Abstract
The CD43 lymphocyte surface receptor is involved in the regulation of lymphocyte adhesion and activation. Many CD43 functions remain controversial or unclear, and it is not known to which extent CD43 signaling pathways are shared with or distinct from those used by the T-cell receptor (TCR). Here, we systematically compared signaling events and target gene expression induced by CD43 or T-cell costimulation in primary human peripheral T cells. These studies identify nuclear factor-κB (NF-κB) p65 serine 468 as a novel inducible phosphorylation site strongly induced by T-cell costimulation and only weakly triggered by CD43 ligation. We also identified CD43 as a novel Jun N-terminal kinase (JNK) activator and a comprehensive analysis of further signaling events suggests that both stimuli use overlapping but also distinct signaling pathways. Microarray analysis of inflammatory genes shows 1 group of genes coregulated by both stimuli and 2 further groups of target genes affected solely by costimulation or primarily by CD43. (Blood. 2004;104:3302-3304)
Introduction
CD43 (leukosialin, sialophorin) is expressed at the membrane of hematopoietic cells except resting B cells and erythrocytes. The physiologic role of CD43 does not yield a coherent picture. It was suggested to have antiadhesive functions,1 but also a positive role for adhesion mediated by integrins.2 Similarly, CD43 was implicated in antiapoptotic3 and proapoptotic4 signaling.
In human T lymphocytes, CD43 engagement triggers the association of the tyrosine kinases Lck and Fyn to the cytoplasmic tail of CD435,6 and tyrosine phosphorylation of Vav, phospholipase C-γ2 (PLCγ2), and the adapter proteins Shc and SLP-76 (SH2 domain-containing leukocyte protein 76).7 These early signals enable the generation of second messengers such as diacylglycerol and Ca2+ mobilization and result in the activation of the mitogen-activated protein kinases (MAPKinases) p38 and ERK (extracellular signal-regulated kinase).8,9 CD43 engagement also induces the DNA binding activity of transcription factors NF-AT (nuclear factor of activated T cells) and NF-κB (nuclear factor-κB),10 but the affected target genes are largely unknown. NF-κB is regulated upon association with inhibitory IκB proteins and also by modulatory phosphorylations of the DNA-binding subunits, including the transactivating p65 subunit.11 NF-κB activity is also elicited by T-cell costimulation that is achieved by simultaneous stimulation of the T-cell receptor (TCR) with coreceptors such as CD28. TCR- and CD43-mediated signaling pathways are interlaced, because CD43 potentiates proliferation of TCR-stimulated T cells independent from the presence of the costimulatory CD28 receptor.12 In addition, T-cell costimulation-induced human immunodeficiency (HI) virus transcription and virus production is further enhanced by CD43 signaling,7 CD43 potentiates TCR-induced proliferation of murine intraepithelial lymphocytes,13 and CD43 recruits the TCR ζ-chain for signal transduction.14
Given the incoherent picture of CD43 function and the question to which extent CD43-mediated signaling routes are shared with those used by T-cell costimulation, we systematically compared CD43- and CD3/CD28-mediated signaling events and gene expression patterns.
Study design
Cell culture and T-cell activation
Blood samples were collected under study protocols approved by the Institutional Review Board of the University of Bern, and all subjects gave informed consent in accordance with the Declaration of Helsinki. Peripheral blood T cells were isolated from donor blood (Central Laboratory of the Swiss Red Cross, Bern, Switzerland) by Ficoll-Paque (Axis Shield, Oslo, Norway) gradient centrifugation. The mononuclear cells were resuspended in RPMI 1640 medium supplemented with 5% (vol/vol) fetal calf serum, 1% (vol/vol) l-glutamine, and 1% (vol/vol) penicillin/streptomycin. Monocytes were depleted by plastic adherence, and nonadherent cells were loaded on a nylon wool column pre-equilibrated with supplemented RPMI. The resultant purified T cells were rested for 24 hours in supplemented RPMI. Stimulation of purified T cells was performed in a final volume of 1 mL by adding αCD43 L10 (Caltag Laboratories, Burlingame, CA), or αCD3 (clone OKT3) and αCD28 (clone 15E8) antibodies at a final concentration of 1 μg/mL together with 1 μg protein A from Staphylococcus aureus for cross-linking.
Cell extracts and Western blotting
Stimulation was terminated upon the addition of ice-cold phospate-buffered saline (PBS) to the cells, and cell extracts were prepared as described.15 Phosphospecific antibodies were from Cell Signaling Technology (Beverly, MA); antibodies recognizing the p65 and c-Jun proteins were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Bound antibodies were detected by horseradish peroxidase-coupled secondary antibodies and enhanced chemiluminescence according to the instructions of the manufacturer (Pharmacia-Amersham, Piscataway, NJ).
DNA microarray experiments and real time PCR
Total cellular RNA was purified using the RNeasy mini kit (Qiagen, Hilden, Germany), and fluorescent cRNAs were prepared by oligo dT-T7-primed reverse transcription followed by in vitro transcription. The labeled cRNAs were hybridized to microarrays containing validated oligonucleotide probes for 110 inflammatory genes15 ; scanning was performed on an Affymetrix 428 array scanner (Santa Clara, CA) at increasing photomultiplier tube (PMT) voltage settings. Means of ratios of replicate experiments were calculated from log2-transformed values. For clarity, log2 values are re-transformed into the numerical values. mRNA expression levels for interleukin 8 (IL-8), interferon-γ (IFNγ), and actin were confirmed using Assays on Demand (Applied Biosystems, Applera Corporation, Foster City, CA) and an ABI 7000 real-time polymerase chain reaction (PCR) instrument according to the manufacturer's instructions.
Results and discussion
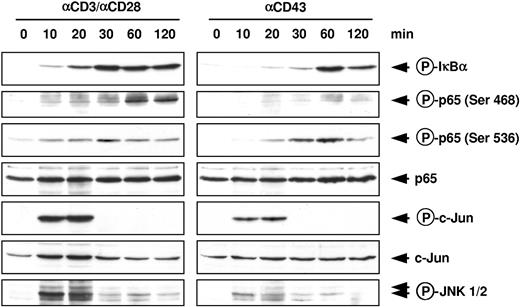
To systematically compare the signaling pathways used by T-cell costimulation and the CD43 receptor, isolated primary human peripheral T cells were stimulated for various periods with agonistic αCD43 or αCD3 (TCR) and αCD28 antibodies. Cell extracts were tested for the induction of signaling pathways by Western blotting using phosphospecific antibodies (Figure 1). Western blotting revealed that phosphorylation of the endogenous IκBα protein started already 10 minutes after T-cell costimulation and was still detectable after 2 hours. In contrast, CD43 ligation triggered IκBα phosphorylation with a slower kinetics, peaking at 1 hour after stimulation, and declining thereafter. A novel phosphospecific antibody allowed to detect a previously undescribed phosphorylation site at NF-κB p65 serine 468. This phosphorylation was strongly triggered by costimulation, but only faintly by CD43. These data show a clear difference between CD43 and costimulation-mediated signaling and also identify serine 468 as a novel, not yet identified p65 phosphorylation site. Because this serine is located within transactivation domain 2, it is tempting to speculate that this modification affects the ability of p65 to direct target gene expression. NF-κB p65 serine 536 phosphorylation precedes modification of serine 468,16 raising the possibility that both sites are modified by different kinases. A hallmark of T-cell costimulation is the activation of JNK,17 and accordingly phosphorylation of the JNK substrate c-Jun at serine 73 was observed at early time points (10 and 20 minutes) after T-cell costimulation. Also activation from the CD43 receptor triggered phosphorylation of c-Jun with a similar kinetics, but to a lesser extent. Because c-Jun can be modified by several kinases, we further investigated the involvement of the JNK pathway by monitoring the phosphorylation and thus activation of JNK1 and JNK2. At least 2 JNK isoforms were phosphorylated either by T-cell costimulation or CD43 ligation, thus identifying the JNK signaling pathway as a novel target for CD43-induced signaling. In summary, these data reveal overlapping and distinct phosphorylation targets, signal intensities, and kinetics used by the 2 pathways.
Comparative analysis of CD43-and T-cell costimulation-induced signaling pathways in human peripheral T lymphocytes. Cells were stimulated for the indicated periods with agonistic αCD43 or αCD3/CD28 antibodies and lysed. Equal amounts of protein contained in total cell extracts were analyzed by immunoblotting. The occurrence of p65 and c-Jun was revealed with specific antibodies; phosphorylation of endogenous proteins was revealed by phosphospecific antibodies detecting phosphorylated forms of p65, IκBα, c-Jun, and Jun N-terminal kinase 1 and 2 (JNK1/2).
Comparative analysis of CD43-and T-cell costimulation-induced signaling pathways in human peripheral T lymphocytes. Cells were stimulated for the indicated periods with agonistic αCD43 or αCD3/CD28 antibodies and lysed. Equal amounts of protein contained in total cell extracts were analyzed by immunoblotting. The occurrence of p65 and c-Jun was revealed with specific antibodies; phosphorylation of endogenous proteins was revealed by phosphospecific antibodies detecting phosphorylated forms of p65, IκBα, c-Jun, and Jun N-terminal kinase 1 and 2 (JNK1/2).
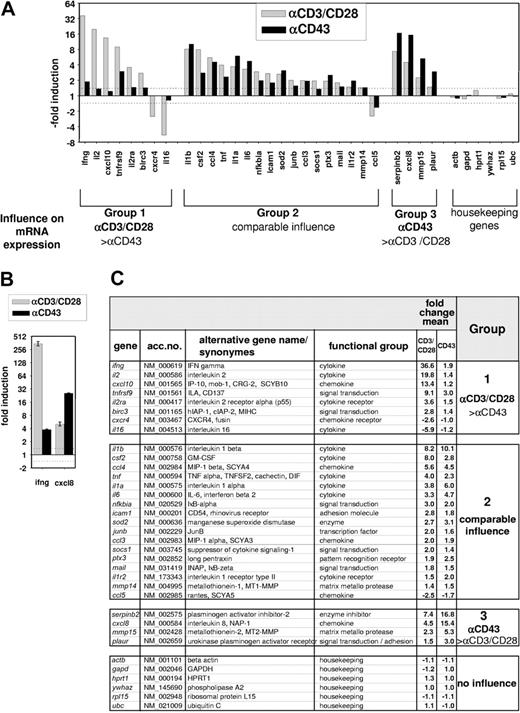
To compare the target genes induced by T-cell costimulation or CD43 triggering on a systematic basis, T cells were stimulated for 4 hours with αCD43 or αCD3/CD28 antibodies, and gene expression was monitored in microarray experiments. We used a fully validated inflammatory array containing 110 human genes known to be strongly regulated during inflammation.15 These assays revealed 29 genes regulated by T-cell costimulation and 25 genes affected by CD43 (Figure 2A). According to their induction pattern in response to the inducing signals, these genes fell into 3 groups: The group 1 genes IFNγ, IL-2, IP-10, CD137, IL-2Rα, IAP, Cxcr4, and IL-16 show strong regulation by T-cell costimulation but are not significantly affected by CD43. Group 2 comprises 17 genes that are comparably affected by either pathway. These include cytokines such as IL-1β, the chemokines MIP-1β (macrophage inflammatory protein 1β) and RANTES (regulated on activation normal T expressed and secreted), and also further inflammatory proteins. Genes primarily regulated by CD43 form group 3, which contains PAI-2, uPAR, MT2-MMP, and IL-8, all of which are involved in the regulation of cell migration and motility. The differential induction of group 1 and group 3 genes by CD43 or CD3/CD28 stimulation was confirmed by real-time PCR (Figure 2B). The complete set of data is displayed in Figure 2C. Collectively, the results presented in Figures 1 and 2 allow several conclusions: (1) the classic T-cell costimulation-triggered cytokines IL-2 and IFNγ are no main targets of CD43, at least at the time point analyzed. (2) T-cell costimulation and CD43 never trigger opposite effects on a given target gene. (3) The strength of gene induction is comparable between both stimuli, establishing CD43 as an inducer of gene expression also in the absence of further signals. (4) CD43- and CD3/CD28-triggered target genes show a significant overlap. All these findings are in accordance with our data showing that the receptors use shared and distinct signaling pathways.
Comparative microarray analysis of gene expression induced by CD43 or T-cell costimulation. Human peripheral T cells were either left untreated or stimulated for 4 hours with αCD43 or αCD3/CD28 antibodies. Gene expression of 110 inflammatory genes was analyzed by DNA microarrays. (A) The average induction factor obtained from 2 independent experiments is shown for the indicated genes. According to the relative potency of T-cell costimulation compared with CD43 ligation to induce gene expression, the genes are placed into 3 groups.1-3 Only genes that were regulated more than 1.4-fold are displayed; this threshold is also indicated by the dashed line. Group 1 or group 3 genes are induced by 1 stimulus 2-fold or more in comparison to the other stimulus. (B) mRNA expression of the group 1 gene IFNγ and the group 3 gene IL-8 was determined by real-time PCR and normalized for the expression of β-actin. Data are expressed as -fold change relative to the unstimulated control. Bars represent means ± SD from triplicate determinations. (C) Complete set of results obtained from the 2 donors. Gene names and accession numbers are taken from the RefSeq database, numerical values of induction factors are given. The numbers in the right column refer to the groups of genes as defined in panel A.
Comparative microarray analysis of gene expression induced by CD43 or T-cell costimulation. Human peripheral T cells were either left untreated or stimulated for 4 hours with αCD43 or αCD3/CD28 antibodies. Gene expression of 110 inflammatory genes was analyzed by DNA microarrays. (A) The average induction factor obtained from 2 independent experiments is shown for the indicated genes. According to the relative potency of T-cell costimulation compared with CD43 ligation to induce gene expression, the genes are placed into 3 groups.1-3 Only genes that were regulated more than 1.4-fold are displayed; this threshold is also indicated by the dashed line. Group 1 or group 3 genes are induced by 1 stimulus 2-fold or more in comparison to the other stimulus. (B) mRNA expression of the group 1 gene IFNγ and the group 3 gene IL-8 was determined by real-time PCR and normalized for the expression of β-actin. Data are expressed as -fold change relative to the unstimulated control. Bars represent means ± SD from triplicate determinations. (C) Complete set of results obtained from the 2 donors. Gene names and accession numbers are taken from the RefSeq database, numerical values of induction factors are given. The numbers in the right column refer to the groups of genes as defined in panel A.
Prepublished online as Blood First Edition Paper, July 27, 2004; DOI 10.1182/blood-2004-04-1536.
Supported by grants from the Deutsche Forschungsgemeinschaft to (Schm 1417/3-1, SFB566, Kr1143/4-1) (L.S. and M.K.) and European Union (EU) project (QLK3-CT-2000-00463) sponsored by the Bundesamt für Bildung und Wissenschaft (BBW), Oncosuisse, Schweizerischer Nationalfonds, and Association for International Cancer Research.
M.L. is employed by Cell Signaling Inc. whose product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank the Central Laboratory of the Swiss Red Cross (Bern, Switzerland) for kindly delivering blood from donors and Heike Schneider for technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal