Abstract
Defects in red blood cell (RBC) membrane skeleton components cause hereditary spherocytosis (HS). Clinically, HS varies significantly even among individuals with identical gene defects, illustrating the profound effects of genetic background on disease severity. We exploited a new spontaneous mouse model, wan, which arose on the inbred C3H/HeJ strain, to identify quantitative trait loci (QTL) that modify the HS phenotype. Homozygous wan mice have severe HS due to a complete deficiency of erythroid band 3. A QTL analysis of RBC count, hemoglobin, hematocrit, mean corpuscular volume (MCV), and mean corpuscular hemoglobin content (MCHC) was performed in wan/wan mice from an F2 intercross between C3H/HeJ+/wan and CAST/Ei+/+ F1 hybrids. Hematologic and survival data from C3H, CAST/Ei F2 wan homozygotes support the hypothesis that genetic modifiers significantly influence the band-3 null HS phenotype. Significant QTL were identified for the MCV trait only, suggesting that RBC membrane characteristics are a target for modifier gene action. The most significant quantitative trait locus, Hsm1 (hereditary spherocytosis modifier 1), localizes to mouse Chromosome 12 and is dominant. The peak LOD score was obtained with a marker for Spnb1 encoding erythroid β-spectrin, an obvious candidate gene. (Blood. 2004;103: 3233-3240)
Introduction
Hereditary spherocytosis (HS) is the most common inherited hemolytic anemia in people of Northern European descent, occurring with a frequency of approximately 1 in 2000.1 HS is caused by defects in the red blood cell (RBC) membrane skeleton, a multiprotein structure located just beneath the lipid bilayer that imparts mechanical strength and elasticity to the RBC membrane. The major component of the membrane skeleton, spectrin, is present as tetramers of α- and β-subunits cross-linked into a 2-dimensional array by short actin filaments.1-3 There are 2 major interactions between membrane skeleton components and integral membrane proteins that attach the spectrin array to the plasma membrane: (1) band 3–spectrin–ankyrin–protein 4.2 and (2) protein 4.1–p55–glycophorin C linkages.4-8
The band 3–spectrin–ankyrin–protein 4.2 interactions are critical in the pathogenesis of HS. Defects in all of these proteins cause HS in humans and in mice.1,9,10 Recent estimates are that approximately 50% of the mutations causing HS in European populations are in ankyrin, 20% in band 3, 20% in β-spectrin, 5% in protein 4.2, and less than 5% in α-spectrin.1 Depending on the exact genetic defect, the hematologic phenotype varies from asymptomatic to life-threatening hemolysis requiring transfusion therapy and/or splenectomy. Secondary complications of HS include jaundice, gallstones, aplastic crises, and, more rarely, extramedullary hematopoietic masses and leg ulcers.1 In the ankyrin-deficient nb mouse, progressive ataxia due to Purkinje cell degeneration occurs.11 In other mouse models of HS, thrombosis and infarction are significant complicating factors.12
Many specific molecular defects that cause HS in humans have been described.1 Notably, as in other heritable syndromes, the clinical presentation varies significantly even among individuals with identical gene defects, illustrating the profound effects of genetic background on disease severity.10 Identifying genetic modifiers by linkage analysis in humans is often difficult, if not impossible, due to environmental influences, genetic diversity, and small population (linkage group) size. Inbred mice, however, provide powerful tools for complex trait analysis. Moreover, as concordance between quantitative trait loci (QTL) in the mouse and human has been demonstrated for several diseases, complex trait analysis in the mouse has significant biomedical relevance.13,14
Here, we describe a new spontaneous HS mutation in the mouse, wan, which arose on the C3H/HeJ (C3H) inbred strain, and identify QTL that modify the severity of the HS phenotype. Homozygous wan mice are severely anemic. A premature stop codon in the gene encoding erythroid band 3, Slc4a1 (solute carrier family 4 [anion exchanger], member 1; formerly Ae1, anion exchanger 1), results in complete deficiency of band 3 in homozygotes. In hybrid wan/wan newborns derived from F2 intercrosses between C3Hwan/+ and Mus musculus castaneus (CAST/Ei), marked phenotypic variation is observed, suggesting the presence of segregating modifier alleles. Analysis of these F2 intercross progeny failed to detect significant QTL using RBC count, hemoglobin (Hgb), hematocrit (Hct), or mean corpuscular hemoglobin content (MCHC) values as quantitative traits. However, using mean corpuscular volume (MCV), 2 significant QTL were identified, 1 on chromosome (Chr) 6 and 1 on Chr 12. The Chr 6 quantitative trait locus is very broad, spanning more than 40 cM (centimorgans), and likely represents several, closely linked QTL. The Chr 12 quantitative trait locus, designated Hsm1 (hereditary spherocytosis modifier 1), spans 12 cM. Notably, the peak LOD score was obtained using a marker for erythroid β-spectrin (Spnb1), a strong candidate gene.
Materials and methods
Mice
The autosomal recessive mutation wan arose on the C3H/HeJ (C3H) inbred strain at The Jackson Laboratory (Bar Harbor, ME). As all C3Hwan homozygotes die within 72 hours of birth, all studies were performed on fetuses and newborns unless otherwise noted. C3H+/wan mice appear overtly normal. The C3Hwan strain was initially maintained by intercrossing progeny-tested heterozygotes and, once molecularly identified, by intercrossing typed heterozygotes (see “Genotyping of progeny”). Mice were housed in humidity- and temperature-controlled rooms with a 12-hour light cycle and free access to acidified water and chow (NIH 5K52). All protocols were approved by The Jackson Laboratory Animal Care and Use Committee. The Jackson Laboratory is fully accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC).
Complete blood counts
Whole blood (∼ 30 μL) was collected from decapitated newborns in EDTA (ethylenediaminetetraacetic acid)–coated microhematocrit tubes, transferred to a clean eppendorf tube containing 1 μL 20% EDTA in murine phosphate-buffered saline (PBS; 10 mM NaCl, 155 mM KCl, 10 mM glucose, 1 mM MgCl2, 2.5 mM KHPO4, pH 7.4), and mixed. The additional anticoagulant is critical in preventing clot formation. Following dilution in murine PBS (1:10), complete blood counts were determined using an Advia 120 Multi-species whole blood analyzer (Bayer, Tarrytown, NY). Adult whole blood (∼ 275 μL) was drawn from the retro-orbital sinus through EDTA-coated microhematocrit tubes directly into an eppendorf tube containing 30 μL 20% EDTA in murine PBS and analyzed without dilution. For fetal blood counts, whole blood (∼ 5 μL) was collected and analyzed using a Coulter Counter Model ZBI (Coulter, Hialeah, FL) as previously described.15 Reticulocytes were counted manually after staining with new methylene blue (Sigma, St Louis, MO). Of note, we previously reported an apparent failure of reticulocytosis in wan/wan mice.16 However, this proved to be an artifact; new methylene blue will not stain reticulocytes in homozygous wan/wan blood containing excess EDTA. The protocols above preserve both RBC morphology and reticulocyte staining. Control reticulocytes (+/wan, +/+) do not show this EDTA sensitivity. The basis for the staining differences is unknown.
Electron microscopy
Whole blood was collected as described above and centrifuged 3 minutes at 3000g. The cells were washed in mouse PBS, fixed in 2% glutaraldehyde in cacodylate buffer (pH 7.4, 4°C), and examined using a Jeol JEM-100 Scanning electron microscope (Jeol USA, Peabody, MA) as previously described.17
Production of polyclonal band-3 antibodies
A band-3 cDNA fragment encoding amino acid 8 through amino acid 399 (encompassing nearly the entire cytoplasmic domain) was subcloned into the pGEX-2TK vector and transformed into DH5α Escherichia coli using standard techniques. The recombinant protein was expressed and purified as described.18 Antibody was generated in rabbits by Strategic Biosolutions (Hercules, CA). The antibody was designated anti-cdb3.
Analysis of red cell membrane skeleton proteins
Hemoglobin-depleted RBC ghosts were prepared as described.19 Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 4% Steck or 5% to 10% discontinuous Laemmli gels.20,21 Gels were stained with Coomassie blue or electrophoretically transferred to Hybond enhanced chemiluminescence (ECL) nitrocellulose membrane (Amersham International, Buckinghamshire, England) using a semidry transfer apparatus (Bio-Rad, Oxnard, CA). Immunoblots were probed with previously described rabbit polyclonal antibodies to spectrin,22 ankyrin,23 protein 4.1,24 and protein 4.225 or antipeptide antibodies recognizing amino acids 214 to 22826 or the C-terminal 12 amino acids of mouse band 3,27 and with the polyclonal anti-cdb3 described above. Antibody to human βIΣ2 spectrin28 was kindly provided by Dr Jon S. Morrow (Yale University, New Haven, CT). Bound antibody was detected with goat antirabbit horseradish peroxidase (HRP)–conjugated immunoglobulin G (Bio-Rad) and visualized using ECL detection reagents (Amersham International).
Northern blotting
Identification of the wan gene defect
Genomic DNA was prepared from tail samples (Gentra Systems, Minneapolis, MN) and used as a template for the polymerase chain reaction (PCR). Band 3–specific primers approximately 20 base pairs in length with approximately 50% GC composition were designed flanking each exon (primer sequences available upon request). The PCR was carried out in an MJ Research PTC-200 Thermocycler (MJ Research, Watertown, MA) for 40 cycles (94°C, 15 seconds; 55°C, 30 seconds; 72°C, 1 minute) with a 15-minute incubation at 72°C following the last cycle. The products were subcloned into the pCR 2.1 vector (Invitrogen, Carlsbad, CA) and sequenced using the automated dye termination technique (ABI Systems, Foster City, CA).
Genotyping of progeny
The wan mutation generates an additional BfaI site, allowing genotyping of progeny (see “Results”). Genomic DNA was prepared and amplified as described above. The oligonucleotides (5′ → 3′) GGCCTTTTCTAAGCCTAGCT and CCAGAAAGCCACCTATGCAG were used as forward and reverse primers, respectively. The PCR products were digested overnight and resolved on a 2% NuSieve gel (BioWhittaker Molecular Applications, Rockland, ME).
β-spectrin sequencing
Genomic DNA was obtained from The Jackson Laboratory DNA Resource (http://www.jax.org/dnares/index.html). The PCR was carried out as above using oligonucleotides flanking each exon of the mouse β-spectrin gene (sequences available upon request). The products were sequenced using the automated dye termination technique (ABI Systems).
QTL analyses
To identify QTL influencing the wan phenotype, an F2 intercross between C3Hwan/+ and wild-type Mus musculus castaneus (CAST/Ei) mice was established. Complete blood counts and genotyping of F2 progeny at birth were performed as described above. Simple sequence length polymorphic (sslp) markers spaced at 10- to 30-cM intervals throughout the genome were typed by the PCR using multiplexed fluorescent markers and ABI 3700 instrumentation. After identification of QTL, additional markers spaced at 2- to 9-cM intervals on Chr 12 were added. Primers were purchased from Research Genetics (MapPairs, Huntsville, AL).
Genome-wide scans were performed using RBC count, Hgb, Hct, MCV, and MCHC as quantitative traits. Results were analyzed by forward stepwise multiple regression analysis using sex and body weight as additive covariates.30 Genome-wide scans were performed using Pseudomarker software (source codes available at http://www.jax.org/staff/churchill/labsite/software/pseudomarker/index.html).31 Significance thresholds were determined by permutation analysis in which the regression analysis is performed on randomized genetic marker and phenotypic data.32 The significant LOD score is that giving a P value of .05, whereas suggestive is a P value of .10. Confidence intervals were determined by computing the region of the posterior density curve containing 95% of the total area as described by Sen and Churchill.31 A simultaneous search for pairs among QTL was performed as described.13,31 Pairwise genome scans can detect QTL that affect a phenotype by interacting with another quantitative trait locus (including those instances where neither quantitative trait locus alone reaches the significance threshold).
Results
Wan mice have severe HS
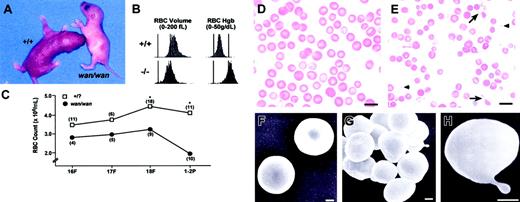
Homozygous wan mice are recognizable at birth by their extreme pallor (Figure 1A). Newborns are severely anemic (Table 1) and smaller than normal littermates. The RBC count, Hgb, Hct, MCV, and mean corpuscular hemoglobin (MCH) are significantly decreased compared with wild-type (+/+) littermates (Table 1). The MCHC is significantly increased. RBC volume and RBC hemoglobin concentration histograms reveal a significant population of microcytic and dehydrated cells (Figure 1B). Reticulocytes are increased nearly 2-fold (56.6 ± 8.0% [X ± SD] [n = 5] in wan homozygotes vs 30.6 ± 9.7% [n = 8] in +/+ littermates; P < .001). Male and female wan/wan mice do not differ significantly from each other in any of these hematologic parameters. Values for heterozygotes (+/wan) do not differ significantly compared with wild type except for the MCV, which is significantly lower (85.0 ± 4.0, n = 19, P < .001; see +/+ data in Table 1).
Hematologic status of C3H/HeJwan homozygotes. (A) A C3H/HeJwan/wan newborn with a wild-type (+/+) littermate. Note the extreme pallor of the wan homozygote. (B) RBC volume and hemoglobin concentration histograms of whole blood from normal (+/+) and wan/wan (-/-) newborn mice. Significant populations of small (left panels) and dehydrated (right panels) RBCs are present in mutant blood compared with controls. (C) RBC counts are decreased in wan/wan fetuses compared with normal littermates. The differences are significant (*P < .001) by fetal day 18 and at birth. (D-E) Wright-stained peripheral blood smear from a newborn C3H+/+ mouse (D) and a mutant C3Hwan/wan mouse (E). Many RBC fragments (arrowheads) and membrane projections (arrows) are evident in mutant blood smears. Bar = 10 μM. (F-H) Scanning electron photomicrographs of normal (F) and mutant (G-H) RBCs. Mutant RBCs are predominantly spherocytic (G) with frequent membrane projections (H). Bar = 1 μM.
Hematologic status of C3H/HeJwan homozygotes. (A) A C3H/HeJwan/wan newborn with a wild-type (+/+) littermate. Note the extreme pallor of the wan homozygote. (B) RBC volume and hemoglobin concentration histograms of whole blood from normal (+/+) and wan/wan (-/-) newborn mice. Significant populations of small (left panels) and dehydrated (right panels) RBCs are present in mutant blood compared with controls. (C) RBC counts are decreased in wan/wan fetuses compared with normal littermates. The differences are significant (*P < .001) by fetal day 18 and at birth. (D-E) Wright-stained peripheral blood smear from a newborn C3H+/+ mouse (D) and a mutant C3Hwan/wan mouse (E). Many RBC fragments (arrowheads) and membrane projections (arrows) are evident in mutant blood smears. Bar = 10 μM. (F-H) Scanning electron photomicrographs of normal (F) and mutant (G-H) RBCs. Mutant RBCs are predominantly spherocytic (G) with frequent membrane projections (H). Bar = 1 μM.
Hematologic parameters in newborn mice
Genotype . | n . | RBC count, × 1012/L . | Hgb, g/L . | Hct . | MCV, fL . | MCH, pg . | MCHC, g/dL . |
|---|---|---|---|---|---|---|---|
| c3H/HeJ+/+ | 9 | 3.6 ± 0.5 (2.9-4.4) | 111 ± 15 (90-140) | 0.332 ± 0.035 (0.27-0.38) | 91.5 ± 2.9 (87.1-96.0) | 30.1 ± 2.4 (27.0-33.6) | 32.9 ± 2.1 (29.4-36.3) |
| C3H/HeJwan/wan | 18 | 1.8 ± 0.5* (1.0-2.6) | 48 ± 15* (20-70) | 0.133 ± 0.042* (0.06-0.19) | 71.9 ± 6.0* (59.6-83.7) | 25.8 ± 3.4† (18.2-30.5) | 36.4 ± 3.5‡ (28.5-41.7) |
Genotype . | n . | RBC count, × 1012/L . | Hgb, g/L . | Hct . | MCV, fL . | MCH, pg . | MCHC, g/dL . |
|---|---|---|---|---|---|---|---|
| c3H/HeJ+/+ | 9 | 3.6 ± 0.5 (2.9-4.4) | 111 ± 15 (90-140) | 0.332 ± 0.035 (0.27-0.38) | 91.5 ± 2.9 (87.1-96.0) | 30.1 ± 2.4 (27.0-33.6) | 32.9 ± 2.1 (29.4-36.3) |
| C3H/HeJwan/wan | 18 | 1.8 ± 0.5* (1.0-2.6) | 48 ± 15* (20-70) | 0.133 ± 0.042* (0.06-0.19) | 71.9 ± 6.0* (59.6-83.7) | 25.8 ± 3.4† (18.2-30.5) | 36.4 ± 3.5‡ (28.5-41.7) |
Complete blood counts were obtained using an Advia 120 Multi-species hematology analyzer. All values are ± SD, and values in parentheses represent ranges.
P < .001.
P < .01.
P < .05 C3H/HeJwan/wan versus C3H/HeJ+/+ (2-tailed Student t-test).
Crosses between heterozygotes yield the expected Mendelian ratio of anemic offspring, indicating no significant fetal loss. However, anemia is present in utero. Decreased RBC counts are apparent by fetal day 16 (Figure 1C). By fetal day 18, RBC counts are significantly lower than in normal littermates. RBC counts decrease temporarily in normal neonates.15 In wan/wan mice, this drop is precipitous; newborn counts decrease to 50% of the levels seen on fetal day 18. In comparison, in normal neonates, the RBC count decreases much less, to 90% of its value on fetal day 18.
Examination of neonatal wan peripheral blood smears reveals marked anisocytosis and spherocytosis (Figure 1D-E). By scanning electron microscopy, wan RBCs are severely spherocytic and membrane protrusions are evident (Figure 1F-H). The hematologic data and RBC morphology are consistent with the phenotype of severe HS.
Band 3 and protein 4.2 are absent in wan red cell membrane skeletons
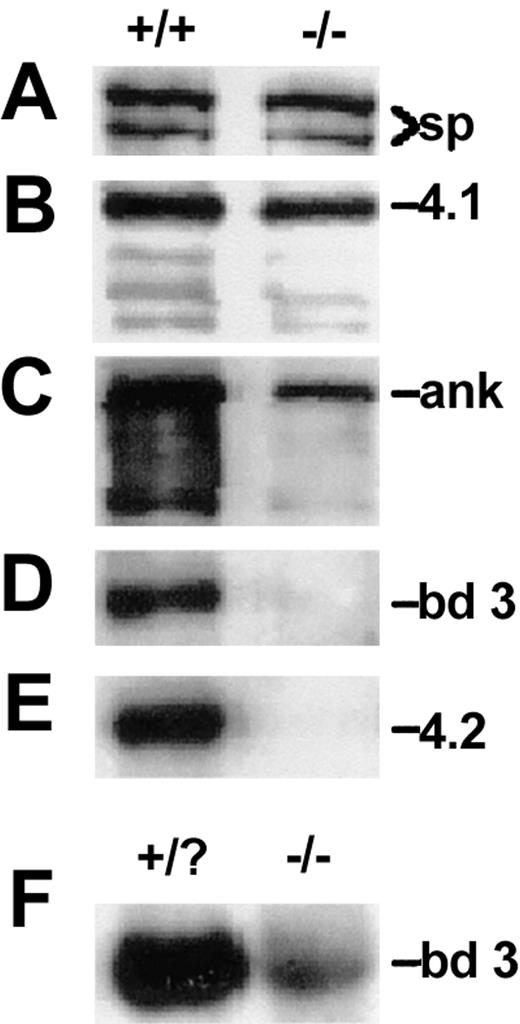
Analysis of RBC ghost membranes by SDS-PAGE and Western blotting reveals normal or near normal levels of spectrin and protein 4.1 in wan homozygotes (Figure 2A-B). Ankyrin is substantially decreased, whereas band 3 and protein 4.2 are absent (Figure 2C-E). As remarkably similar phenotypes were observed in 2 previously described targeted band-3 null mouse models,33,34 we intercrossed +/wan and targeted band-3 heterozygotes generated in our laboratory.34 The crosses yielded the expected Mendelian frequency of anemic offspring. Together, the data indicate that the wan gene defect lies within the erythroid band-3 gene. Consistent with these findings, wan reticulocyte band-3 mRNA levels were decreased to approximately 10% of normal (Figure 2F).
Membrane skeleton proteins in wan homozygotes. (A-E) Western blots showing levels of spectrin (sp, A), protein 4.1 (4.1, B), ankyrin (ank, C), band 3 (bd 3, D), and protein 4.2 (4.2, E) in normal (+/+) and wan/wan (-/-) RBC ghosts. Note the absence of band 3 and protein 4.2 and the decrement of ankyrin in mutant RBCs. In panel D, a peptide antibody raised to amino acids 214 to 228 of the cytoplasmic domain of band 3 was used. The same result was obtained using a pan-cytoplasmic domain band-3 antibody as well as a peptide antibody recognizing the C-terminal 12 amino acids of the membrane spanning domain, confirming that wan homozygotes are band-3 null. (F) Northern blot showing the severe decrement in band-3 mRNA in wan homozygotes. Each lane contains 2 μg total RNA purified from 2 pooled mutant (-/-) and 4 phenotypically normal (+/?) newborn peripheral blood samples whose genotypes were subsequently confirmed by PCR (see “Materials and methods”).
Membrane skeleton proteins in wan homozygotes. (A-E) Western blots showing levels of spectrin (sp, A), protein 4.1 (4.1, B), ankyrin (ank, C), band 3 (bd 3, D), and protein 4.2 (4.2, E) in normal (+/+) and wan/wan (-/-) RBC ghosts. Note the absence of band 3 and protein 4.2 and the decrement of ankyrin in mutant RBCs. In panel D, a peptide antibody raised to amino acids 214 to 228 of the cytoplasmic domain of band 3 was used. The same result was obtained using a pan-cytoplasmic domain band-3 antibody as well as a peptide antibody recognizing the C-terminal 12 amino acids of the membrane spanning domain, confirming that wan homozygotes are band-3 null. (F) Northern blot showing the severe decrement in band-3 mRNA in wan homozygotes. Each lane contains 2 μg total RNA purified from 2 pooled mutant (-/-) and 4 phenotypically normal (+/?) newborn peripheral blood samples whose genotypes were subsequently confirmed by PCR (see “Materials and methods”).
Sequencing reveals a premature stop codon in the wan Ae1 gene
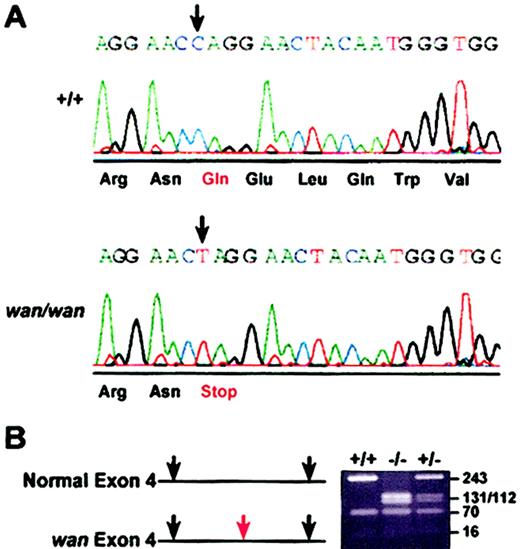
Band-3 exons from homozygous wan mice and C3H+/+ controls were amplified from genomic DNA, subcloned, and sequenced. A single base transition (C>T) at nucleotide 3998 (GenBank accession no. J02756) in wan was identified. The change introduces a premature stop codon (TAG) in place of glutamine (Gln) (CAG) at amino acid 85, which lies within the cytoplasmic domain of band 3, close to the N-terminus (Figure 3A). No other nucleotide changes in band-3 exons or splice junctions were found in C3Hwan versus C3H+/+ genomic DNA. The transition at nucleotide 3998 generates a BfaI restriction site, which was exploited for genotyping purposes (Figure 3B).
Premature stop codon in the wan band-3 gene. (A) Chromatogram showing the sequence of Slc4a1 encoding erythroid band-3 in control (+/+) and wan/wan genomic DNA. In wan homozygotes, a C>T transition (arrows) results in a premature stop codon at amino acid 85 within the cytoplasmic domain of band 3. (B) BfaI restriction digest of PCR-amplified products from control (+/+), homozygous (-/-), and heterozygous (+/-) mice resolved on a 2% NuSieve gel. The transition in the band-3 gene in wan creates an additional BfaI site (red arrow) that was exploited to genotype mice.
Premature stop codon in the wan band-3 gene. (A) Chromatogram showing the sequence of Slc4a1 encoding erythroid band-3 in control (+/+) and wan/wan genomic DNA. In wan homozygotes, a C>T transition (arrows) results in a premature stop codon at amino acid 85 within the cytoplasmic domain of band 3. (B) BfaI restriction digest of PCR-amplified products from control (+/+), homozygous (-/-), and heterozygous (+/-) mice resolved on a 2% NuSieve gel. The transition in the band-3 gene in wan creates an additional BfaI site (red arrow) that was exploited to genotype mice.
Evidence for segregating modifier genes in band-3 null mice
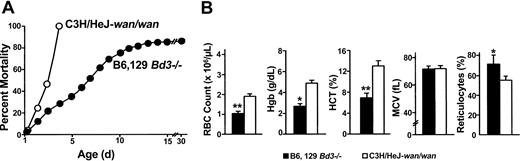
Band-3 null mice maintained on different genetic backgrounds show striking phenotypic differences. All homozygous C3Hwan inbred mice die within 72 hours of birth. In contrast, 75% of the knock-out band-3 null mice (B6,129 Bd3-/-) maintained on a hybrid C57BL/6J (B6) and 129/Sv (129) genetic background survive beyond 72 hours, with 15% surviving to adulthood (Figure 4A).34 Notably, more than 99% of B6 congenic Bd3-/- mice (n = 15) die in utero. This is remarkable given the molecular similarities of the 2 models. No band-3 protein is produced in red cells of either the knock-out or the spontaneous model, and the kidney transcript, which uses a separate start codon upstream of the wan premature stop codon,35 is disrupted in both. Both mutations result in severe anemia. Surprisingly, B6,129 Bd3-/- newborns appear to be more severely anemic than wan homozygotes despite their overall improved survival. RBC counts, Hgb, and Hct values are significantly lower and the reticulocyte percentage is significantly higher compared with wan/wan newborns (Figure 4B).
Phenotypic differences between band-3 null strains maintained on different genetic backgrounds. (A) Of inbred band-3 null C3H/HeJwan/wan mice, 100% die within 72 hours of birth (○), whereas a targeted strain of band-3 null mice maintained on a hybrid (B6, 129) genetic background show significantly less early mortality, with approximately 15% surviving to adulthood (•). (B) Hematologic analysis of newborn mice obtained using an Advia Multi-species whole-blood analyzer reveals severe anemia in both band-3 null strains, although significant differences are seen. *P < .01; **P < .001. Error bars indicate standard error.
Phenotypic differences between band-3 null strains maintained on different genetic backgrounds. (A) Of inbred band-3 null C3H/HeJwan/wan mice, 100% die within 72 hours of birth (○), whereas a targeted strain of band-3 null mice maintained on a hybrid (B6, 129) genetic background show significantly less early mortality, with approximately 15% surviving to adulthood (•). (B) Hematologic analysis of newborn mice obtained using an Advia Multi-species whole-blood analyzer reveals severe anemia in both band-3 null strains, although significant differences are seen. *P < .01; **P < .001. Error bars indicate standard error.
We also noted that some C3H, CAST/Ei F2 wan/wan newborns lacked significant pallor and could be identified with confidence only by genotyping. Moreover, some survived well beyond the 72-hour maximum observed for inbred C3H wan mice (up to 5 months). To examine this more closely, we allowed a subset of F1 mating pairs to give birth and rear their young undisturbed. Of 188 weaned (3-4 weeks of age) offspring, 10 wan/wan mice were identified, well below the number of wan homozygotes expected based on Mendelian inheritance of a recessive allele (25%). Assuming that homozygosity at the wan locus is the only genetically determined cause of mortality in all offspring prior to weaning, we can infer the number of wan/wan animals that died prior to weaning. Using the number of live +/+ (65) and +/wan (113) animals and the expected 25%:50%:25% ratio of +/+-to-+/wan-to-wan/wan F2 offspring, we predict that a total of 60 wan/wan mice were born. Hence, as in the band-3 knock-out model, a mixed genetic background markedly improves survivability in wan homozygotes, from 0% on C3H to 17%. Taken together, the observations of the various band-3 null strains indicate that genetic background profoundly influences the band-3 null HS phenotype in terms of survival and basic hematologic parameters.
Identification of genetic loci that modify the band-3 null phenotype
Complete blood counts were performed in a total of 670 C3H, CAST/Ei F2 wan/wan mice. Of these, 183 (27%) were homozygous wan with the expected ratio of males and females (53% female). As in the inbred C3Hwan strain, the RBC count, Hgb, Hct, MCV, and MCH are significantly decreased, and the MCHC is significantly increased, in homozygous C3H, CAST F2 wan mice compared with F2 wild-type mice (Table 2). Notably, F2 wan/wan newborns show an extremely broad range of values for all parameters, and these values significantly overlap the normal ranges seen in F2 wild-type mice (Table 2). The F2 hematologic data further confirm the profound effect of genetic background, indicating that the CAST/Ei parent is contributing an allele(s) that significantly influences the band-3 null HS phenotype.
Hematologic parameters in newborn C3H, CAST F2 mice
Genotype . | n . | RBC count, × 1012/L . | Hgb, g/L . | Hct . | MCV, fL . | MCH, pg . | MCHC, g/dL . |
|---|---|---|---|---|---|---|---|
| C3H, CAST+/+ | 148 | 3.7 ± 0.6 (2.2-5.4) | 111 ± 20 (64-160) | 0.347 ± 0.054 (0.2-0.52) | 95.1 ± 5.9 (80.7-111.2) | 30.2 ± 2.6 (23.7-38.4) | 31.8 ± 2.6 (25.0-37.5) |
| C3H, CAST-wan/wan | 183 | 2.2 ± 0.7* (0.7-5.2) | 62 ± 20* (16-130) | 0.174 ± 0.058* (0.048-0.4) | 79.9 ± 6.9* (62.3-95.3) | 28.2 ± 3.6* (17.8-36.4) | 35.3 ± 3.4* (25.0-46.2) |
Genotype . | n . | RBC count, × 1012/L . | Hgb, g/L . | Hct . | MCV, fL . | MCH, pg . | MCHC, g/dL . |
|---|---|---|---|---|---|---|---|
| C3H, CAST+/+ | 148 | 3.7 ± 0.6 (2.2-5.4) | 111 ± 20 (64-160) | 0.347 ± 0.054 (0.2-0.52) | 95.1 ± 5.9 (80.7-111.2) | 30.2 ± 2.6 (23.7-38.4) | 31.8 ± 2.6 (25.0-37.5) |
| C3H, CAST-wan/wan | 183 | 2.2 ± 0.7* (0.7-5.2) | 62 ± 20* (16-130) | 0.174 ± 0.058* (0.048-0.4) | 79.9 ± 6.9* (62.3-95.3) | 28.2 ± 3.6* (17.8-36.4) | 35.3 ± 3.4* (25.0-46.2) |
Complete blood counts were obtained using an Advia 120 Multi-species hematology analyzer. All values are ± SD, and values in parentheses represent ranges. *P <.001 (2-tailed Student t test).
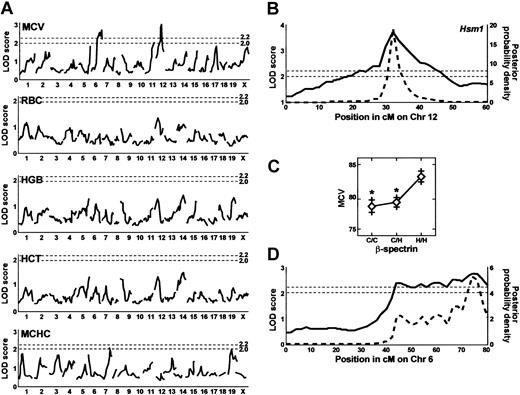
To identify modifiers of the wan phenotype, we performed genome-wide scans of F2 wan/wan mice using RBC count, Hgb, Hct, and MCV as quantitative traits with sex and weight as additive covariates. A total of 96 markers was genotyped. In an initial scan of 94 F2 animals, a single suggestive quantitative trait locus on Chr 12 was identified using MCV as trait (not shown). No other suggestive or significant QTL were identified using RBC count, Hgb, or Hct as traits. These data suggested that the QTL could influence the HS phenotype by modifying RBC membrane characteristics such as the extent of membrane vesiculation and/or the rate of cation and water loss, both of which would be expected to alter the MCV value. The marker D12Mit156, located 32 cM distal to the centromere, gave the peak LOD score. Notably, the gene encoding erythroid β-spectrin, Spnb1, maps to this region of mouse Chr 12.35 Spectrin could influence membrane characteristics in a number of ways and is an obvious candidate gene. Therefore, we generated additional F2 animals and added several polymorphic markers mapping to this region of Chr 12, including β-spectrin (see “Materials and methods”), and repeated the analyses using the same traits as above plus the MCHC value. Analysis of 183 F2 animals identified 2 significant QTL using MCV as trait—the previously suspected Chr 12 quantitative trait locus and a second quantitative trait locus on Chr 6 (Figure 5A). Again, no significant QTL were identified using the RBC count, Hgb, or Hct as trait in this analysis. No sex differences were seen. A simultaneous search for pairs to detect interacting QTL was negative. The maximum LOD score for the Chr 12 MCV quantitative trait locus (3.7) was obtained with the β-spectrin marker and exceeds the minimum LOD score for significance (2.2) (Figure 5A-B). The 95% confidence limits for the QTL span from 28 to 40 cM on Chr 12 (Figure 5B). As this quantitative trait locus is a modifier of hereditary spherocytosis, we have designated the locus Hsm1 (hereditary spherocytosis modifier 1). The allele effects of Hsm1 on red cell volume are shown in Figure 5C; the presence of 1 or 2 CAST alleles at the β-spectrin locus significantly lowers the MCV, suggesting Hsm1 is dominant. Hsm1 accounts for 9% of the total variance of the trait.
QTL analysis of F2 wan homozygotes. (A) Genome-wide scan of MCV, RBC count, Hgb, Hct, and MCHC. Significant QTL (minimum LOD score of 2.2, indicated by the top dashed line) were detected on Chr 6 and Chr 12 using MCV as the quantitative trait. No suggestive (LOD score of 2.0, indicated by the bottom dashed line) or significant LOD scores were obtained using RBC count, Hgb, or Hct as traits, but 2 suggestive QTL are seen for the MCHC. (B) Interval map of the Chr 12 MCV quantitative trait locus (solid line) showing the position (cM) of the markers. This quantitative trait locus was designated Hsm1. The peak LOD score for Hsm1 was obtained using a marker for the erythroid β-spectrin gene (Spnb1). The posterior probability density curve corresponding to the 95% confidence interval (bold dashed line) was computed by the method of Sen and Churchill.31 This analysis shows that the 95% confidence interval for Hsm1 is between 28 and 40 cM. (C) Allele effects of Hsm1 using the peak LOD score marker (Spnb1, β-spectrin) indicates that it acts as a dominant and lowers the MCV. C indicates CAST/Ei Spnb1 allele; H, C3H Spnb1 allele. *P < .01. Error bars indicate standard error. (D) Interval map of the Chr 6 MCV quantitative trait locus reveals multiple shoulder peaks, indicating that several QTL are likely present in the interval. The 95% confidence interval is between 40 and 80 cM. The peak LOD score was obtained with D6Mit15.
QTL analysis of F2 wan homozygotes. (A) Genome-wide scan of MCV, RBC count, Hgb, Hct, and MCHC. Significant QTL (minimum LOD score of 2.2, indicated by the top dashed line) were detected on Chr 6 and Chr 12 using MCV as the quantitative trait. No suggestive (LOD score of 2.0, indicated by the bottom dashed line) or significant LOD scores were obtained using RBC count, Hgb, or Hct as traits, but 2 suggestive QTL are seen for the MCHC. (B) Interval map of the Chr 12 MCV quantitative trait locus (solid line) showing the position (cM) of the markers. This quantitative trait locus was designated Hsm1. The peak LOD score for Hsm1 was obtained using a marker for the erythroid β-spectrin gene (Spnb1). The posterior probability density curve corresponding to the 95% confidence interval (bold dashed line) was computed by the method of Sen and Churchill.31 This analysis shows that the 95% confidence interval for Hsm1 is between 28 and 40 cM. (C) Allele effects of Hsm1 using the peak LOD score marker (Spnb1, β-spectrin) indicates that it acts as a dominant and lowers the MCV. C indicates CAST/Ei Spnb1 allele; H, C3H Spnb1 allele. *P < .01. Error bars indicate standard error. (D) Interval map of the Chr 6 MCV quantitative trait locus reveals multiple shoulder peaks, indicating that several QTL are likely present in the interval. The 95% confidence interval is between 40 and 80 cM. The peak LOD score was obtained with D6Mit15.
The second MCV quantitative trait locus, located on Chr 6, is extremely broad, spanning approximately 40 cM (Figure 5D). This quantitative trait locus likely represents several closely linked loci, as revealed by the distinct “shoulders” seen in the genome scan (Figure 5D), that together account for 7% of the variance of the trait. No significant QTL were identified using the MCHC as the quantitative trait. However, suggestive QTL were identified on Chr 7 and 19 (Figure 5A). Further analyses using additional markers to saturate the relevant chromosomal intervals and increased numbers of F2 mice will be required to determine the exact number of QTL on Chr 6 and to determine if the MCHC QTL reach statistical significance.
Red cell morphology is markedly pleiotropic in C3H, CAST F2 mice
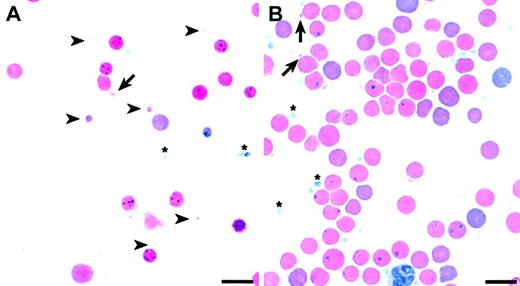
The data suggest that changes in RBC membrane characteristics, which are reflected in the MCV value, may modify the HS phenotype. If membrane characteristics are indeed influenced by a segregating modifier in our F2 intercross, one would expect to see widely varying RBC morphology among the F2 wan homozygotes, as is the case with other hematologic parameters (Table 2). Peripheral blood smears confirm this expectation. A subset of F2 wan/wan mice shows extremely aberrant red cell morphology (Figure 6A) similar to that seen in inbred C3Hwan homozygotes (Figure 1E), that is, striking anisocytosis with extreme microcytosis. Indeed, many RBC fragments smaller than platelets and frequent cells with tubular membrane extensions are present. Other F2 wan homozygotes, however, show markedly improved overall red cell morphology (Figure 6B). It should be emphasized that the smears presented in Figure 6 represent the extremes of RBC morphologies seen in the F2 population. The majority of slides examined revealed morphologic characteristics intermediate to these 2 extremes.
Pleiotropic RBC morphologies in F2 wan homozygotes. (A) Wright-stained peripheral blood smear showing extreme microspherocytosis and tubular membrane extensions (arrow). Note that some microcytes (arrowheads) are smaller than platelets (asterisks). (B) Peripheral blood smear showing significantly improved overall RBC morphology. Evidence of some membrane loss, however, persists (arrows). Bar = 10 μM.
Pleiotropic RBC morphologies in F2 wan homozygotes. (A) Wright-stained peripheral blood smear showing extreme microspherocytosis and tubular membrane extensions (arrow). Note that some microcytes (arrowheads) are smaller than platelets (asterisks). (B) Peripheral blood smear showing significantly improved overall RBC morphology. Evidence of some membrane loss, however, persists (arrows). Bar = 10 μM.
Spnb1 encoding β-spectrin is a candidate gene for Hsm1
Spnb1 encoding erythroid β-spectrin is located within the QTL interval at 33 cM.36 No other RBC membrane proteins or obvious candidate genes map within the Hsm1 critical interval on mouse Chr 12. We sequenced the entire Spnb1 cDNA in the parental strains, C3H and CAST/Ei, as well as in 2 additional inbred strains, B6 and 129. The amino acid sequence was identical in C3H, B6, and 129. There were 3 amino acid differences found in the CAST/Ei sequence: M1003V (C3H → CAST), corresponding to amino acid 19 of repeat 7; M1238K, amino acid 80 of repeat 9; and R1571C, amino acid 103 of repeat 12 (numbering according to Genbank sequence NM013675). None of these changes suggests an obvious mechanism whereby the CAST/Ei allele might influence membrane characteristics. In addition, no differences in β-spectrin protein levels between the parental strains, C3H and CAST/Ei, are detectable by Western blotting of mature RBC membrane ghosts, nor are there any differences in the amount of spectrin retained following low salt extraction (data not shown). Reverse transcriptase-PCR of fetal liver RNA and Western blotting of RBC ghosts failed to detect the presence of the pleckstrin homology (PH) domain in either strain (not shown), suggesting that strain differences in the regulation of the alternatively spliced “muscle” spectrin isoform (β1Σ2)28 are not present. Nevertheless, β-spectrin remains a candidate for Hsm1 for reasons discussed below.
Discussion
To dissect modifier genes critical to the pathogenesis of band 3-deficient HS, we mapped QTL influencing the wan phenotype. No significant QTL were detected when RBC count, Hgb, Hct, or MCHC values were used as quantitative traits, suggesting that these RBC characteristics are not significant targets for modifying genes in our cross. However, 2 significant QTL were identified using MCV as trait. It is noteworthy that the MCV is a directly measured trait on the Advia 120 hematology analyzer. Moreover, as the F2 wan homozygotes had similarly elevated reticulocyte counts, varying degrees of reticulocytosis are unlikely to have significantly influenced the MCV in the F2 population.
The first MCV quantitative trait locus spans a 40-cM interval on the distal half of Chr 6 and probably represents several QTL. The only RBC membrane skeleton gene on mouse Chr 6 is Add2 encoding β-adducin.36 Add2 is located proximal to the QTL interval at 35.5 cM, making it an unlikely candidate gene. The size of the interval precludes meaningful analysis of other potential candidate genes at this time. To identify all the QTL within the interval and candidate genes for each will require additional F2 animals and sufficient markers to saturate the relevant portion of Chr 6.
The second significant QTL interval, which we have designated Hsm1, is a single peak that spans a 12-cM interval on mouse Chr 12. Analysis of allele effects indicates that Hsm1 acts in a dominant fashion. Interestingly, both the Chr 6 modifying gene(s) and Hsm1 were identified using the MCV as the quantitative traiting suggest that RBC membrane characteristics are a target for modifier gene action in band-3 null HS. Moreover, heterozygous C3H+/wan mice do not differ from wild-type C3H+/+ controls in any hematologic parameter except the MCV, suggesting that the MCV value is the most sensitive surrogate for band-3 deficiency.
The peak LOD score on Chr 12 is obtained with the Spnb1 gene encoding erythroid β-spectrin. Of the 3 β-spectrin polymorphisms identified between the C3H and CAST/Ei inbred strains, none occurs in domains known to influence the binding of spectrin to the membrane, for example, the ankyrin binding domain bridging spectrin repeats 14 and 15 and the membrane association domain 1 (MAD1) in repeat 1, which mediates ankyrin- and protein 4.1-independent spectrin binding to the membrane.37,38 In addition, we were unable to detect any gross differences between the parental strains in RBC β-spectrin protein levels or differences in the amount of spectrin remaining in resealed RBC ghosts following low salt extraction by Western blotting. Such preliminary data seem to argue against β-spectrin as a legitimate candidate for Hsm1. However, homozygosity at the Hsm1 locus per se is not associated with hemolysis in either C3H or CAST/Ei wild-type mice. Hence, Hsm1 polymorphisms are likely to be significant only in the presence of the wan defect; differences in the parental strains would therefore not be expected. Such epistatic gene interactions between mutant genes and QTL have been described previously in several other mouse mutations.39,40 Alternatively, the application of more sensitive testing methods may detect subtle differences in C3H versus CAST spectrin function. It is noteworthy that many spectrin repeats with no known function are highly conserved across species, suggesting that spectrin likely participates in other, as yet undefined, processes within the red cell.1
That 17% of C3H, CAST/Ei F2 wan/wan mice survived to adulthood is telling evidence of the strong influence of genetic modifiers. Clearly, however, genetic factors other than Hsm1 are also influencing the band-3 null phenotype as judged by 2 criteria. First, we identified additional QTL on Chr 6. Second, the β-spectrin sequence in B6 and 129 mice is identical to that of C3H. Despite this, B6, 129 hybrid and congenic B6 (n = 15) band-3 knock-out mice have significantly different survival rates compared with each other and with C3Hwan homozygotes.
By what mechanism(s) might a genetic modifier of band 3-deficient HS influence RBC membrane characteristics? Band 3 is hypothesized to stabilize the lipid bilayer in 2 ways, skeleton anchoring and lipid anchoring.1 According to the skeleton anchoring hypothesis, the bilayer is stabilized by direct interactions with the underlying spectrin array. As a major attachment site for the spectrin array, band 3 plays a pivotal role in anchoring the skeleton in place and ensuring a normal membrane spectrin content. In the “lipid anchoring” hypothesis, the bilayer is stabilized by interactions between lipids and integral membrane proteins within the horizontal plane of the bilayer. The approximately 106 copies of band 3 per RBC, each consisting of a transmembrane domain that traverses the lipid bilayer 14 times, are predicted to contribute significantly to lipid anchoring.41 The primary evidence for lipid anchoring comes from the observation that band-3 deficiency in humans and mice is associated with membrane loss despite an intact membrane skeleton with normal spectrin content.1,34 Of note, these 2 mechanisms are not mutually exclusive and likely not all inclusive. A recent study suggests that both are important and can compensate for each other to reduce the impact of membrane skeleton defects while pointing out that many observations cannot be accounted for by either mechanism, alone or in combination, indicating that other factors must contribute to the overall integrity and stability of the membrane.42
Thus, polymorphisms in β-spectrin could influence membrane stability in a number of ways. In both mice and humans, defects in spectrin are associated with membrane instability as well as altered cation transporter activities.1,9,43,44 Either mechanism could potentially alter the MCV. Other mechanisms are also possible, however. For example, spectrin is known to interact with phosphatidylserine (PS) at multiple sites, and this interaction is mediated by the β-subunit.45,46 Hence, β-spectrin polymorphisms could enhance spectrin-PS interactions. Alternatively, polymorphic β-spectrin alleles could bind other, currently unknown, integral membrane proteins with differing affinities. Finally, spectrin self-association, ankyrin binding, and band-3 binding have been shown to be coupled in a positively cooperative manner.47 Loss of band 3, therefore, is expected to weaken spectrin self-association. β-spectrin polymorphisms may influence this positive cooperativity in such a way that it is less sensitive to the loss of band 3.
Hsm1 acts in a dominant manner to decrease the MCV. It seems intuitive that a beneficial quantitative trait locus would increase the MCV toward a normal level. Hence, a legitimate question is whether the Hsm1-wan interaction is detrimental rather than beneficial. The present data do not allow for a definitive assignment. The presence of CAST/Ei alleles is clearly associated with improved survival, RBC morphology, and hematologic status in a subset of the F2 hybrid animals. As several QTL are segregating, it is likely that a combination of modifier alleles, which individually may act to exacerbate or ameliorate the wan phenotype, acts in concert to produce the phenotypic variation seen in the hybrid wan homozygotes.
The definitive identification of Hsm1 and its function will require construction of reciprocal recombinant congenic strains and bioinformatic approaches to further decrease the QTL interval and, ultimately, a transgenic or knock-out/knock-in approach to verify the most likely candidate gene.48 To define the precise mechanism of Hsm1 function, studies such as ektacytometry, which provide independent measurements of surface area and volume, and membrane lipid analysis will be required. Comparing congenic/targeted strains with the original C3H-wan mice in these and other parameters will clarify the mechanism of Hsm1 modification of HS severity and shed light on basic mechanisms of membrane stabilization.
Classical twin studies have shown that there is a substantial genetic component underlying Hgb, RBC, white blood cell (WBC), and platelet numbers in humans.49 The normal phenotypic variation seen among the inbred strains for these and other hematologic values, including the MCV, is clear evidence for genetic determinants in mice as well,50 and we have identified QTL for MCV in crosses between normal inbred strains (L.L.P and G.A.C., unpublished data, November 2003). Moreover, variation in hematologic parameters is known to affect disease severity. For example, the baseline WBC count directly correlates with severity of disease symptoms in sickle cell patients.51,52 It is clear, therefore, that basic hematologic parameters are strongly influenced by genetic factors, which in turn affect disease outcome. The determination of the genes underlying Hsm1 and the Chr 6 quantitative trait locus, and the identification of other QTL as well, will ultimately identify genetic interactions critical to RBC stability and/or survival and the pathogenesis of HS.
Prepublished online as Blood First Edition Paper, December 30, 2003; DOI 10.1182/blood-2003-08-2813.
Supported by National Institutes of Health (NIH) grants HL64885 (L.L.P.), HL32262 and DK34083 (S.E.L.), and RR01183 (The Jackson Laboratory Mouse Mutant Resource); The March of Dimes (L.L.P.); and grant no. CA34196 (The Jackson Laboratory Cancer Core).
Presented in part at the 43rd Annual Meeting of the American Society of Hematology, Orlando, FL, December 7-11, 2001.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Beverly J. Paigen and Jane E. Barker for critical review of the manuscript, and Eva M. Eicher and Linda L. Washburn for identifying the first wan mutants in their breeding colony.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal