reduced the precipitation of C/EBPβ and Homer-3 (Figure 7E, right lane).
Discussion
In the present study, we have demonstrated a novel role for Homer 3 in early T-cell signaling. In resting T cells, Homer-3 is a cytosolic protein and, on TCR engagement, is recruited to the immunologic synapse. Furthermore, we have shown that Homer-3 is translocated into the nucleus following actin, Ezrin, and WASP accumulation at the immunologic synapse. The synaptic distribution depends on the CCD of Homer-3, which shares 40% to 45% homology with CCDs found in motor molecules such as myosin heavy chain, kinesin heavy chain, and dynactin.22 Although the specific mechanism that drives the distribution of Homer-3 has not been identified, the CCD could be involved in the recruitment to the immunologic synapse. Additional studies are in progress to examine the functional implications of Homer-3 at the immunologic synapse.
We also present the unanticipated findings that Homer-3 is translocated into the nucleus via its EVH1 domain and that the nuclear translocation of Homer-3 has functional implications in regulation of SRE activation. The SRE is known to be essential for the transcription of c-fos, a gene induced early in T-cell activation.48,60 SRE activation by OKT3 or PMA was inhibited by overexpression of Homer-3 in Jurkat cells. As predicted, the inhibition of SRE activation was also observed with overexpression of the EVH1 domain fragment. On the other hand, SRE activation was elevated when Homer-3 expression was down-regulated using SiRNA. The overexpression of full-length or EVH1 domain fragments of Homer-3 reduced SRE activation induced by constitutively active Ha-RasG12V, suggesting that Homer-3 may regulate SRE activation downstream of the Ras pathway via its EVH1 domain. Our database analysis has found that C/EBPβ has a consensus motif binding to Homer family EVH1 domain (PPxxFr; r is lysine in human, and arginine in mouse and rat C/EBPβ) in the N-terminal region, where the transactivation domain exists. Descombes and Schibler58 have demonstrated that C/EBPβ, which is called liver-enriched transcriptional activator protein in their paper, contains the basic DNA-binding domain and the leucine zipper dimerization helix in the C-terminal region and the transactivation domain in the N-terminal region. Hanlon et al52,61 have demonstrated that C/EBPβ transactivates the c-fos SRE downstream of the Ras pathway. We found that overexpressed Homer-3 coprecipitated with endogenous C/EBPβ and reduced its transcriptional activation through the EVH1 domain, whereas the interaction of Homer-3 with C/EBPβ did not impair binding of C/EBPβ to the consensus sequence. Taken together, these findings suggest a novel role for Homer-3 in T-cell activation.
Homer-3 is recruited to immunologic synapse and subsequently translocated to the nucleus. Homer-3, by regulating transcription, invites analogies with calmodulin-dependent serine protein kinase (CASK), a scaffolding protein in neuronal synaptic signaling, or beta catenin, a cadherin-binding protein that is associated with adherence junctions, which can also function as a nuclear mediator by binding T-box brain protein 1 (Tbr1) or T-cell factor/lymphoid enhancer factor (TCF/LEF) transcription factor, respectively.62,63 In addition to transcription factor regulation, synapse to nucleus translocation has been implicated in cell survival, circadian rhythm generation, and long-term memory.64 These studies further support the existence of shared cell biologic machinery between neuronal, junctional, and immunologic synapses.
Prepublished online as Blood First Edition Paper, November 26, 2003; DOI 10.1182/blood-2003-08-2671.
Supported by grants from the National Institutes of Health (NIH) and Crohn's Colitis Foundation of America (CCFA) (R.X.). K.I. was supported by the Fellowship of Japan Foundation for Aging and Health and Sankyo Foundation of Life Science.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Brian Seed, in whose laboratory these experiments were initiated, for insightful comments and generously sharing reagents. We also thank Nils Jacobson, Ian Rosenberg, and Daniel Podolsky for critical reading of the manuscript and Joe Avruch, Marco Lopez, Dan Billadeau, Joel Habener, Michael White, and Paul Worley for generous provision of research materials.

![Figure 1. Ezrin, WASP, and Homer-3 localization in Jurkat cells stimulated with anti-CD3/CD28 antibody–coated beads. (A) Antibody-coated bead T-cell conjugates demonstrated redistribution of endogenous Homer-3 following T-cell receptor activation. F-actin accumulation (phalloidin-Alexa Fluor 594, red) in the contact area was induced by stimulation with anti-CD3/CD28 antibody–coated beads for 10 minutes, whereas no increase in actin accumulation was present at 2 minutes in Jurkat cells. Ezrin (green) and WASP (green) were recruited to the contact area along with actin accumulation after 10 minute stimulations. In contrast, Homer-3 (green) was recruited to the contact area after 2 minutes' stimulation, where increased actin accumulation was not observed. After actin accumulation in the contact area, Homer-3 was translocated into the nucleus (DAPI [4,6 diamidino-2-phenylindole], blue). Representative images showed that neither recruitment to the contact area nor nuclear translocation of Homer-3 was observed in the cells stimulated with anti-CD19 antibody–coated beads. (B) Jurkat cells were electroporated with Myc construct expressing coiled-coil domain (CCD) or EVH1 domain fragment of Homer-3. Twenty-four hours later, the cells were stimulated with anti-CD3/CD28 antibody-coated beads for 2 minutes and then stained with anti-Myc antibody. The CCD fragment of Homer-3 was recruited to the contact area, whereas the EVH1 domain fragment was not. Representative images were presented from experiments performed at least 3 times. Original magnification × 1000.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/103/6/10.1182_blood-2003-08-2671/6/m_zh80060458570001.jpeg?Expires=1767773200&Signature=kisUBYi0zWS5Y9r5GM7ZbfvzZ5YFJCi20FT8h3UM0pckuFmvKTQAtsz5Njl6J4W6THtkqIn1jmIRTbz6hmI83E2JcOpo3Io1RQftx560UAkAHi9GsOHQ43NFBMlKGsnAMZYkVup0GrYXB8NDrLF2MpKTqdmMsSQahGd3zKwr2XkiY2oWj4~3C6C~f9w3zmL~pM-XDWA8UIW6sBLP3cM2uekY9AJSLddT3vfgu~IZjAaZGoEgFtZkw6iqIyMRurWjkqF31OisqXrTvVSNtHGdYFEJgaULMff5jDto1QgI-3Wnpen2EPXEjB0yLzu~ZV6JR0ZVd-FalAs7ecX4FAqXIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal