Abstract
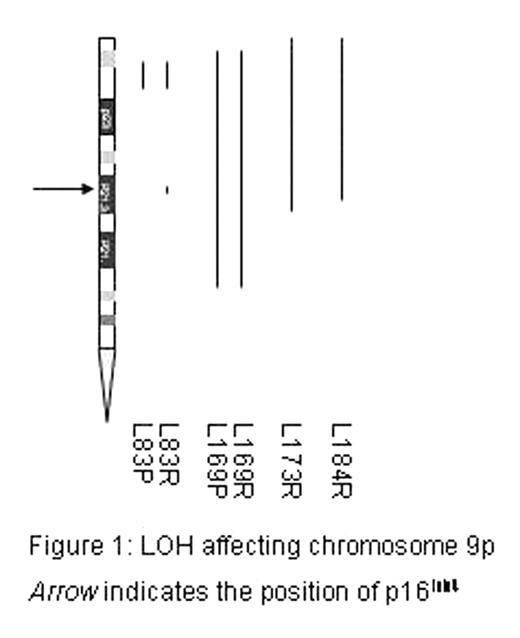
Loss of heterozygosity (LOH) is detectable in many forms of malignancy, including leukaemia, using techniques such as microsatellite analysis and comparative genomic hybridisation. However, these techniques are laborious and require the use of relatively large amounts of DNA if the whole genome is to be examined. Here we describe the use of oligonucleotide microarrays to characterise single nucleotide polymorphisms (SNPs) in lymphoblasts isolated from children with acute lymphoblastic leukaemia for the pan-genomic mapping of LOH with a resolution of 100–200kb. Results were compared with DNA obtained during remission and on relapse. Abnormalities were seen in 8 of 10 cases. The two cases with no abnormalities and one case which showed identical changes affecting whole chromosomes at relapse and presentation remain in remission 1–9 years following retreatment. The 7 cases which showed LOH not affecting entire chromosomes died following relapse, suggesting that partial LOH may be associated with a poor prognosis. In 4 cases LOH was only detectable at relapse suggesting that progressive LOH may be a cause of disease progression and/or drug resistance. This was supported by detailed analysis of one case in which LOH involving the glucocorticoid receptor (GR) was associated with mutation of the remaining allele. In cell line models the loss of a functional GR is associated with profound resistance to steroids. The most frequent abnormality detected in this series involved chromosome 9p. In each of the four cases where this was observed LOH included the INK4 locus. In three of the four cases INK4 loss was only observed at relapse (see figure), suggesting that this abnormality may be commonly associated with treatment failure, supporting previous reports that 9p abnormailities are associated with a poor prognosis. One case was reported as showing monosomy 20 as the sole cytogenetic aberration but LOH analysis identified 9p LOH and loss of 20q, with retention of heterozygocity for 20p. These findings strongly implicate unbalanced translocation der(9)t(9;20),-20 as described by Clark et al (Leukaemia, 2000, 14:241). Our observations demonstrate that SNP array analysis is a powerful new tool for the analysis of allelic imbalance and unbalanced translocations in leukaemic blasts.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal