Abstract
Human leukocyte antigen (HLA) class I antigen defects may have a negative impact on the growing application of T-cell–based immunotherapeutic strategies for treatment of leukemia. Therefore in the present study, taking advantage of a large panel of HLA class I allele–specific human monoclonal antibodies, we have compared HLA class I antigen expression on leukemic cells with that on autologous and allogeneic normal cells. Down-regulation of HLA-A and/or -B allospecificities was present in the majority of the patients studied. However, down-regulation did not affect all HLA class I alleles uniformly, but was almost exclusively restricted to HLA-A allospecificities and to HLA-B allospecificities which belong to the HLA-Bw6 group. The latter allospecificities, at variance from those that belong to the HLA-Bw4 group, do not modulate the interactions of leukemic cells with natural killer (NK) cells. Therefore, our results suggest that the selective down-regulation of HLA-A and HLA-Bw6 allospecificities associated with HLA-Bw4 preservation provides leukemic cells with an escape mechanism not only from cytotoxic T lymphocytes (CTLs), but also from NK cells. As a result T-cell–based immunotherapeutic strategies for leukemia should utilize HLA-Bw4 alloantigens as restricting elements since a selective HLA-Bw4 allele loss would provide leukemic cells with an escape mechanism from CTLs, but would increase their susceptibility to NK cell–mediated lysis. (Blood. 2004;103:3122-3130)
Introduction
The classic human leukocyte antigen (HLA) class I molecules (HLA-A, -B, and -C) are expressed on the surface of most nucleated cells as a trimolecular complex composed of a 45-kDa polymorphic heavy chain (α-chain), a 12-kDa monomorphic light chain (β2-microglobulin), and a 9– to 11–amino acid long peptide. HLA class I antigens play an important role in the interaction of virus-infected and malignant cells with effector cells. They mediate recognition of target cells by cytotoxic T lymphocytes (CTLs) by presenting viral- and tumor antigen–derived 8- to 11–amino acid long peptides to the T-cell receptor.1 Furthermore, HLA class I antigens modulate activity of natural killer (NK) cells by interacting with inhibitory and activating killer immunoglobulin-like receptors expressed on NK cells.2
Analysis of a large number of cell lines and of surgically removed lesions has convincingly shown that defects in HLA class I antigen expression and/or function occur in almost every type of solid tumor, although the frequency of these abnormalities varies markedly among the various types of tumors (for review, see Garrido et al3 and Marincola et al4 ). Defects in HLA class I antigen expression have been known for some time to have clinical significance, since they are associated with the clinical course of the disease in many types of malignant diseases (for review, see Marincola et al4 ). Nevertheless, HLA class I defects in tumor cells have been of limited interest among tumor immunologists for many years. However, in recent years interest in the characterization of HLA class I antigen expression in malignant lesions and of the molecular mechanisms underlying HLA class I defects has been rekindled by the potential negative impact of HLA class I abnormalities on the outcome of T-cell–based immunotherapy. The latter is being applied with increasing frequency for the treatment of solid tumors thanks to the availability of well-defined tumor antigens to be used as immunogens in clinical trials.5,6
During the last few years numerous leukemia-specific translocations or overexpressed genes have been described both in myeloid and lymphoid leukemia cells.7 The resulting gene products may eventually be utilized to implement T-cell–based immunotherapy of leukemia, especially since a spontaneous or induced immune response to some of these moieties has already been described.8,9 These possibilities have stimulated interest in the characterization of HLA class I antigen expression by leukemic cells. Contrary to solid tumors, HLA class I antigen expression has been investigated only to a limited extent in hematologic malignancies such as in B-cell and Hodgkin lymphomas,10,11 chronic lymphoblastic leukemia (CLL),12 acute lymphoblastic leukemia (ALL), and acute myeloid leukemia (AML).13-15 Defects in HLA class antigen expression have been found only in a low percentage of the samples analyzed. Total HLA class I antigen loss has been associated with an aggressive clinical course of the disease in non-Hodgkin lymphoma16 and with an increased expansion of the leukemic clone at diagnosis in AML.17
The scanty information about HLA class I antigen expression has a negative impact on the selection and design of immunotherapeutic strategies for the treatment of leukemia. Therefore in the present study, taking advantage of a large panel of HLA class I allele–specific human monoclonal antibodies (mAbs), we have compared the level of HLA class I antigen expression on acute and chronic leukemic cells with that on autologous and allogeneic normal lymphocytes.
Materials and methods
Cell lines
Cultured human lymphoid cells K562 (HLA-A11,-A31,-B18,-B40), KARPAS-299 (HLA-A03,-A11,-B07,-B35), ML-2 (HLA-A02,-B44,-B51), MONO-MAC-6 (HLA-A03,-BO7,-B51), NALM-6 (HLA-A01,-A02,-B08,-B15), NB-4 (HLA-A11,-B35,-B40), RPMI-8402 (HLA-A01,-A29,-B07,-B38), and 697 (HLA-A02,-A25,-B07,-B15) were maintained in RPMI 1640 medium supplemented with 5% fetal calf serum, antibiotics, and l-glutamine at 37°C in a humidified 5% CO2 incubator.
Monoclonal and polyclonal antibodies
Anti-CD2 (clone leu 5b), anti-CD3 (clone leu 4), anti-CD5 (clone leu1), anti-CD16 (clone leu 11), anti-CD19 (clone leu 12), anti-CD45 (clone leukocyte), and anti-CD56 (clone leu19) mAbs were purchased from Becton Dickinson (Erembodegem, Belgium). Anti-CD13 (clone My7) and anti-CD33 (clone My9) mAbs were purchased from Analis (Namur, Belgium).
The mouse mAb W6/32,18 which recognizes a conformational determinant expressed on β2-microglobulin (β2m)–associated HLA-A, -B, and -C heavy chains, the mouse mAb LGIII-147.4.1,19 which recognizes a conformational determinant expressed on all β2m-associated HLA-A heavy chains except HLA-A9, the mouse mAb B1.23.2,20 which recognizes a conformational determinant expressed on all β2m-associated HLA-B and -C heavy chains, and the mouse anti-β2m mAb NAMB-121 were developed and characterized as described. The mouse anti–HLA-DR (clone HLA-DR) and anti–HLA-DR, -DQ, -DP (clone Tu-39) mAbs were purchased from Becton Dickinson.
Human mAbs (hu-mAbs) are secreted by Epstein Barr virus (EBV)–transformed B lymphocytes isolated from multiparous women who had developed HLA antigen–specific antibodies during pregnancy. HLA antibody–producing EBV cell lines were stabilized by electrofusion and rigorous cloning.22-25 The HLA specificity of mAbs was determined by testing with a large (n > 240) panel of HLA-typed peripheral blood mononuclear cell suspensions in the conventional complement-dependent microcytotoxicity (CDC) assay. For flow cytometry application, HLA hu-mAbs were used as undiluted hybridoma culture supernatants.
Fluorescein isothiocyanate (FITC)–conjugated F(ab1)2 fragments of goat antimouse immunoglobulin G (IgG) antibodies and FITC-conjugated F(ab1)2 fragments of rabbit antihuman Ig antibodies were purchased from Prosan (Merelbeke, Belgium).
Leukemia cells
EDTA (ethylenediaminetetraacetic acid) anticoagulated blood or bone marrow samples were obtained from 64 patients with leukemia at diagnosis. The only selection criterion was CD19+ and CD33+ membrane expression for lymphoid and myeloid leukemias, respectively. All patients' samples were obtained from the Clinical Hematology Department of the Academic Hospital of the Vrije Universiteit Brussel, and the Université Catholique de Louvain, Brussels, Belgium.
Control hematopoietic cells
Normal blood or bone marrow was obtained from blood donors and from allogeneic bone marrow donors at the time of collection. The ethics committee of the Academic Hospital-Vrije Universiteit Brussel (Brussels, Belgium) approved this study. These samples were used to isolate normal CD19+ B cells and normal CD33+ progenitors to serve as reference material for the flow cytometry studies. Mononuclear cells from peripheral blood and bone marrow were separated by Ficoll-Hypaque density gradient centrifugation (Lucron Bioproducts, De Pinte, Belgium) The isolated cells were further washed and diluted in phosphate-buffered saline (PBS) Facsflow (Becton Dickinson) supplemented with 0.5% bovine serum albumin (BSA).
Flow cytometry
Indirect immunofluorescence (IIF) staining of cells was performed as follows. Twenty-five microliters of a cell suspension (4 × 106 cells/mL PBS supplemented with 5% BSA) was incubated for 20 minutes at room temperature with 25 μL mAb preparation. Following 3 washings with PBS supplemented with 1% BSA, 500 μL FACSflow solution was added (Becton Dickinson) and samples were analyzed. Data were acquired, analyzed, and displayed by a Coulter Epics II–MCC cytometer using the Coulter System II software v 3.0 (Coulter, Miami, FL). The mean fluorescence intensity (MFI) from samples was noted and used for further analysis.
Magnetic cell sorting system (MACS)
The MACS and the microbeads were obtained from Miltenyi (Sanvertech, Boechout, Belgium) and applied as described by the manufacturer. In brief, a starting population of mononuclear cells was incubated for 15 minutes with the selected mouse anti-CD antibodies, as indicated further in this section. Cells were then washed in buffer and incubated for 15 minutes on ice in 100 μL of a 1:10 dilution of the antimouse IgG–coated magnetic microbeads.
The cell suspension was then applied to a MIDIMACS column in a magnetic collar. The column was washed thrice to obtain untouched cells. Bound cells (touched fraction) were recovered by removing the magnetic collar and passing 5 mL buffer through the column using a plunger.
Negative (untouched) isolated cell suspensions were utilized in all experiments to study HLA antigen expression. This strategy was utilized to exclude the possible interference of antibody-coated magnetic beads retained on cells in additional assays. Moreover, HLA antigen expression on leukemic cells and autologous normal cells could be studied under identical experimental conditions.
Therefore, 2 individual magnetic bead separations were performed on each peripheral blood and bone marrow sample to obtain untouched normal and leukemic cell fractions. Different microbeads were used depending on the tumor type.
To isolate leukemic B cells from CLL-B or ALL-B samples, the patients' normal cells were magnetically labeled by a cocktail of anti-CD2, -CD4, -CD11b, -CD16 and -CD36 mAbs. Unlabeled CD19+ B cells were recovered by washing. Patients' normal cells were obtained after the malignant B cells were retained on the column by anti-CD19 microbeads. In the case of myeloid tumors, patients' normal cells were isolated after tumor cells were bound to the column through anti-CD33 microbeads. Untouched CD33+ tumor cells were obtained by retaining normal cells on the column by a combination of anti-CD2 and anti-CD19 microbeads. The purity of isolated cells, as tested by flow cytometry, was always higher than 95%.
All touched, positive fractions (patients' leukemic and normal cells) were cryopreserved in complete medium containing 10% dimethyl sulfoxide (DMSO) for eventual DNA isolation purposes. An identical procedure was used to isolate normal B cells and normal T cells from peripheral blood and bone marrow from healthy donors. No attempt was made in this study to isolate the small fraction of NK cells (5%) out of the major T-cell fraction (95%). These cell preparations are called “normal cells” in the present study.
Purified, untouched cell preparations were preferentially used for flow cytometry studies. The remaining cell preparations were used for CDC assays and in part cryopreserved for staining experiments with HLA class I allele–specific mAb.
Serologic HLA typing
The HLA-A and HLA-B phenotypes were established on the different isolated cell fractions using the conventional CDC assay. Each typing plate contained at least 2 sera identifying individual HLA class I allospecificities. When weak or extra reactions were obtained, cell preparations were tested with additional commercially available typing plates (Biotest Seralc, Kortenberg, Belgium). All microcytotoxicity assays were supervised by the same experienced technician ensuring reproducibility and reliability of the assays during the study.
Molecular HLA typing
In all cases in which typing by serology identified homozygosity for HLA-A or -B antigens, molecular typing was performed utilizing the reverse hybridization technique (Innogenetics, Ghent, Belgium). DNA was isolated using a routine salting-out method (DNA E-Z Prepkit; Orchid Diagnostics Europe, St Katelijne Waver, Belgium).
In the cases in which total HLA class I antigen loss was identified on leukemic cells by CDC assay, DNA was extracted from cryopreserved normal and leukemic cells (touched fractions). A 2-kb polymerase chain reaction (PCR) amplicon spanning exons 1 to 5 from HLA-A and -B antigens was generated using a Gene Amp PCR 9700 (Applied Biosystems, Foster City, CA). The PCR product was cleaned up using Centricon YM-100 filters (Millipore, Brussels, Belgium) and visualized in agarose gel electrophoresis using ethidiumbromide. Sequencing reactions were performed separately for exons 2, 3, and 4 both in forward and reverse directions for each sample using Big Dye Terminator Chemistry (Applied Biosystems). Sample electrophoresis was done on an ABI Prism 310 sequencer. Sequencing reactions were analyzed using ABI Match Tools PPC v 1.0. Both the PCR amplification and sequencing reagents were purchased from Applied Biosystems.
Statistics
The nonparametric (2-sided) Mann-Whitney U test was used to calculate probabilities between groups. A P value less than or equal to .05 was considered significant.
Results
Specificity of human anti–HLA class I mAb
Preliminary experiments analyzed the specificity of the panel of human mAbs to be utilized in this study by testing with a panel of HLA class I–typed peripheral blood lymphocytes and cultured lymphoid cells. The results are shown separately for HLA locus A (Table 1) and locus B (Table 2) antigen-specific mAbs. The reactivity of mAbs with cells expressing the corresponding HLA class I allospecificity was 1- to 3-log stronger than that with negative controls in flow cytometry. Representative results are shown in Figure 1A-B. Overlapping profiles were not observed with any of the mAbs. Some mAbs recognize determinants expressed by more than one HLA class I allospecificity. For example, mAb SN66E3 recognizes a determinant shared by HLA-A2 and A28 allospecificities. In these cases, target cells were selected for expressing only one of these allospecificities. Other mAbs, such as clone VDK1D12, are restricted in the recognition of only one HLA class I allospecificity. The overall results showed that all mAbs, except mAb BR011F6, reacted as predicted by the HLA type of all the cells under study confirming their specificity. mAb BR011F6 did not recognize HLA-A1104. All mAbs did not stain the HLA class I–negative K562 cells.
Reactivity of HLA-A-specific humAbs with a panel of HLA-typed peripheral blood lymphocytes and cell lines
mAb . | Isotype . | Specificity . | PBLs . | Cell lines . | Flow cytometry . |
|---|---|---|---|---|---|
| VDK1D12 | IgM, κ | A1 | 10* | 2* | +++ |
| SN66E3 | IgM, κ | A2 | 22 | 3 | ++/+++ |
| SN66E3 | IgM, κ | A28 | 3 | 0 | +++ |
| OK2F3 | IgM, κ | A3 | 11 | 2 | +++ |
| GV5D1 | IgG1, λ | A1 | 10 | 2 | +++ |
| GV5D1 | IgG1, λ | A9 | 8 | 0 | +++ |
| NIE44B8 | IgM, κ | A10 | 4 | 1 | +/++ |
| BRO11F6 | IgG1, λ | A11 | 4δ | 3 | -/+++ |
| HDG6B6 | IgM, λ | A29, 31, 32, 33 (19) | 12 | 2 | + |
| SN230G6 | IgG1, λ | A2 | 22 | 3 | +++ |
| OK5A3 | IgM, λ | A3 | 8 | 2 | +++ |
| OK5A3 | IgM, λ | A11 | 3 | 3 | + |
| OK5A3 | IgM, λ | A1 | 10 | 2 | + |
| OK4F9 | IgM, κ | A1 | 8 | 2 | +++ |
| OK4F9 | IgM, κ | A3 | 7 | 2 | +++ |
| OK4F9 | IgM, κ | A11 | 3 | 3 | +++ |
| HDG2G7 | IgG1, κ | A29, 31, 32, 33 (19) | 16 | 2 | +++ |
| OK1C9 | IgM, λ | A3 | 11 | 2 | +++ |
| OK1C9 | IgM, λ | A11 | 3 | 2 | + |
| OK1C9 | IgM, λ | A33 (19) | 2 | 0 | + |
| OK1C9 | IgM, λ | A31 (19) | 2 | 1 | + |
mAb . | Isotype . | Specificity . | PBLs . | Cell lines . | Flow cytometry . |
|---|---|---|---|---|---|
| VDK1D12 | IgM, κ | A1 | 10* | 2* | +++ |
| SN66E3 | IgM, κ | A2 | 22 | 3 | ++/+++ |
| SN66E3 | IgM, κ | A28 | 3 | 0 | +++ |
| OK2F3 | IgM, κ | A3 | 11 | 2 | +++ |
| GV5D1 | IgG1, λ | A1 | 10 | 2 | +++ |
| GV5D1 | IgG1, λ | A9 | 8 | 0 | +++ |
| NIE44B8 | IgM, κ | A10 | 4 | 1 | +/++ |
| BRO11F6 | IgG1, λ | A11 | 4δ | 3 | -/+++ |
| HDG6B6 | IgM, λ | A29, 31, 32, 33 (19) | 12 | 2 | + |
| SN230G6 | IgG1, λ | A2 | 22 | 3 | +++ |
| OK5A3 | IgM, λ | A3 | 8 | 2 | +++ |
| OK5A3 | IgM, λ | A11 | 3 | 3 | + |
| OK5A3 | IgM, λ | A1 | 10 | 2 | + |
| OK4F9 | IgM, κ | A1 | 8 | 2 | +++ |
| OK4F9 | IgM, κ | A3 | 7 | 2 | +++ |
| OK4F9 | IgM, κ | A11 | 3 | 3 | +++ |
| HDG2G7 | IgG1, κ | A29, 31, 32, 33 (19) | 16 | 2 | +++ |
| OK1C9 | IgM, λ | A3 | 11 | 2 | +++ |
| OK1C9 | IgM, λ | A11 | 3 | 2 | + |
| OK1C9 | IgM, λ | A33 (19) | 2 | 0 | + |
| OK1C9 | IgM, λ | A31 (19) | 2 | 1 | + |
HLA-A allele–specific mAbs were tested with HLA class I–typed peripheral blood lymphocytes (PBLs) and cell lines in indirect immunofluorescence (IIF). Two isotype-matched controls were included in every experiment. With the exception of mAb BRO11F6, all mAbs stained all cell samples expressing the corresponding HLA class I allospecificity. Reactivity is expressed as 1 log (+), 2 logs (++), or 3 logs (+++) stronger than the negative controls used. - indicates negative reaction. Numbers in parentheses indicate broad specificity.
Number of samples tested. - indicates negative reaction. Numbers in parentheses indicate broad specificity.
mAb BRO11F6 stained only 2 of the 4 HLA-A 1104–positive PBL samples.
Reactivity of HLA-B-specific humAbs with a panel of HLA-typed lymphocytes and cell lines
mAb . | Isotype . | Specificity . | PBLs . | Cell lines . | Flow cytometry . |
|---|---|---|---|---|---|
| 13E12 | IgM, κ | B12 | 7* | 0* | +++ |
| GK31F12 | IgM, κ | B13 | 4 | 0 | +++ |
| HA2C10B12 | IgM, κ | B60 (40) | 7 | 2 | +++ |
| VTM3A1 | IgG1, κ | B7 | 7 | 4 | +++ |
| VTM4D9 | IgG1, κ | B7 | 7 | 4 | ++ |
| DMS4G2 | IgG1, λ | B62 (15) | 2 | 2 | ++/+++ |
| DMS2G2 | IgG1, λ | B35 | 7 | 2 | +++ |
| KAL3D5 | IgG1, λ | B51 (5) | 5 | 2 | +++ |
| HDG8D9 | IgG1, λ | B51 (5) | 5 | 2 | +++ |
| GR5B3 | IgM, λ | B62 (15) | 2 | 2 | ++ |
| BVK5B10 | IgM, κ | B8 | 7 | 1 | +++ |
| AE9D9 | IgM, λ | B8 | 7 | 1 | + |
| AE9D9 | IgM, λ | B14 | 3 | 0 | 0 |
| OK6H10 | IgM, κ | B15 | 1 | 2 | +++ |
| OK6H10 | IgM, κ | B35 | 7 | 2 | +++ |
| SN230G6 | IgG1, λ | B17 | 3 | 0 | +++ |
| KG30A7 | IgM, λ | B12 | 7 | 1 | ++ |
| KG30A7 | IgM, λ | B14 | 4 | 0 | +++ |
| GVK2F8 | IgM, λ | B18 | 4 | 1 | +++ |
| GVK2F8 | IgM, λ | B39 (16) | 3 | 0 | +++ |
| HDG2G7 | IgG1, κ | B57 (17) | 2 | 0 | +++ |
| OK6H10 | IgM, κ | B35 | 7 | 2 | +++ |
| OK6H10 | IgM, κ | B15 | 1 | 2 | +++ |
| OK6H10 | IgM, κ | B15 | 1 | 2 | +++ |
| OK6H10 | IgM, κ | B35 | 7 | 2 | +++ |
| FVS4G4 | IgM, κ | B35 | 7 | 2 | +++ |
| FVS4G4 | IgM, κ | B17 | 6 | 0 | + |
| FVS4G4 | IgM, κ | B62 (15) | 2 | 2 | +++ |
| FVS4G4 | IgM, κ | B51 (5) | 5 | 2 | +++ |
| FVS4G4 | IgM, κ | B14 | 5 | 0 | +++ |
| FVS4G4 | IgM, κ | B18 | 4 | 1 | ++/+++ |
| FVS4G4 | IgM, κ | B38 (16) | 3 | 1 | +++ |
mAb . | Isotype . | Specificity . | PBLs . | Cell lines . | Flow cytometry . |
|---|---|---|---|---|---|
| 13E12 | IgM, κ | B12 | 7* | 0* | +++ |
| GK31F12 | IgM, κ | B13 | 4 | 0 | +++ |
| HA2C10B12 | IgM, κ | B60 (40) | 7 | 2 | +++ |
| VTM3A1 | IgG1, κ | B7 | 7 | 4 | +++ |
| VTM4D9 | IgG1, κ | B7 | 7 | 4 | ++ |
| DMS4G2 | IgG1, λ | B62 (15) | 2 | 2 | ++/+++ |
| DMS2G2 | IgG1, λ | B35 | 7 | 2 | +++ |
| KAL3D5 | IgG1, λ | B51 (5) | 5 | 2 | +++ |
| HDG8D9 | IgG1, λ | B51 (5) | 5 | 2 | +++ |
| GR5B3 | IgM, λ | B62 (15) | 2 | 2 | ++ |
| BVK5B10 | IgM, κ | B8 | 7 | 1 | +++ |
| AE9D9 | IgM, λ | B8 | 7 | 1 | + |
| AE9D9 | IgM, λ | B14 | 3 | 0 | 0 |
| OK6H10 | IgM, κ | B15 | 1 | 2 | +++ |
| OK6H10 | IgM, κ | B35 | 7 | 2 | +++ |
| SN230G6 | IgG1, λ | B17 | 3 | 0 | +++ |
| KG30A7 | IgM, λ | B12 | 7 | 1 | ++ |
| KG30A7 | IgM, λ | B14 | 4 | 0 | +++ |
| GVK2F8 | IgM, λ | B18 | 4 | 1 | +++ |
| GVK2F8 | IgM, λ | B39 (16) | 3 | 0 | +++ |
| HDG2G7 | IgG1, κ | B57 (17) | 2 | 0 | +++ |
| OK6H10 | IgM, κ | B35 | 7 | 2 | +++ |
| OK6H10 | IgM, κ | B15 | 1 | 2 | +++ |
| OK6H10 | IgM, κ | B15 | 1 | 2 | +++ |
| OK6H10 | IgM, κ | B35 | 7 | 2 | +++ |
| FVS4G4 | IgM, κ | B35 | 7 | 2 | +++ |
| FVS4G4 | IgM, κ | B17 | 6 | 0 | + |
| FVS4G4 | IgM, κ | B62 (15) | 2 | 2 | +++ |
| FVS4G4 | IgM, κ | B51 (5) | 5 | 2 | +++ |
| FVS4G4 | IgM, κ | B14 | 5 | 0 | +++ |
| FVS4G4 | IgM, κ | B18 | 4 | 1 | ++/+++ |
| FVS4G4 | IgM, κ | B38 (16) | 3 | 1 | +++ |
HLA-B allele–specific mAbs were tested with HLA class I–typed peripheral blood lymphocytes (PBLs) and cell lines in indirect immunofluorescence (IIF). Two isotype-matched controls were included in every experiment. All mAbs stained all cell samples expressing the corresponding HLA class I allospecificity. Reactivity is expressed as 1 log (+), 2 logs (++), or 3 logs (+++) stronger than the negative controls used; 0 indicates no reaction. Numbers in parentheses indicate broad specificity.
Number of samples tested.
HLA antigen expression by cell lines. Flow cytometric analysis of NALM-6 (A) and MONO-MAC-6 (B) cell lines stained with HLA class I and class II antigen–specific mAbs (black). Irrelevant, isotype-matched mAbs were used as controls (white).
HLA antigen expression by cell lines. Flow cytometric analysis of NALM-6 (A) and MONO-MAC-6 (B) cell lines stained with HLA class I and class II antigen–specific mAbs (black). Irrelevant, isotype-matched mAbs were used as controls (white).
Changes in HLA class I antigen level on leukemic cells
In an initial study, the HLA class I antigen level on leukemic cells from 17 patients with CLL-B, 9 with ALL-B, and 20 with AML was analyzed by flow cytometry utilizing the anti–HLA-A, -B, -C mAb W6/32, the anti–HLA-A mAb LGIII-147.4.2, and the anti–HLA-B mAb B1.23.2. The expression level of HLA class I antigens on untouched leukemic B cells was compared with that on HLA-matched B cells from healthy donors. The total HLA class I antigen level, as measured with the mAb W6/32, was significantly lower (P = .0067) on CLL-B cells than on normal B cells (Table 3). A difference was also found when cells were stained with HLA locus–specific mAbs. However, this difference did not reach the level of statistical significance. The total HLA class II antigen level was also significantly lower (P = .0134) on malignant B cells than on normal B cells.
HLA antigen expression on CLL-B and ALL-B cells
. | MFI of normal B cells (SD) . | MFI of CLL-B cells (SD) . | P . | MFI of ALL-B cells (SD) . | P . |
|---|---|---|---|---|---|
| HLA-A, -B, -C | 87 (33) | 55 (39) | .0067 | 32 (24) | .0012 |
| HLA-A | 28 (8) | 22 (16) | NS | 16 (8) | .0002 |
| HLA-B | 33 (14) | 32 (14) | NS | 16 (10) | .005 |
| β2m | 69 (32) | 52 (37) | NS | 24 (15) | NS |
| HLA-DR, -DQ, -DP | 30 (7) | 17 (13) | .0134 | 22 (23) | NS |
| HLA-DR | 29 (2) | 19 (15) | .0452 | 26 (20) | NS |
. | MFI of normal B cells (SD) . | MFI of CLL-B cells (SD) . | P . | MFI of ALL-B cells (SD) . | P . |
|---|---|---|---|---|---|
| HLA-A, -B, -C | 87 (33) | 55 (39) | .0067 | 32 (24) | .0012 |
| HLA-A | 28 (8) | 22 (16) | NS | 16 (8) | .0002 |
| HLA-B | 33 (14) | 32 (14) | NS | 16 (10) | .005 |
| β2m | 69 (32) | 52 (37) | NS | 24 (15) | NS |
| HLA-DR, -DQ, -DP | 30 (7) | 17 (13) | .0134 | 22 (23) | NS |
| HLA-DR | 29 (2) | 19 (15) | .0452 | 26 (20) | NS |
CLL-B and ALL-B cells at diagnosis and peripheral blood B cells isolated from healthy donors were stained with anti-HLA mAbs and analyzed by flow cytometry. All cells tested were untouched fractions derived from immunomagnetic isolation procedures. Results are expressed as mean fluorescence intensity (MFI) values and standard deviation (SD). Results obtained with leukemic and normal B cells were compared using the nonparametric Mann-Whitney U test. Populations are as follows: MFI of normal B cells, n = 10; MFI of CLL-B cells, n = 17; and MFI of ALL-B cells, n = 9. NS indicates not significant.
The decrease in total HLA class I antigen level on ALL-B cells was even more pronounced than on CLL-B cells. In all individual cases (data not shown) a severe decrease of HLA-A and -B antigens was observed. Furthermore, in all CLL-B and ALL-B cases, malignant cells expressed β2m. On the other hand, no significant changes were noted for HLA class II antigen expression.
In contrast, the HLA class I antigen level on CD33+ AML cells was significantly increased when compared with HLA-matched CD33+ bone marrow–derived progenitors from healthy donors (Table 4). Also in this case, the difference between leukemic cells and normal counterparts did not reach the level of statistical significance when cells were stained with HLA locus–specific mAbs. In all AML cases, malignant cells expressed β2m. HLA class II antigen expression, and especially HLA-DR antigen expression, was increased significantly.
HLA antigen expression on bone marrow AML and CD33+ healthy donor progenitors
. | MFI of AML cells (SD) . | MFI of CD33+ normal progenitors (SD) . | P . |
|---|---|---|---|
| HLA-A, -B, -C | 53(32) | 15(11) | .0032 |
| HLA-A | 32(22) | 19(10) | NS |
| HLA-B | 41(26) | 26(15) | NS |
| β2m | 62(52) | 21(19) | .0119 |
| HLA-DR, -DQ, -DP | 28(31) | 7(2) | .0329 |
| HLA-DR | 33(31) | 6(2) | .0013 |
. | MFI of AML cells (SD) . | MFI of CD33+ normal progenitors (SD) . | P . |
|---|---|---|---|
| HLA-A, -B, -C | 53(32) | 15(11) | .0032 |
| HLA-A | 32(22) | 19(10) | NS |
| HLA-B | 41(26) | 26(15) | NS |
| β2m | 62(52) | 21(19) | .0119 |
| HLA-DR, -DQ, -DP | 28(31) | 7(2) | .0329 |
| HLA-DR | 33(31) | 6(2) | .0013 |
AML cells at diagnosis and HLA-matched bone marrow–derived CD33+ progenitors from healthy donors were stained with anti-HLA mAbs and analyzed by flow cytometry. All cells tested were untouched CD33+ cells obtained from immunomagnetic isolation procedures. Results obtained with AML cells and progenitors from healthy donors were compared using the nonparametric Mann-Whitney U test. Populations are as follows: MFI of AML cells, n = 20; and MFI of CD33+ normal progenitors, n = 10. NS indicates not significant.
Selective HLA class I antigen loss by leukemic cells
Immunomagnetically separated normal and leukemic cells from 37 leukemic samples were analyzed for HLA class I antigen expression by CDC assay (Table 5). Representative results obtained with 2 samples are shown in Figures 2 and 3. HLA class I antigen defects were found in 5 samples. Specifically, HLA-A locus antigen loss was found in one sample, loss of one HLA-A allospecificity in 2 samples, and loss of one HLA-B allospecificity in 2 samples.
Selective HLA class I antigen loss identified by testing leukemic cells with HLA class I antigen-typing sera in CDC assay
Case no. . | Leukemic subtype . | Sample type . | Normal cells . | . | . | . | Tumor cells . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | . | A . | A . | B . | B . | A . | A . | B . | B . | ||||||
| 1 | CLL | BL | 10 | 31 | 38 | 40 | NR* | NR* | 38 | 40 | ||||||
| 2 | AML | BL | 24 | 31 | 60 | H | 24 | 31 | 60 | H | ||||||
| 3 | CLL | BL | 2 | 3 | 7 | 51 | 2 | 3 | 7 | 51 | ||||||
| 4 | CLL | BL | 1 | 2 | 5 | 8 | 1 | 2 | 5 | 8 | ||||||
| 5 | CLL | BL | 3 | 10 | 35 | 44 | 3 | 10 | 35 | 44 | ||||||
| 6 | AML | BL | 24 | 32 | 39 | 57 | 24 | 32 | 39 | 57 | ||||||
| 7 | CLL | BL | 2 | 10 | 8 | 14 | 2 | 10 | NR* | 14 | ||||||
| 8 | ALL | BL | 2 | 3 | 60 | H | 2 | 3 | 60 | H | ||||||
| 9 | CLL | BL | 1 | 3 | 35 | 40 | 1 | 3 | 35 | 40 | ||||||
| 11 | AML | BL | 1 | 2 | 7 | 60 | 1 | 2 | 7 | 60 | ||||||
| 12 | CLL | BL | 3 | 23 | 7 | 44 | 3 | 23 | 7 | 44 | ||||||
| 13 | CLL | BL | 1 | 24 | 8 | 18 | 1 | 24 | 8 | 18 | ||||||
| 14 | CLL | BL | 24 | 32 | 35 | 51 | 24 | 32 | 35 | 51 | ||||||
| 15 | AML | BL | ND | ND | ND | ND | 3 | 11 | 35 | 38 | ||||||
| 16 | CLL | BL | 2 | 24 | 13 | 18 | 2 | 24 | 13 | 18 | ||||||
| 17 | ALL | BL | 1 | H | 5 | 7 | 1 | H | 5 | 7 | ||||||
| 18 | ALL | BL | 2 | 33 | 7 | 14 | 2 | 33 | 7 | 14 | ||||||
| 21 | ALL | BL | ND | ND | ND | ND | 10 | 28 | 52 | H | ||||||
| 23 | AML | BL | 2 | 32 | 8 | 17 | 2 | 32 | 8 | 17 | ||||||
| 26 | CLL | BL | 2 | H | 7 | 44 | 2 | H | 7 | 44 | ||||||
| 27 | CLL | BL | 2 | H | 8 | 44 | 2 | H | 8 | 44 | ||||||
| 28 | CLL | BL | 2 | 32 | 18 | 35 | 2 | 32 | 18 | 35 | ||||||
| 29 | AML | BL | 2 | 28 | 13 | 18 | 2 | NR* | 13 | 18 | ||||||
| 30 | AML | BL | 2 | H | 17 | 42 | 2 | H | 17 | 42 | ||||||
| 31 | CLL | BL | 11 | 33 | 35 | 57 | 11 | 33 | 35 | 57 | ||||||
| 32 | AML | BL | 2 | 3 | 39 | 51 | 2 | 3 | 39 | 51 | ||||||
| 33 | AML | BL | 2 | 23 | 35 | 44 | 2 | 23 | 35 | 44 | ||||||
| 34 | AML | BM | 1 | 2 | 13 | 60 | 1 | 2 | 13 | 60 | ||||||
| 38 | CLL | BL | 1 | 2 | 8 | 12 | NR* | 2 | 8 | 12 | ||||||
| 40 | AML | BL | 3 | 29 | 44 | 55 | 3 | 29 | 44 | 55 | ||||||
| 41 | CLL | BL | 3 | 29 | 51 | 60 | 3 | 29 | 51 | 60 | ||||||
| 42 | CLL | BL | 2 | 19 | 39 | H | 2 | 19 | 39 | H | ||||||
| 43 | AML | BL | ND | ND | ND | ND | 11 | 24 | 35 | 62 | ||||||
| 44 | AML | BL | 2 | 29 | 35 | 51 | 2 | 29 | NR* | 51 | ||||||
| 46 | AML | BL | ND | ND | ND | ND | 3 | 25 | 22 | 35 | ||||||
| 47 | CLL | BL | 1 | 3 | 7 | 8 | 1 | 3 | 7 | 8 | ||||||
| 48 | AML | BL | ND | ND | ND | ND | 1 | 2 | 8 | 60 | ||||||
Case no. . | Leukemic subtype . | Sample type . | Normal cells . | . | . | . | Tumor cells . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
. | . | . | A . | A . | B . | B . | A . | A . | B . | B . | ||||||
| 1 | CLL | BL | 10 | 31 | 38 | 40 | NR* | NR* | 38 | 40 | ||||||
| 2 | AML | BL | 24 | 31 | 60 | H | 24 | 31 | 60 | H | ||||||
| 3 | CLL | BL | 2 | 3 | 7 | 51 | 2 | 3 | 7 | 51 | ||||||
| 4 | CLL | BL | 1 | 2 | 5 | 8 | 1 | 2 | 5 | 8 | ||||||
| 5 | CLL | BL | 3 | 10 | 35 | 44 | 3 | 10 | 35 | 44 | ||||||
| 6 | AML | BL | 24 | 32 | 39 | 57 | 24 | 32 | 39 | 57 | ||||||
| 7 | CLL | BL | 2 | 10 | 8 | 14 | 2 | 10 | NR* | 14 | ||||||
| 8 | ALL | BL | 2 | 3 | 60 | H | 2 | 3 | 60 | H | ||||||
| 9 | CLL | BL | 1 | 3 | 35 | 40 | 1 | 3 | 35 | 40 | ||||||
| 11 | AML | BL | 1 | 2 | 7 | 60 | 1 | 2 | 7 | 60 | ||||||
| 12 | CLL | BL | 3 | 23 | 7 | 44 | 3 | 23 | 7 | 44 | ||||||
| 13 | CLL | BL | 1 | 24 | 8 | 18 | 1 | 24 | 8 | 18 | ||||||
| 14 | CLL | BL | 24 | 32 | 35 | 51 | 24 | 32 | 35 | 51 | ||||||
| 15 | AML | BL | ND | ND | ND | ND | 3 | 11 | 35 | 38 | ||||||
| 16 | CLL | BL | 2 | 24 | 13 | 18 | 2 | 24 | 13 | 18 | ||||||
| 17 | ALL | BL | 1 | H | 5 | 7 | 1 | H | 5 | 7 | ||||||
| 18 | ALL | BL | 2 | 33 | 7 | 14 | 2 | 33 | 7 | 14 | ||||||
| 21 | ALL | BL | ND | ND | ND | ND | 10 | 28 | 52 | H | ||||||
| 23 | AML | BL | 2 | 32 | 8 | 17 | 2 | 32 | 8 | 17 | ||||||
| 26 | CLL | BL | 2 | H | 7 | 44 | 2 | H | 7 | 44 | ||||||
| 27 | CLL | BL | 2 | H | 8 | 44 | 2 | H | 8 | 44 | ||||||
| 28 | CLL | BL | 2 | 32 | 18 | 35 | 2 | 32 | 18 | 35 | ||||||
| 29 | AML | BL | 2 | 28 | 13 | 18 | 2 | NR* | 13 | 18 | ||||||
| 30 | AML | BL | 2 | H | 17 | 42 | 2 | H | 17 | 42 | ||||||
| 31 | CLL | BL | 11 | 33 | 35 | 57 | 11 | 33 | 35 | 57 | ||||||
| 32 | AML | BL | 2 | 3 | 39 | 51 | 2 | 3 | 39 | 51 | ||||||
| 33 | AML | BL | 2 | 23 | 35 | 44 | 2 | 23 | 35 | 44 | ||||||
| 34 | AML | BM | 1 | 2 | 13 | 60 | 1 | 2 | 13 | 60 | ||||||
| 38 | CLL | BL | 1 | 2 | 8 | 12 | NR* | 2 | 8 | 12 | ||||||
| 40 | AML | BL | 3 | 29 | 44 | 55 | 3 | 29 | 44 | 55 | ||||||
| 41 | CLL | BL | 3 | 29 | 51 | 60 | 3 | 29 | 51 | 60 | ||||||
| 42 | CLL | BL | 2 | 19 | 39 | H | 2 | 19 | 39 | H | ||||||
| 43 | AML | BL | ND | ND | ND | ND | 11 | 24 | 35 | 62 | ||||||
| 44 | AML | BL | 2 | 29 | 35 | 51 | 2 | 29 | NR* | 51 | ||||||
| 46 | AML | BL | ND | ND | ND | ND | 3 | 25 | 22 | 35 | ||||||
| 47 | CLL | BL | 1 | 3 | 7 | 8 | 1 | 3 | 7 | 8 | ||||||
| 48 | AML | BL | ND | ND | ND | ND | 1 | 2 | 8 | 60 | ||||||
Untouched leukemic cells and autologous lymphocytes were isolated using immunomagnetic procedure. HLA class I allospecificity expression was assessed by testing cells with HLA class I-typing sera in complement-dependent microcytotoxicity (CDC) assay.
H indicates a homozygous sample; ND, not done or not enough normal cells available; and NR, no reaction with the respective anti–HLA class I antisera.
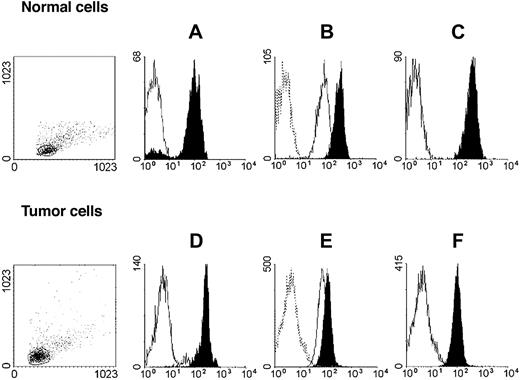
HLA class I antigen expression by immunomaganetic isolated lymphoid leukemic and autologous normal lymphocytes. Untouched autologous normal cells (top row) were stained with anti-CD3 (black) and anti-CD19 (white) mAbs (A); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (B); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (C). Untouched leukemic cells from a patient with CLL-B (bottom row) were stained with anti-CD3 (white) and anti-CD19 (black) mAbs (D); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (E); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (F). Forward/side scatter dot plots of the isolated cell populations are shown on the left.
HLA class I antigen expression by immunomaganetic isolated lymphoid leukemic and autologous normal lymphocytes. Untouched autologous normal cells (top row) were stained with anti-CD3 (black) and anti-CD19 (white) mAbs (A); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (B); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (C). Untouched leukemic cells from a patient with CLL-B (bottom row) were stained with anti-CD3 (white) and anti-CD19 (black) mAbs (D); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (E); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (F). Forward/side scatter dot plots of the isolated cell populations are shown on the left.
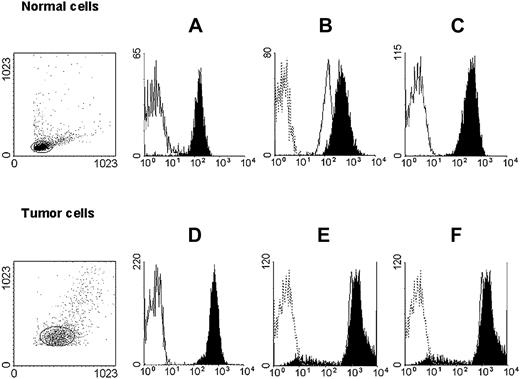
HLA class I antigen expression by immunomagnetic isolated myeloid leukemic and autologous normal myeloid cells. Untouched autologous normal cells (top row) were stained with anti-CD3 (black) and anti-CD33 (white) mAbs (A); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (B); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (C). Untouched leukemic cells from a patient with AML (bottom row) were stained with anti-CD3 (white) and anti-CD33 (black) mAbs (D); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (E); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (F). Forward/side scatter dot plots of the isolated cell populations are shown on the left.
HLA class I antigen expression by immunomagnetic isolated myeloid leukemic and autologous normal myeloid cells. Untouched autologous normal cells (top row) were stained with anti-CD3 (black) and anti-CD33 (white) mAbs (A); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (B); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (C). Untouched leukemic cells from a patient with AML (bottom row) were stained with anti-CD3 (white) and anti-CD33 (black) mAbs (D); anti–HLA-A mAb LGIII-147.4.2 (open, solid line), anti–HLA-B mAb B1.23.2 (black), and control mAb (open, dotted line) (E); and with anti–HLA-A, -B, -C mAb W6/32 (black) and control mAb (white) (F). Forward/side scatter dot plots of the isolated cell populations are shown on the left.
To determine whether the detection of only one HLA-B allospecificity in case 21 reflected homozygosity at the HLA-B locus or loss of one HLA-B allospecificity, molecular typing of leukemic cells was performed. This sample was found to be homozygous at the HLA-B locus. In 5 samples, too few normal cells could be isolated to perform serologic HLA typing.
Confirmation of HLA-A and -B allelic losses on leukemic cells by flow cytometry
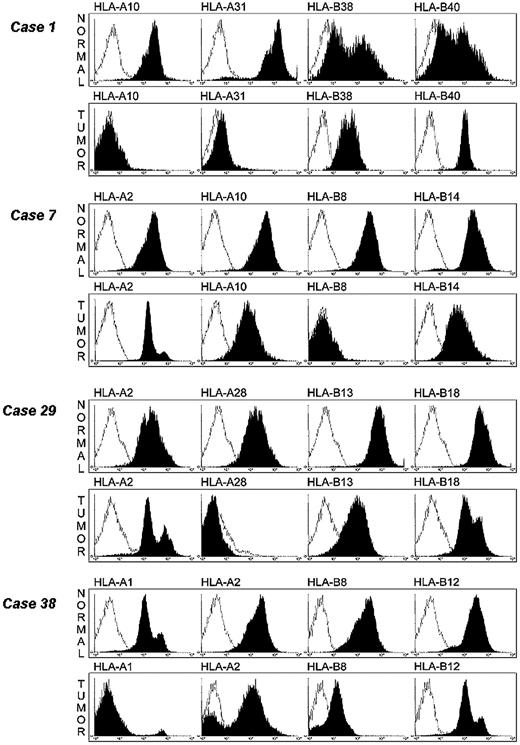
The availability of HLA class I allele–specific mAbs allowed us to study the HLA antigen expression on leukemic cells by flow cytometry. Cryopreserved normal and leukemic cells were available from 4 samples in which single HLA class I allele loss had been detected by CDC assay. mAbs recognizing all the HLA-A and -B allospecificities expressed in the 4 samples were available to us and were therefore utilized to stain leukemic cells isolated from the 4 samples. As shown in Figure 4, HLA-A10 and HLA-A31 alleles were not detected in sample 1 and HLA-B8, HLA-A28, and HLA-A1 alleles were not detected in samples 7, 29, and 38, respectively. The contour plots are identical to those from the negative isotype controls. Furthermore, HLA-B8 allele was barely detectable by flow cytometry on leukemic cells from sample 38, although it was easily detectable by CDC assay. This finding illustrates the high sensitivity of CDC assay for HLA typing and that the latter methodology is useful to detect total and selective HLA class I losses, but is not useful to measure HLA class I antigen down-regulation. HLA-B8 allele expression is approximately 10 times lower (1 log) on leukemic cells than on autologous normal cells. It is noteworthy that in this sample HLA-A2 allospecificity was not detectable on a small fraction of leukemic cells. This finding may reflect the emergence of a variant leukemic clone with HLA-A2 antigen loss. Total HLA class I antigen losses were not detected on patients' normal cells, emphasizing the association of HLA class I antigen defects with malignant transformation of cells.
Detection of selective HLA class I allelic losses on leukemic cells by flow cytometry analysis. Leukemic and autologous normal cells were stained with HLA class I allele–specific mAb (black) and analyzed by flow cytometry. Cells stained with irrelevant isotype-matched mAb were used as controls (white).
Detection of selective HLA class I allelic losses on leukemic cells by flow cytometry analysis. Leukemic and autologous normal cells were stained with HLA class I allele–specific mAb (black) and analyzed by flow cytometry. Cells stained with irrelevant isotype-matched mAb were used as controls (white).
Characterization of mutations underlying selective HLA-A and -B allele losses in leukemic cells
To define the molecular mechanism(s) underlying HLA class I allele losses by leukemic cells, the HLA-A genes in samples 1, 29, and 38 and the HLA-B gene in sample 7 were cloned from leukemic and autologous normal cells. Exons 2, 3, and 4 of each gene were sequenced in forward and reverse direction. No mutation was detected in the 3 exons in the 4 samples analyzed.
HLA class I allele expression levels on leukemic cells
Additional experiments took advantage of the panel of HLA class I allele–specific human mAbs available to us to compare the expression level of HLA class I alleles on leukemic and autologous normal cells. In these experiments the level of HLA class I alleles on leukemic cells was not compared with that on allogeneic normal lymphoid cells to avoid the interference of individual genetically determined variability in HLA class I antigen expression with the interpretation of the results. The results obtained with leukemic and normal cells are expressed as MFI. They are presented separately for the gene products of HLA-A and -B loci in Table 6. A ratio R value was calculated utilizing the formula R = MFI leukemic cells/MFI T cells. An HLA class I allele was classified as down-regulated when the R value was lower than 1. T cells and not total lymphocytes were used for comparison since in preliminary studies total HLA class I antigen expression as well as individual HLA class I allele expression was found to be lower on T cells than on autologous B cells isolated from healthy donors. The mean MFI value obtained with B cells was approximately double that obtained with autologous T cells.
HLA-A and -B allospecificity expression on leukemic cells
. | . | HLA locus A . | . | . | . | HLA locus B . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Leukemic subtype . | HLA allele . | Normal T cells . | Malignant cells . | R . | HLA allele . | Normal T cells . | Malignant cells . | R . | ||||||
| 3 | CLL-B | A2 | 41.0 | 69.3 | 1.69 | B7 | 63.8 | 57.7 | 0.90* | ||||||
| 3 | CLL-B | A3 | 92.5 | 184.9 | 1.99 | B51 | 120.4 | 156.2 | 1.29 | ||||||
| 4 | CLL-B | A1 | 20.7 | 19.8 | 0.95* | B5 | ND | ND | — | ||||||
| 4 | CLL-B | A2 | 39.9 | 33.9 | 0.84* | B8 | 59.3 | 57.7 | 0.97* | ||||||
| 13 | CLL-B | A1 | 23.9 | 42.4 | 1.77 | B8 | 83.1 | 43.7 | 0.52* | ||||||
| 13 | CLL-B | A24 | 29.2 | 38.6 | 1.32 | B18 | 24.5 | 93.4 | 3.81 | ||||||
| 27 | CLL-B | A2 | 30 | 18.2 | 0.60* | B8 | 56.7 | 37.2 | 0.65* | ||||||
| 27 | CLL-B | H | — | — | — | B44 | ND | ND | — | ||||||
| 28 | CLL-B | A2 | 29.2 | 31.9 | 1.09 | B18 | 6.14 | 28 | 4.56 | ||||||
| 28 | CLL-B | A32 | ND | ND | — | B35 | 57.2 | 36 | 0.62* | ||||||
| 29 | AML | A2 | 21.1 | 14.1 | 0.66* | B13 | 10.5 | 10.7 | 1.01 | ||||||
| 29 | AML | A28 | 18.2 | 2.83 | 0.15* | B18 | 2.14 | 1.97 | 0.92* | ||||||
| 30 | AML | A2 | 16.8 | 10.9 | 0.64* | B17 | 27.9 | 16.7 | 0.59* | ||||||
| 30 | AML | H | — | — | — | B42 | ND | ND | — | ||||||
| 31 | CLL-B | A1 | 25.7 | 29.0 | 1.12 | B35 | 17.7 | 17.3 | 0.97* | ||||||
| 31 | CLL-B | A11 | 42.7 | 46.7 | 1.09 | B57 | 38.6 | 39.1 | 1.01 | ||||||
| 32 | AML | A2 | 29.1 | 29.6 | 1.01 | B39 | 4.9 | 7.1 | 1.44 | ||||||
| 32 | AML | A3 | 12.2 | 17.6 | 1.44 | B51 | 34.7 | 47.3 | 1.36 | ||||||
| 33 | AML | A2 | 7.6 | 13.9 | 1.82 | B35 | 14.9 | 25.8 | 1.73 | ||||||
| 33 | AML | H | — | — | — | H | — | — | — | ||||||
| 34 | AML | A1 | 57.3 | 78.1 | 1.36 | B13 | 44.6 | 58.7 | 1.31 | ||||||
| 34 | AML | A2 | 87.7 | 126.6 | 1.44 | B60 | 12.1 | 20.1 | 1.66 | ||||||
| 43 | AML | A3 | 6.46 | 7.33 | 1.13 | B35 | 14.8 | 4.63 | 0.31* | ||||||
| 43 | AML | A11 | 5.53 | 7.45 | 1.34 | B62 | 15.4 | 6.85 | 0.44* | ||||||
| 45 | AML | A1 | 19.2 | 5.98 | 0.31* | B7 | 11.8 | 8.33 | 0.70* | ||||||
| 45 | AML | A2 | 34.6 | 6.05 | 0.17* | B60 | 9.46 | 8.18 | 0.86* | ||||||
| 50 | ALL-B | A3 | 4.91 | 17.7 | 3.60 | B7 | 12.1 | 23.0 | 1.9 | ||||||
| 50 | ALL-B | A26 | 5.07 | 17.9 | 3.53 | B38 | ND | ND | — | ||||||
| 54 | CLL-B | A1 | 20.1 | 6.79 | 0.33* | B8 | 8.31 | 10.2 | 1.22 | ||||||
| 54 | CLL-B | A28 | 12.5 | 6.57 | 0.52* | B49 | ND | ND | — | ||||||
| 55 | CLL-B | A3 | 8.53 | 11.7 | 1.37 | B7 | 19.3 | 10.8 | 0.55* | ||||||
| 55 | CLL-B | A33 | ND | ND | — | B35 | 32.0 | 29.3 | 0.91* | ||||||
| 56 | CLL-B | A2 | 32.5 | 4.75 | 0.14* | B7 | 16.9 | 5.24 | 0.30* | ||||||
| 56 | CLL-B | A33 | ND | ND | — | B60 | 12.9 | 10.5 | 0.81* | ||||||
| 57 | AML | A3 | 7.05 | 7.89 | 1.11 | B35 | 5.84 | 7.94 | 1.35 | ||||||
| 57 | AML | A11 | 8.25 | 9.18 | 1.11 | B39 | ND | ND | — | ||||||
| 61 | CLL-B | A2 | 67.4 | 41.4 | 0.61* | B13 | 70.9 | 64.0 | 0.90* | ||||||
| 61 | CLL-B | A3 | 15.9 | 7.8 | 0.49* | B61 | ND | ND | — | ||||||
| 62 | CLL-B | A32 | 6.1 | 10.5 | 1.72 | B62 | 52.6 | 45.5 | 0.86* | ||||||
| 62 | CLL-B | H | — | — | — | H | — | — | — | ||||||
| 63 | CLL-B | A2 | 37.4 | 22.1 | 0.59* | B44 | 7.4 | 7.1 | 0.96* | ||||||
| 63 | CLL-B | A31 | 3.7 | 3.1 | 0.84* | B18 | 5.2 | 6.1 | 1.17 | ||||||
| 64 | CLL-B | A2 | 50.3 | 37.3 | 0.74* | B15 | 35.4 | 39.9 | 1.13 | ||||||
| 64 | CLL-B | A24 | 49.2 | 34.1 | 0.69* | H | — | — | — | ||||||
| 65 | CLL-B | A32 | 37.2 | 61.0 | 1.64 | B13 | 46.8 | 103.8 | 2.21 | ||||||
| 65 | CLL-B | A9 | 11 | 26.6 | 2.42 | B39 | 2.8 | 13.3 | 4.75 | ||||||
| 67 | CLL-B | A2 | 26.5 | 35.5 | 1.34 | B8 | 50.5 | 212.5 | 4.21 | ||||||
| 67 | CLL-B | A24 | 20.6 | 137.5 | 6.67 | B44 | 7.1 | 117.6 | 16.56 | ||||||
| 68 | CLL-B | A28 | 45.1 | 40.8 | 0.90* | B53 | ND | ND | — | ||||||
| 68 | CLL-B | A32 | 4.7 | 27.3 | 5.81 | B7 | 23.5 | 43.1 | 1.83 | ||||||
| 69 | CLL-B | A3 | 22.3 | 27.8 | 1.25 | B35 | 17.1 | 28.6 | 1.67 | ||||||
| 69 | CLL-B | A10 | 4.2 | 5.9 | 1.40 | B44 | 5.4 | 5.9 | 1.09 | ||||||
| 70 | CLL-B | A26 | 90.1 | 64.4 | 0.71* | B18 | ND | ND | — | ||||||
| 70 | CLL-B | H | — | — | — | B55 | ND | ND | — | ||||||
| 71 | CLL-B | A1 | 31.9 | 43.9 | 1.38 | B50 | 33.9 | 61.3 | 1.81 | ||||||
| 71 | CLL-B | A24 | 51.3 | 81.9 | 1.60 | B62 | 59.9 | 87.9 | 1.47 | ||||||
| 72 | CLL-B | A2 | 30.5 | 33.3 | 1.09 | B35 | 34.4 | 49.6 | 1.44 | ||||||
| 72 | CLL-B | A3 | 22.8 | 25.0 | 1.10 | B60 | 12.3 | 20.6 | 1.67 | ||||||
| 73 | CLL-B | A11 | 43.1 | 25.2 | 0.58* | B14 | 60.5 | 75.3 | 1.24 | ||||||
| 73 | CLL-B | A19 | 36.6 | 39.4 | 1.08 | B51 | 30.2 | 36.5 | 1.21 | ||||||
| 74 | CLL-B | A1 | 25.8 | 29.4 | 1.14 | B8 | 55.9 | 57.0 | 1.02 | ||||||
| 74 | CLL-B | A2 | 36.4 | 34.2 | 0.94* | B51 | 41.4 | 42.0 | 1.01 | ||||||
| 75 | CLL-B | A2 | 20.3 | 29.6 | 1.46 | B39 | 6.9 | 19.6 | 2.84 | ||||||
| 75 | CLL-B | A31 | 3.8 | 8.8 | 2.32 | H | — | — | — | ||||||
. | . | HLA locus A . | . | . | . | HLA locus B . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Leukemic subtype . | HLA allele . | Normal T cells . | Malignant cells . | R . | HLA allele . | Normal T cells . | Malignant cells . | R . | ||||||
| 3 | CLL-B | A2 | 41.0 | 69.3 | 1.69 | B7 | 63.8 | 57.7 | 0.90* | ||||||
| 3 | CLL-B | A3 | 92.5 | 184.9 | 1.99 | B51 | 120.4 | 156.2 | 1.29 | ||||||
| 4 | CLL-B | A1 | 20.7 | 19.8 | 0.95* | B5 | ND | ND | — | ||||||
| 4 | CLL-B | A2 | 39.9 | 33.9 | 0.84* | B8 | 59.3 | 57.7 | 0.97* | ||||||
| 13 | CLL-B | A1 | 23.9 | 42.4 | 1.77 | B8 | 83.1 | 43.7 | 0.52* | ||||||
| 13 | CLL-B | A24 | 29.2 | 38.6 | 1.32 | B18 | 24.5 | 93.4 | 3.81 | ||||||
| 27 | CLL-B | A2 | 30 | 18.2 | 0.60* | B8 | 56.7 | 37.2 | 0.65* | ||||||
| 27 | CLL-B | H | — | — | — | B44 | ND | ND | — | ||||||
| 28 | CLL-B | A2 | 29.2 | 31.9 | 1.09 | B18 | 6.14 | 28 | 4.56 | ||||||
| 28 | CLL-B | A32 | ND | ND | — | B35 | 57.2 | 36 | 0.62* | ||||||
| 29 | AML | A2 | 21.1 | 14.1 | 0.66* | B13 | 10.5 | 10.7 | 1.01 | ||||||
| 29 | AML | A28 | 18.2 | 2.83 | 0.15* | B18 | 2.14 | 1.97 | 0.92* | ||||||
| 30 | AML | A2 | 16.8 | 10.9 | 0.64* | B17 | 27.9 | 16.7 | 0.59* | ||||||
| 30 | AML | H | — | — | — | B42 | ND | ND | — | ||||||
| 31 | CLL-B | A1 | 25.7 | 29.0 | 1.12 | B35 | 17.7 | 17.3 | 0.97* | ||||||
| 31 | CLL-B | A11 | 42.7 | 46.7 | 1.09 | B57 | 38.6 | 39.1 | 1.01 | ||||||
| 32 | AML | A2 | 29.1 | 29.6 | 1.01 | B39 | 4.9 | 7.1 | 1.44 | ||||||
| 32 | AML | A3 | 12.2 | 17.6 | 1.44 | B51 | 34.7 | 47.3 | 1.36 | ||||||
| 33 | AML | A2 | 7.6 | 13.9 | 1.82 | B35 | 14.9 | 25.8 | 1.73 | ||||||
| 33 | AML | H | — | — | — | H | — | — | — | ||||||
| 34 | AML | A1 | 57.3 | 78.1 | 1.36 | B13 | 44.6 | 58.7 | 1.31 | ||||||
| 34 | AML | A2 | 87.7 | 126.6 | 1.44 | B60 | 12.1 | 20.1 | 1.66 | ||||||
| 43 | AML | A3 | 6.46 | 7.33 | 1.13 | B35 | 14.8 | 4.63 | 0.31* | ||||||
| 43 | AML | A11 | 5.53 | 7.45 | 1.34 | B62 | 15.4 | 6.85 | 0.44* | ||||||
| 45 | AML | A1 | 19.2 | 5.98 | 0.31* | B7 | 11.8 | 8.33 | 0.70* | ||||||
| 45 | AML | A2 | 34.6 | 6.05 | 0.17* | B60 | 9.46 | 8.18 | 0.86* | ||||||
| 50 | ALL-B | A3 | 4.91 | 17.7 | 3.60 | B7 | 12.1 | 23.0 | 1.9 | ||||||
| 50 | ALL-B | A26 | 5.07 | 17.9 | 3.53 | B38 | ND | ND | — | ||||||
| 54 | CLL-B | A1 | 20.1 | 6.79 | 0.33* | B8 | 8.31 | 10.2 | 1.22 | ||||||
| 54 | CLL-B | A28 | 12.5 | 6.57 | 0.52* | B49 | ND | ND | — | ||||||
| 55 | CLL-B | A3 | 8.53 | 11.7 | 1.37 | B7 | 19.3 | 10.8 | 0.55* | ||||||
| 55 | CLL-B | A33 | ND | ND | — | B35 | 32.0 | 29.3 | 0.91* | ||||||
| 56 | CLL-B | A2 | 32.5 | 4.75 | 0.14* | B7 | 16.9 | 5.24 | 0.30* | ||||||
| 56 | CLL-B | A33 | ND | ND | — | B60 | 12.9 | 10.5 | 0.81* | ||||||
| 57 | AML | A3 | 7.05 | 7.89 | 1.11 | B35 | 5.84 | 7.94 | 1.35 | ||||||
| 57 | AML | A11 | 8.25 | 9.18 | 1.11 | B39 | ND | ND | — | ||||||
| 61 | CLL-B | A2 | 67.4 | 41.4 | 0.61* | B13 | 70.9 | 64.0 | 0.90* | ||||||
| 61 | CLL-B | A3 | 15.9 | 7.8 | 0.49* | B61 | ND | ND | — | ||||||
| 62 | CLL-B | A32 | 6.1 | 10.5 | 1.72 | B62 | 52.6 | 45.5 | 0.86* | ||||||
| 62 | CLL-B | H | — | — | — | H | — | — | — | ||||||
| 63 | CLL-B | A2 | 37.4 | 22.1 | 0.59* | B44 | 7.4 | 7.1 | 0.96* | ||||||
| 63 | CLL-B | A31 | 3.7 | 3.1 | 0.84* | B18 | 5.2 | 6.1 | 1.17 | ||||||
| 64 | CLL-B | A2 | 50.3 | 37.3 | 0.74* | B15 | 35.4 | 39.9 | 1.13 | ||||||
| 64 | CLL-B | A24 | 49.2 | 34.1 | 0.69* | H | — | — | — | ||||||
| 65 | CLL-B | A32 | 37.2 | 61.0 | 1.64 | B13 | 46.8 | 103.8 | 2.21 | ||||||
| 65 | CLL-B | A9 | 11 | 26.6 | 2.42 | B39 | 2.8 | 13.3 | 4.75 | ||||||
| 67 | CLL-B | A2 | 26.5 | 35.5 | 1.34 | B8 | 50.5 | 212.5 | 4.21 | ||||||
| 67 | CLL-B | A24 | 20.6 | 137.5 | 6.67 | B44 | 7.1 | 117.6 | 16.56 | ||||||
| 68 | CLL-B | A28 | 45.1 | 40.8 | 0.90* | B53 | ND | ND | — | ||||||
| 68 | CLL-B | A32 | 4.7 | 27.3 | 5.81 | B7 | 23.5 | 43.1 | 1.83 | ||||||
| 69 | CLL-B | A3 | 22.3 | 27.8 | 1.25 | B35 | 17.1 | 28.6 | 1.67 | ||||||
| 69 | CLL-B | A10 | 4.2 | 5.9 | 1.40 | B44 | 5.4 | 5.9 | 1.09 | ||||||
| 70 | CLL-B | A26 | 90.1 | 64.4 | 0.71* | B18 | ND | ND | — | ||||||
| 70 | CLL-B | H | — | — | — | B55 | ND | ND | — | ||||||
| 71 | CLL-B | A1 | 31.9 | 43.9 | 1.38 | B50 | 33.9 | 61.3 | 1.81 | ||||||
| 71 | CLL-B | A24 | 51.3 | 81.9 | 1.60 | B62 | 59.9 | 87.9 | 1.47 | ||||||
| 72 | CLL-B | A2 | 30.5 | 33.3 | 1.09 | B35 | 34.4 | 49.6 | 1.44 | ||||||
| 72 | CLL-B | A3 | 22.8 | 25.0 | 1.10 | B60 | 12.3 | 20.6 | 1.67 | ||||||
| 73 | CLL-B | A11 | 43.1 | 25.2 | 0.58* | B14 | 60.5 | 75.3 | 1.24 | ||||||
| 73 | CLL-B | A19 | 36.6 | 39.4 | 1.08 | B51 | 30.2 | 36.5 | 1.21 | ||||||
| 74 | CLL-B | A1 | 25.8 | 29.4 | 1.14 | B8 | 55.9 | 57.0 | 1.02 | ||||||
| 74 | CLL-B | A2 | 36.4 | 34.2 | 0.94* | B51 | 41.4 | 42.0 | 1.01 | ||||||
| 75 | CLL-B | A2 | 20.3 | 29.6 | 1.46 | B39 | 6.9 | 19.6 | 2.84 | ||||||
| 75 | CLL-B | A31 | 3.8 | 8.8 | 2.32 | H | — | — | — | ||||||
HLA class I allelic expression levels were determined by flow cytometry on leukemic cells and on autologous normal T cells. All cells tested were untouched cell fractions obtained by immunomagnetic procedures. The results are expressed as MFI after background correction. The ratio R was derived from MFI malignant cells/MFI normal cells. An R value lower than 1 was considered as down-regulation of that particular allele.
H indicates homozygous; ND, not done or no specific antibody available; and —, not available or not appropriate.
An R value less than 1 was considered a down-regulation of that particular allele.
Of the 56 HLA-A and 50 HLA-B alleles analyzed in 32 samples, 20 (35%) and 19 (38%), respectively, were found to be down-regulated on leukemic cells. Combining the results obtained with HLA-A and -B alleles, one or more HLA class I alleles were found to be down-regulated in 21 (65%) of the 32 samples analyzed. This is likely to be an underestimated value, since the expression of some HLA-A and -B alleles present in the patients' HLA class I phenotype could not be investigated because of the lack of availability of mAbs with the appropriate specificity.
Restriction of HLA class I antigen down-regulation to HLA-Bw6 allospecificities
HLA class I allele expression on leukemic cells was further analyzed taking into account whether they belong to the HLA-Bw4 or HLA-Bw6 group. Out of the 36 HLA-B alleles belonging to the HLA-Bw6 group, 16 (44%) were down-regulated. In contrast, only 3 (14%) of the HLA-A and HLA-B alleles belonging to the HLA-Bw4 group were down-regulated. The difference in the frequency of down-regulation between HLA class I alleles belonging to the HLA-Bw6 group and the remaining HLA class I alleles is statistically significant (P < .03).
Down-regulation of HLA-Bw6 antigens, at variance from that of HLA-Bw4 antigens, does not affect the interaction of target cells with NK cells.26 Therefore, our results suggest that emerging tumor cells acquire an HLA class I antigen phenotype which allows them to escape not only from CTLs, but also from NK cells.
Discussion
The present study has analyzed for the first time HLA class I antigen expression on leukemic cells utilizing 3 types of probes. They include mouse mAbs, which recognize framework or locus-specific determinants of HLA class I antigens and a large panel of conventional HLA typing sera, and hu-mAbs, which recognize HLA class I alleles. Both CDC assay and flow cytometry have been used as assays. In agreement with the scanty information available in the literature,13,14 HLA class I antigen loss has been detected in a very low percentage of leukemic samples. This finding is at variance with what has been found in most every type of solid tumor analyzed.3,4 We believe that this difference is likely to reflect the shorter time between malignant transformation and diagnosis in leukemia than in solid tumors. A short time interval between the onset of leukemia and diagnosis may not be sufficient for cells to acquire mutations in the gene(s) involved in HLA class I antigen expression and for selective pressure to facilitate the expansion of malignant cells with HLA class I antigen abnormalities. In our study we could not detect any abnormality in the sequence of the polymorphic exons of the lost HLA-A and -B antigens. The other exons and introns of these antigens could not be analyzed because of the lack of sufficient DNA that could be isolated from each patient. Therefore we cannot exclude abnormalities in these regions of the lost HLA class I genes. However, the available information in the literature13,27 suggests that defects in regulatory mechanisms controlling HLA class I antigen expression and not structural abnormalities of HLA class I genes may be the underlying mechanism of HLA class I antigen loss in leukemic cells. Brouwer et al13 did not detect abnormalities in genes encoding HLA class I antigens lost by leukemic cells. Moreover, in their experiments, IFN-γ could restore HLA class I antigen expression by leukemic cells as well as T-cell recognition and lysis of the target cells. Furthermore, Real et al27 showed that expression of a lost HLA class I allospecificity could be restored by treating leukemic cells with cytokines. We could not test whether the lost HLA class I allospecificities could be restored in leukemic samples by cytokines, since additional blood samples could not be obtained from the patients with a complete HLA class I allelic loss. All of them died shortly after diagnosis. Whether this association is a fortuitous one or reflects an aggressive phenotype of leukemic cells with HLA class I antigen loss and/or an escape of leukemic cells from CTL recognition and destruction remains to be determined, since the number of patients with this HLA class I phenotype we have identified in our studies is too small to draw conclusions.
The availability of a large panel of hu-mAbs recognizing HLA class I allospecificities has allowed us to analyze for the first time the level of HLA class I alleles on leukemic cells. Comparison of the expression level of 116 HLA class I alleles on 32 leukemic samples and autologous normal cells has detected down-regulation of HLA-A and HLA-B alleles in 35% and 38%, respectively, of the samples analyzed. As a result, down-regulation of HLA-A and/or HLA-B allospecificities is present in the majority of the leukemic patients investigated. How these findings compare with the HLA phenotypes in solid tumors cannot be assessed at present, since the methodology available at present is not suitable to measure HLA class I antigen level in solid tumors.
In our study, down-regulation does not affect all HLA class I alleles with similar frequency, but appears to be preferential for HLA class I alleles expressing the HLA-Bw6 determinant. This finding is intriguing, because of the differential interaction of HLA-Bw4 and HLA-Bw6 alleles with KIR receptors expressed on NK cells. In contrast to HLA-C allospecificities, not all HLA-B alleles inhibit NK cell cytotoxicity. Only the HLA-B alleles that carry the HLA-Bw4 epitope protect target cells from NK cell attack. Therefore, it is tempting to speculate that the selective HLA-Bw6 down-regulation provides leukemic cells with an escape mechanism from CTL attack. If this interpretation is correct, immunotherapeutic strategies should use HLA-Bw4 alloantigens as restricting elements since escape of leukemic cells from CTLs, because of an HLA-Bw4 allele loss, would render them susceptible to NK cell–mediated lysis.
Prepublished online as Blood First Edition Paper, December 4, 2003; DOI 10.1182/blood-2003-07-2500.
Supported by a grant from the “Fonds voor Wetenschappelijk Onderzoek Vlaanderen “ (FWO-Vlaanderen; no. 1.5.157.99) and by Public Health Services grants CA37959, CA67108, and P30 CA16056 awarded by the National Cancer Institute, Department of Health and Human Services.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Mrs Brigitte Guns, Ms Charlene DeMont, Ms Celeste Ross, and Mr Tom Spence for preparation of the manuscript. We also wish to thank biomedical student Tina Lamberts for help in cytometry analysis, and Marrie Kardol, Marry Franke-Van Dijk, and Chantal Eijsink for monoclonal antibody preparation.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal