Although phospholipid scramblase 1 (PLSCR1) was originally identified based on its capacity to promote transbilayer movement of membrane phospholipids, subsequent studies also provided evidence for its role in cell proliferation, maturation, and apoptosis. In this report, we investigate the potential role of PLSCR1 in leukemic cell differentiation. We show that all-trans retinoic acid (ATRA), an effective differentiation-inducing agent of acute promyelocytic leukemic (APL) cells, can elevate PLSCR1 expression in ATRA-sensitive APL cells NB4 and HL60, but not in maturation-resistant NB4-LR1 cells. ATRA- and phorbol 12-myristate 13-acetate (PMA)–induced monocytic differentiation is accompanied by increased PLSCR1 expression, whereas only a slight or no elevation of PLSCR1 expression is observed in U937 cells differentiated with dimethyl sulfoxide (DMSO), sodium butyrate, or vitamin D3. Cell differentiation with ATRA and PMA, but not with vitamin D3 or DMSO, results in phosphorylation of protein kinase Cδ (PKCδ), and the PKCδ-specific inhibitor rottlerin nearly eliminates the ATRA- and PMA-induced expression of PLSCR1, while ectopic expression of a constitutively active form of PKCδ directly increases PLSCR1 expression. Finally, decreasing PLSCR1 expression with small interfering RNA inhibits ATRA/PMA-induced differentiation. Taken together, these results suggest that as a protein induced upon PKCδ activation, PLSCR1 is required for ATRA- and PMA-triggered leukemic cell differentiation.
Introduction
Human phospholipid scramblase 1 (PLSCR1),1,2 also known as MmTRA1b (Mm-1 cell–derived transplantability-associated gene 1b),3 is a multiply palmitoylated, calcium-binding, lipid raft–associated endofacial plasma membrane protein.4 PLSCR1 was originally identified based on its capacity to promote rapid transbilayer movement of phospholipids (PLs) in response to the elevation of Ca2+ and has been proposed to play a role in the cell-surface exposure of phosphatidylserine (PtdSer) following cell activation, injury, or apoptosis.1-3 However, its role in membrane PL scrambling is controversial. While overexpression of PLSCR1 was reported to increase cell-surface exposure of PtdSer or to induce apoptosis in a variety of cells,5-7 an increase in PL scrambling activity with elevated expression of PLSCR1 was not observed in other studies.8,9 Blood platelets from PLSCR1-/- mice also showed normal capacity to expose PtdSer upon cell activation.10 Although the precise biologic function(s) of PLSCR1 remains to be determined, recent studies provide strong evidence for its role in cell signaling and in cell maturation. PLSCR1 has been reported to be a substrate of several kinases that participate in cell proliferation, differentiation, or apoptotic responses, including c-Abl, c-Src, and protein kinase Cδ (PKCδ).11-14 In response to growth factors such as epidermal growth factor (EGF), tyrosine phosphorylation of PLSCR1 by c-Src resulted in association of phosphorylated PLSCR1 with the adaptor protein Shc and the activated EGF receptor complex, and, in cells deficient in PLSCR1, signaling through the EGF receptor is markedly attenuated.12,13 Furthermore, PLSCR1 itself is transcriptionally up-regulated by a number of selective growth factors and cytokines, including interferon.9,15 Interestingly, whereas palmitoylation of PLSCR1 is required for its anchoring in the plasma membrane, PLSCR1 was found in the nucleus after transcriptional induction by cytokines and in circumstances in which palmitoylation is prevented. Such nuclear localization of PLSCR1 was shown to be actively mediated by import through a nuclear localization signal within the polypeptide, and once imported, the protein was shown to bind to genomic DNA, implying a possible effect on gene transcription.16,17
Proliferation and terminal differentiation of myeloid precursor cells in response to selective growth factors are impaired in PLSCR1-/- mice, and in both monocytic and granulocytic lineages, the expression of PLSCR1 markedly increases upon terminal differentiation into neutrophils and macrophages.10 Conversely, de novo expression of a mutant mRNA encoding a truncated form of murine PLSCR1 (also known as MmTRA1a, deleting the proline-rich segment between codons 1-128) was identified in a monocytic leukemia cell line, and this mutation was found to correlate with the ability of these cells to proliferate in vivo.3 By contrast, the expression of full-length PLSCR1 induced differentiation of these leukemic cells to macrophages.3 Furthermore, Nakamaki et al18 reported that PLSCR1 mRNA was specifically induced during granulocytic differentiation of acute promyelocytic leukemia (APL) cells by all-trans retinoic acid (ATRA), a widely studied potent inducer of cell differentiation and growth arrest of malignant cells in vitro and in vivo.19 Finally, a recent study performed in patients with acute myelogenous leukemia (AML) showed that higher levels of PLSCR1 mRNA were associated with significantly longer overall survival, particularly in patients of the AML-M4 subtype, independent of chromosomal aberrations such as t(8;21) and inv(16), suggesting PLSCR1 mRNA level as a new prognostic factor for AML.20
In this study, we investigate effects of known differentiation-inducing agents on PLSCR1 expression in leukemic cell lines in order to gain insight into a potential role of PLSCR1 in leukemic cell differentiation. We show that PLSCR1 is significantly up-regulated during both granulocytic and monocytic differentiation induced by ATRA and phorbol 12-myristate 13-acetate (PMA), and that activation of PKCδ is required for this process. Furthermore, by ectopic expression of a constitutively active form of PKCδ, we demonstrate for the first time that PKCδ can directly induce PLSCR1 expression. Finally, using small interfering RNA (siRNA) to decrease cellular PLSCR1 expression, we provide evidence that PLSCR1 is required for ATRA- and PMA-induced leukemic cell differentiation.
Materials and methods
Cells and cell treatment
Leukemia cell lines used in this study included chromosomal translocation t(15;17)–positive and ATRA-sensitive human APL cell line NB421 and NB4-derived maturation-resistant, ATRA-responsive cell line NB4-LR122 ; t(15;17)-negative ATRA-sensitive APL cell line HL60; human acute monocytic leukemia cell line U937; and human T-lymphocytic leukemia cell line Jurkat. In addition, adherent cell line Cos-7 was also used. All cell lines were cultured in RPMI-1640 medium (Sigma-Aldrich, St Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Logan, UT) in a 5% CO2–95% air humidified atmosphere at 37°C. For experiments, cells were seeded at a concentration of 2 × 105 cells/mL into a Falcon 6-well plate or plastic incubating flask (Becton Dickinson, San Jose, CA), and were treated with the indicated concentrations of ATRA, PMA, dimethyl sulfoxide (DMSO), vitamin D3 (VD3), sodium butyrate (SB), and/or rottlerin, all of which were purchased from Sigma-Aldrich (St Louis, MO) except for rottlerin (BIOMOL, Plymouth, PA). During the treatment with these compounds, cell viability was at least 90% by trypan-blue exclusion assay.
Plasmids and transient transfection
Plasmids pEGFP-N1 and pEGFP-PKC-CFδ carrying the catalytic fragment of PKCδ were obtained as a generous gift from Dr Mary E. Reyland (Denver, CO).23 These plasmids were transfected into Cos-7 cells using Polyfect transfection reagent (QIAGEN, Valencia, CA) according to the manufacturer's instructions. pEGFP-N1 plasmid was transfected as a negative control. Transfected cells were analyzed 48 hours after transfection for expression of PLSCR1.
siRNA design and stable expression
The mammalian expression vector pSilencer 3.1-H1 neo (Ambion, Austin, TX) was used for expression of siRNA in U937 cells. SiRNAs to PLSCR1 were designed following the procedure from Ambion. There were 5 target sequences selected: P1, 5′-TCA GCC AGT ATA TAA TCA G-3′; P2, 5′-CTC TGG AGA GAC CAC TAA G-3′; P3, 5′-ATA AGT GGT CCA TGT GTT G-3′; P4, 5′-TTT CCA AGC ACT GGA CTG G-3′; and P5, 5′-AGT CTC CTC AGG AAA TCT G-3′. Each sequence was aligned to the human genome database in a BLAST search to eliminate those with significant homology to other genes. For each target sequence, we designed complementary 55- to 60-mer oligonucleotides with 5′ single-stranded overhangs for ligation into the pSilencer 3.1-H1 neo vector. The oligonucleotides encoded 19-mer hairpin sequences specific to PLSCR1 mRNA target, a loop sequence separating the 2 complementary domains, and a polythymidine tract to terminate transcription. These sequences were synthesized and inserted into the pSilencer 3.1-H1 neo vector according to the manufacturer's instructions (Ambion). U937 cells were transfected using Nucleofector Solution (Amaxa, Gaithersburg, MD) according to the manufacturer's instructions. At 48 hours after transfection, 800 μg/mL G418 was added to the medium to select the stable transfected cells.
Evaluation of cell differentiation
Cell differentiation was evaluated by morphologic characterization and the percentage of mature-related cell-surface differentiation antigens CD11b, CD11c, and CD14. For morphologic observation, cells were collected onto slides by cytospin (Shandon, Runcorn, United Kingdom), stained (Wright staining), and observed by light microscope (Olympus, B×51, Tokyo, Japan). The images were captured with Olympus DP50 digital camera by Image-Pro plus. Differentiation antigens were measured by flow cytometry (Beckman-Coulter, Miami, FL) using fluorescein isothiocyanate (FITC)–labeled or phycoerythrin (PE)–labeled antibodies as previously described.24 Briefly, cells were collected, washed, and incubated with monoclonal mouse antihuman FITC-labeled anti-CD11b/CD11c or PE-labeled anti-CD14 (Immunotech, Marseille, France) for 30 minutes at room temperature. Becton Dickinson Simultest Control r1/r2α was used as a negative control. Fluorescence intensity was analyzed by flow cytometry. Data were based on examination of 10 000 cells/sample selected randomly from 5 × 105 cells.
Semiquantitative reverse transcription–PCR for PLSCR1 mRNA
Total RNA was isolated by Trizol kit (Invitrogen, Paisley, Scotland, United Kingdom) and reverse transcription (RT) was performed by TaKaRa RNA polymerase chain reaction (PCR) kit (Takara, Dalian, China) following the manufacturer's instructions. PCR reactions to amplify PLSCR1 (first described by Sims et al2 ) and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) cDNA were performed in a single tube using the Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) with specific primers for PLSCR1 (sense strand, 5′-CAG CCT CCA TTA AAC TGT CC-3′; antisense strand, 5′-TCT TAG TGG TCT CTC CAG AG-3′) and for G3PDH (sense strand, 5′-TGA AGG TCG GAG TCA ACG GAT TTG G-3′; antisense strand, 5′-ATG TGG GCC ATG AGG TCC ACC AC-3′). PCR consisted of 28 cycles with denaturing at 95°C for 45 seconds, annealing at 55°C for 45 seconds, and extension at 72°C for 60 seconds. Amplification cycles were preceded by a denaturation step (95°C for 5 minutes) followed by an elongation step (72°C for 10 minutes). After amplification, PCR products were analyzed on a 1% agarose gel, and the signal intensities of amplified PLSCR1 fragments were normalized against 983-bp G3PDH using a densitometer (SmartView version 5.0 software from Furi, Shanghai, China).
Quantitative real-time RT-PCR for PLSCR1 mRNA
For quantitative analysis of gene expression, total RNA was isolated by Trizol kit (Invitrogen). RNA was treated with DNase (Promega, Madison, WI). Complementary DNA was synthesized using the cDNA synthesis kit (Applied Biosystem, Foster City, CA) according to the manufacturer's instructions. Fluorescence real-time RT-PCR was performed with the double-stranded DNA dye SYBR Green PCR Core Reagents (PE Biosystems, Warrington, United Kingdom) using the ABI PRISM 7900 system (Perkin-Elmer, Torrance, CA). The reaction of SYBR Green assay contained 1 μL 10 × SYBR Green PCR buffer, 0.8 μL deoxynucleoside triphosphate (dNTP) mixture, 0.1 μL AmpErase UNG (1 U/μL), 0.05 μL AmpliTaq Gold DNA Polymerase (5 U/μL), 1.2 μL MgCl2 (25 mM), 0.1 μL forward and reverse primer (20 μM), 1 μL cDNA, and 5.65 μL double distilled H2O. The following primers were used: PLCSR1 forward, 5′-CTG ACT TCT GAG AAG GTT GC-3′ and reverse, 5′-GAA TGC TGT CGG TGG ATA CTG-3′; and β-actin forward, 5′-CAT CCT CAC CCT GAA GTA CCC-3′ and reverse, 5′-AGC CTG GAT AGC AAC GTA CAT G-3′. PCR was begun with one cycle of 50°C for 2 minutes (UNG incubation) and 95°C for 10 minutes (hot-start PCR) and preceded by 40 cycles with denaturing at 95°C for 30 seconds, annealing at 59°C for 30 seconds, and extension at 72°C for 30 seconds. After PCR amplification cycles, a dissociation curve (melting curve) was constructed in the range of 65°C to 95°C. All amplifications and detections were carried out in a MicroAmp optical 384-well reaction plate with optical adhesive covers (Applied Biosystems, Foster, CA). PCR was done in triplicate and standard deviations representing experimental errors were calculated. All data were analyzed using ABI PRISM SDS 2.0 software (Perkin-Elmer). This software, which is coupled to the instrument, allows the determination of the threshold cycle (Ct) that represents the number of the cycle where the florescence intensity is significantly above the background fluorescence intensity. Using the ΔCt method, β-actin was coamplified to normalize the amount of RNA added to the reaction, and the data were subjected to cycling threshold analysis according to published procedures.25
Western blot
Cells were harvested, washed with ice-cold phosphate-buffered saline (PBS), and lysed with ice-cold lysis buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl [pH 7.4], 150 mM NaCl, 1% nonidet P-40, 2 mM sodium orthovanadate, 5 mM EDTA [ethylenediaminetetraacetic acid], and protease inhibitor cocktail) or phosphorylation lysis buffer (62.5 mM Tris-HCl [pH 6.8], 2% wt/vol sodium dodecyl sulfate [SDS], 10% glycerol, 50 mM dithiothreitol [DTT], and 0.01% bromophenol blue) on ice for 30 minutes. Cell lysates were centrifuged at 20 000g for 10 minutes at 4°C and protein in the supernatants was quantified. Protein extracts were equally loaded on 12% SDS–polyacrylamide gel and electrophoretically transferred to Immobilon–polyvinylidene fluoride (PVDF) membranes (Schleicher & Schuell, Dassel, Germany). After blocking with 5% nonfat milk in Tris-buffered saline (TBS), the membranes were incubated for 2 hours with monoclonal antihuman PLSCR1 4D2 antibody,9 rabbit polyclonal anti–phospho-PKCδ (Ser643) (Cell Signaling, Beverly, MA), and rabbit polyclonal anti-PKCδ antibody (Santa Cruz Biotechnology, Santa Cruz, CA), followed by horseradish peroxidase (HRP)–linked secondary antibodies (Cell Signaling). Detection was performed by chemiluminescence phototope-HRP kit (Cell Signaling) according to the manufacturer's instructions. All blots were stripped and reprobed with mouse monoclonal anti–β-actin antibody (pan, Ab-5; NeoMarkers, Fremont, CA) to ascertain equal loading of protein.
In vitro phosphorylation of PLSCR1 by PKCδ
Recombinant PKCδ (CalBiochem, La Jolla, CA) at a concentration of 20 nM was incubated with either 1.1 μM purified human erythrocyte PLSCR11 or 1.1 μM histone (Sigma-Aldrich, St Louis, MO) in 60 μL reaction mixture of 20 mM Tris-HCl (pH 7.5), 5 mM DTT, 5 mM MgCl2, 20 μM adenosine triphosphate (ATP), 0.6 μCi (2.2 × 104 Bq) [γ-32P] ATP, 6 μg PtdSer, 10 μM PMA, and 0.03% Triton X-100.26 After 2 hours at 30°C, reactions were stopped by addition of hot SDS sample buffer. Proteins were resolved on 18% Tris-glycine gels, transferred to PVDF membranes, and either visualized by autoradiography or immunoblotted for PLSCR1 using antibody 4D2.
Phosphorylation of PLSCR1 in apoptotic Jurkat cells
Jurkat T cells (American Type Culture Collection, Rockville, MD) in exponential growth phase were washed once with phosphate-free RPMI-1640, and incubated for 2 hours at 37°C in phosphate-free RPMI-1640, 10% dialyzed fetal bovine serum supplemented with [γ-32P] ATP (0.2 mCi/mL, 7.4 × 106 Bq/mL), with or without 0.5 μg/mL anti-Fas antibody (Kamiya Biomedical, Seattle, WA) to induce apoptosis. Aliquots of 3 × 107 cells were washed once in PBS and lysed on ice for 30 minutes in lysis buffer (1% Triton X-100 in 150 mM NaCl, 10 mM sodium phosphate [pH 7.2], 2 mM EDTA, 50 mM sodium fluoride, and 100 units/mL aprotinin). After centrifugation at 20 000g for 10 minutes at 4°C, supernatants were diluted 10-fold into detergent-free lysis buffer, and lysates were precleared with normal mouse immunoglobulin G (IgG, 10 μg/mL) and protein G beads (20 μL per mL). PLSCR1 was immunoprecipitated with antibody 4D2 and captured on protein G beads. Normal mouse IgG served as control. Beads were washed 5 times in lysis buffer with 0.1% Triton X-100 and added to hot sample buffer. Samples were separated on 18% Tris-glycine gels, transferred to PVDF membranes, and either visualized by autoradiography or immunoblotted with anti-PLSCR1 antibody. For assessment of apoptosis, cell-surface exposure of PtdSer was quantified by incubation with factor Va light chain, and samples were analyzed by flow cytometry as described.7
Results
ATRA induces PLSCR1 expression in ATRA-sensitive NB4 cells but not in ATRA-resistant NB4-derived LR1 cells
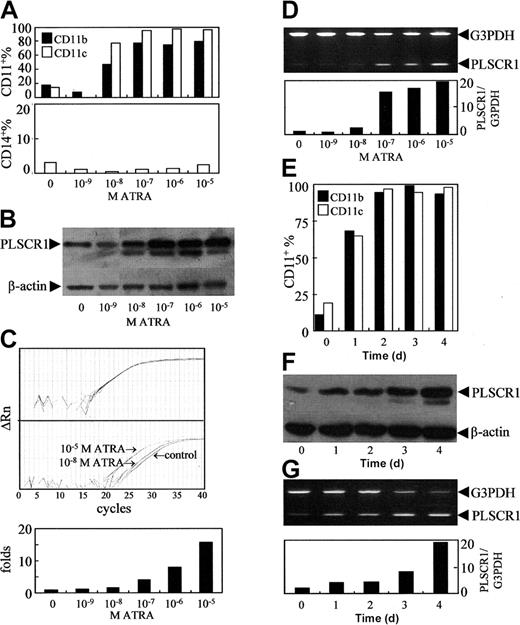
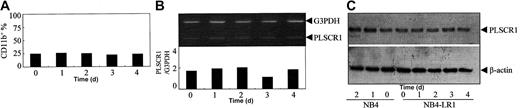
As previously reported,27 ATRA at concentrations of 10-8 to 10-5 M was found to induce NB4 cells to undergo granulocytic differentiation, as assessed by morphology (data not shown) and by analysis of granulocytic differentiation–related antigens CD11b+ and CD11c+/CD14- cells (Figure 1A). Since PLSCR1 has been implicated in the maturation of granulocytes, we evaluated possible alterations of PLSCR1 expression during ATRA-induced differentiation. The results revealed that ATRA at a concentration of 10-9 M, which did not induce differentiation, failed to modulate the expression of PLSCR1 (Figure 1B-D). However, differentiation-inducing concentrations (10-8 to 10-5 M) of ATRA dose-dependently elevated PLSCR1 protein (Figure 1B) as well as PLSCR1 mRNA levels, as evidenced by real-time quantitative PCR (Figure 1C) and semiquantitative RT-PCR (Figure 1D). Moreover, the time course of ATRA-induced expression of PLSCR1 (Figure 1F-G) closely paralleled that of ATRA-induced differentiation of NB4 cells (Figure 1E). Next, we treated NB4-derived maturation-resistant NB4-LR1 cells with 10-6 M ATRA. As shown in Figure 2A, the cell line was resistant to ATRA-induced differentiation.22,28 Furthermore, ATRA also failed to modulate PLSCR1 expression at either the mRNA (Figure 2B) or the protein level (Figure 2C) in these cells, providing further evidence for the association between ATRA-induced PLSCR1 expression and granulocytic differentiation.
Effects of ATRA on PLSCR1 expression in NB4 cells. NB4 cells were treated with the indicated concentrations of ATRA for 3 days (A-D), or with 10-6 M ATRA for days shown (E-G). CD11b+, CD11c+, and/or CD14+ cells (A,E) were measured by flow cytometry as described in “Materials and methods.” PLSCR1 protein (B,F) was detected by Western blot with β-actin as loading control. PLSCR1 mRNA was detected by real-time quantitative PCR and semiquantitative RT-PCR. (C) Typical amplification plots for PLSCR1 and β-actin showing how their relative expression levels can be assayed by real-time RT-PCR using cDNA template derived by reverse-transcriptase treatment of RNA from a single sample. Upper panel shows that amplification plots for β-actin in different multiplex tubes used to assay PLSCR1 are closely superimposed within experimental error. Middle panel shows amplification plots for PLSCR1 in untreated control, and samples treated with 10-5 M or 10-8 M ATRA; the difference between these assays at the cycle threshold detection line represents the ΔΔCt value when the β-actin results superimpose exactly. Lower panel shows the PLSCR1 mRNA level detected by real-time quantitative PCR under the indicated concentrations of ATRA. For semiquantitative RT-PCR (D,G), the signal intensities of amplified PLSCR1 fragments were normalized against 983-bp G3PDH using a densitometer. Each point represents the mean from triplicate samples with a variance of less than 15%. All experiments were repeated at least 3 times with similar results.
Effects of ATRA on PLSCR1 expression in NB4 cells. NB4 cells were treated with the indicated concentrations of ATRA for 3 days (A-D), or with 10-6 M ATRA for days shown (E-G). CD11b+, CD11c+, and/or CD14+ cells (A,E) were measured by flow cytometry as described in “Materials and methods.” PLSCR1 protein (B,F) was detected by Western blot with β-actin as loading control. PLSCR1 mRNA was detected by real-time quantitative PCR and semiquantitative RT-PCR. (C) Typical amplification plots for PLSCR1 and β-actin showing how their relative expression levels can be assayed by real-time RT-PCR using cDNA template derived by reverse-transcriptase treatment of RNA from a single sample. Upper panel shows that amplification plots for β-actin in different multiplex tubes used to assay PLSCR1 are closely superimposed within experimental error. Middle panel shows amplification plots for PLSCR1 in untreated control, and samples treated with 10-5 M or 10-8 M ATRA; the difference between these assays at the cycle threshold detection line represents the ΔΔCt value when the β-actin results superimpose exactly. Lower panel shows the PLSCR1 mRNA level detected by real-time quantitative PCR under the indicated concentrations of ATRA. For semiquantitative RT-PCR (D,G), the signal intensities of amplified PLSCR1 fragments were normalized against 983-bp G3PDH using a densitometer. Each point represents the mean from triplicate samples with a variance of less than 15%. All experiments were repeated at least 3 times with similar results.
Effects of ATRA on PLSCR1 expression in the maturation-resistant leukemic cell line NB4-LR1. NB4-LR1 cells were treated with 10-6 M ATRA for the times indicated. Quantification of CD11b+ cells (A), semiquantitative RT-PCR for PLSCR1 mRNA (B), and Western blot for PLSCR1 (C) were performed as detailed in “Materials and methods.” For Western blot of PLSCR1, NB4 cells treated without or with 10-6 M ATRA for 1 to 2 days were used as controls, demonstrating ATRA-induced up-regulation of PLSCR1 in NB4 cells.
Effects of ATRA on PLSCR1 expression in the maturation-resistant leukemic cell line NB4-LR1. NB4-LR1 cells were treated with 10-6 M ATRA for the times indicated. Quantification of CD11b+ cells (A), semiquantitative RT-PCR for PLSCR1 mRNA (B), and Western blot for PLSCR1 (C) were performed as detailed in “Materials and methods.” For Western blot of PLSCR1, NB4 cells treated without or with 10-6 M ATRA for 1 to 2 days were used as controls, demonstrating ATRA-induced up-regulation of PLSCR1 in NB4 cells.
Increase in PLSCR1 expression is not limited to ATRA-induced granulocytic differentiation
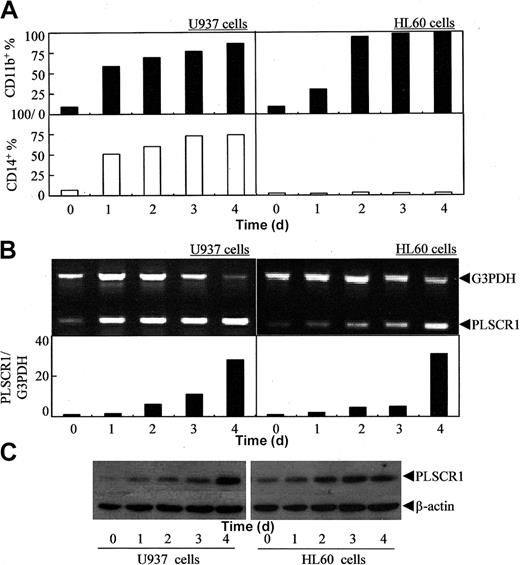
To investigate whether ATRA-induced PLSCR1 expression is specific for ATRA-induced granulocytic differentiation, we treated 2 different AML cell lines, U937 and HL60, with 10-6 M ATRA. Under the conditions of these experiments, U937 cells differentiated to monocytes (represented by CD11b+/CD14+, Figure 3A, left),29 whereas HL60 cells differentiated to granulocytes (represented by CD11b+/CD14-, Figure 3A, right).30 PLSCR1 expression was found to be significantly elevated by ATRA, irrespective of whether ATRA induced granulocytic (HL60) or monocytic (U937) differentiation (Figure 3B-C).
Effects of ATRA on PLSCR1 expression in leukemic cell lines U937 and HL60. U937 and HL60 cells were treated with 10-6 M ATRA for times indicated. CD11b+/CD14+ cells (A), semiquantitative RT-PCR for PLSCR1 mRNA (B), and Western blot for PLSCR1 with β-actin as loading control (C) were performed as detailed in “Materials and methods.”
Effects of ATRA on PLSCR1 expression in leukemic cell lines U937 and HL60. U937 and HL60 cells were treated with 10-6 M ATRA for times indicated. CD11b+/CD14+ cells (A), semiquantitative RT-PCR for PLSCR1 mRNA (B), and Western blot for PLSCR1 with β-actin as loading control (C) were performed as detailed in “Materials and methods.”
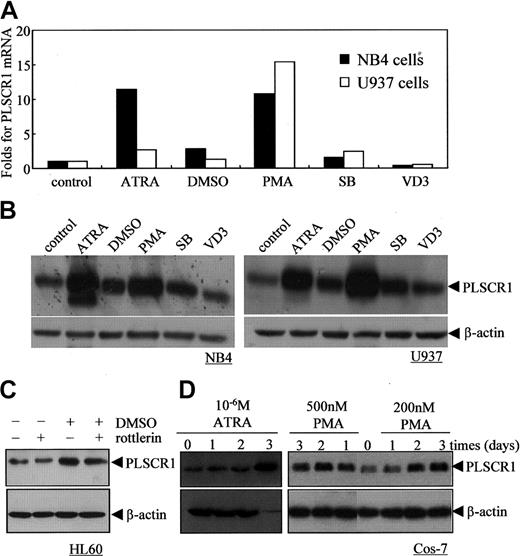
Next, we investigated the effects of other known differentiation-inducing agents, including SB (2 mM), DMSO (1%), PMA (100 nM), and VD3 (2.5 × 10-7 M), on PLSCR1 expression in NB4 and U937 cells (Figure 4A-B). Although SB, a histone deacetylase inhibitor,31 and DMSO32 could effectively induce NB4 and U937 cells to undergo granulocytic differentiation, they only slightly up-regulated PLSCR1 mRNA and protein. VD3, which induced monocytic differentiation,33 did not alter PLSCR1 expression in these cells. By contrast, PMA, which induced monocytic differentiation,34 significantly enhanced PLSCR1 expression comparable with ATRA. Of note, 1% DMSO and 100 nM PMA also increased PLSCR1 expression in HL60 cells (Figure 4C and data not shown). These results suggested that whereas an increase in PLSCR1 expression is observed in ATRA- or PMA-induced cell differentiation, increased PLSCR1 expression might not be necessary when differentiation is induced by other factors in NB4 and U937 cells.
Effects of various differentiation-inducing agents on PLSCR1 expression in leukemic cell lines and Cos-7 cells. (A-B) NB4 and U937 cells were treated with the differentiation inducers 10-6 M ATRA, 1% DMSO, 100 nM PMA, 2 mM SB, or 2.5 × 10-7 M VD3 for 3 days; then real-time PCR (A) and Western blots (B) for PLSCR1 with β-actin as loading control were performed as described in “Materials and methods.” (C) HL60 cells were treated with 1% DMSO and/or 1 μM rottlerin for 24 hours, and PLSCR1 was detected by Western blot with β-actin as loading control. (D) Cos-7 cells were treated with ATRA (10-6 M) or PMA (200 nM and 500 nM) for day(s) shown, and PLSCR1 was detected by Western blot with β-actin as loading control. All experiments were repeated at least 3 times with similar results.
Effects of various differentiation-inducing agents on PLSCR1 expression in leukemic cell lines and Cos-7 cells. (A-B) NB4 and U937 cells were treated with the differentiation inducers 10-6 M ATRA, 1% DMSO, 100 nM PMA, 2 mM SB, or 2.5 × 10-7 M VD3 for 3 days; then real-time PCR (A) and Western blots (B) for PLSCR1 with β-actin as loading control were performed as described in “Materials and methods.” (C) HL60 cells were treated with 1% DMSO and/or 1 μM rottlerin for 24 hours, and PLSCR1 was detected by Western blot with β-actin as loading control. (D) Cos-7 cells were treated with ATRA (10-6 M) or PMA (200 nM and 500 nM) for day(s) shown, and PLSCR1 was detected by Western blot with β-actin as loading control. All experiments were repeated at least 3 times with similar results.
Role of PKCδ in ATRA/PMA-induced PLSCR1 expression
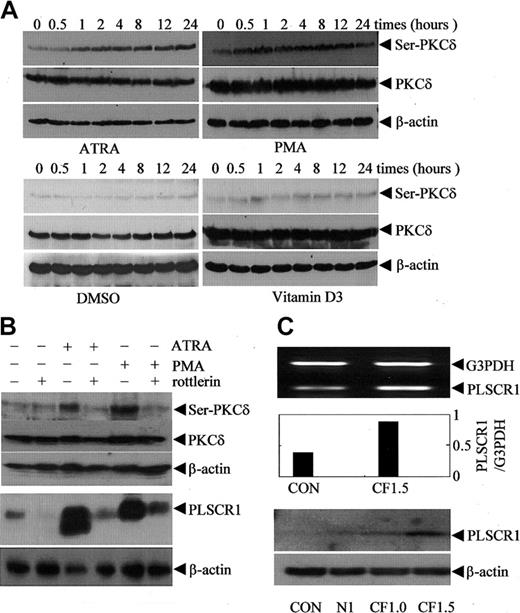
The increase in PLSCR1 expression induced by ATRA and PMA was not restricted to leukemic cells. As shown in Figure 4D, both ATRA at 10-6 M and PMA at 2 to 5 × 10-7 M also increased expression of PLSCR1 in Cos-7 cells. This suggested that ATRA and PMA might regulate PLSCR1 expression via a common mechanism that is independent of the cell type. As depicted in Figure 5A, both PMA and ATRA, but not DMSO and VD3, activated PKCδ, as evidenced by phosphorylation of PKCδ on Serine 643 in NB4 cells (Figure 5A) and U937 cells (data not shown). Thus, we speculated that PKCδ might contribute to the up-regulation of PLSCR1 by ATRA and PMA. Therefore, we treated NB4 and U937 cells with 100 nM PMA or 10-6 M ATRA with or without rottlerin, a specific inhibitor of PKCδ.35 The results revealed that inhibition of PKCδ phosphorylation by rottlerin almost completely abrogated PMA- and ATRA-induced PLSCR1 expression (Figure 5B). It is noteworthy that rottlerin inhibited DMSO-induced PLSCR1 expression in HL60 cells only slightly (Figure 4C), suggesting that PLSCR1 expression was regulated by a mechanism that was predominantly independent of PKCδ under these conditions. To further investigate whether PKCδ is involved in the regulation of PLSCR1 expression, we transfected the catalytic fragment of PKCδ (CFδ)23 into Cos-7 cells, which express low basal levels of PLSCR1. As shown in Figure 5C, ectopic expression of CFδ dose-dependently induced the expression of PLSCR1.
Association of protein kinase Cδ phosphorylation/activation with PLSCR1 expression. (A) After treatment with 10-6 M ATRA, 100 nM PMA, 1% DMSO, or 2.5 × 10-7 M VD3 for the times indicated, NB4 cells were extracted by phosphorylation lysis buffer for Western blots. The blots were probed with antiphospho-PKC-δ (Ser643); then the blots were stripped and reprobed with antibody against PKC-δ. (B, top) After preincubation for 2 hours in the presence or absence of 4 μM rottlerin, NB4 cells were treated with 10-6 M ATRA or 100 nM PMA for 8 hours. After lysis with phosphorylation lysis buffer, equal amounts of total cell lysates were analyzed by Western blot with antiphospho-PKC-δ (Ser643); then the blots were stripped and reprobed with antibody against PKC-δ. (B, bottom) After preincubation for 2 hours in the presence or absence of 1 μM rottlerin, NB4 cells were treated with 10-6 M ATRA or 100 nM PMA for 3 days. After lysis with ice-cold lysis buffer (see “Materials and methods”), equal amounts of total cell lysates were analyzed by Western blot for PLSCR1. In all cases, β-actin served as loading control. Similar results were observed for U937 cells (data not shown). (C) Cos-7 cells were transiently transfected without (CON) or with 1.5 μg empty vector pEGFP-N1 (N1), or 1 μg (CF1, supplemented by 0.5 μg empty vector) and 1.5 μg (CF1.5) pEGFP-CF-PKCδ containing the catalytic fragment of PKCδ. At 48 hours after transfection, semiquantitative RT-PCR (top and middle) and Western blot (bottom) for PLSCR1 were performed.
Association of protein kinase Cδ phosphorylation/activation with PLSCR1 expression. (A) After treatment with 10-6 M ATRA, 100 nM PMA, 1% DMSO, or 2.5 × 10-7 M VD3 for the times indicated, NB4 cells were extracted by phosphorylation lysis buffer for Western blots. The blots were probed with antiphospho-PKC-δ (Ser643); then the blots were stripped and reprobed with antibody against PKC-δ. (B, top) After preincubation for 2 hours in the presence or absence of 4 μM rottlerin, NB4 cells were treated with 10-6 M ATRA or 100 nM PMA for 8 hours. After lysis with phosphorylation lysis buffer, equal amounts of total cell lysates were analyzed by Western blot with antiphospho-PKC-δ (Ser643); then the blots were stripped and reprobed with antibody against PKC-δ. (B, bottom) After preincubation for 2 hours in the presence or absence of 1 μM rottlerin, NB4 cells were treated with 10-6 M ATRA or 100 nM PMA for 3 days. After lysis with ice-cold lysis buffer (see “Materials and methods”), equal amounts of total cell lysates were analyzed by Western blot for PLSCR1. In all cases, β-actin served as loading control. Similar results were observed for U937 cells (data not shown). (C) Cos-7 cells were transiently transfected without (CON) or with 1.5 μg empty vector pEGFP-N1 (N1), or 1 μg (CF1, supplemented by 0.5 μg empty vector) and 1.5 μg (CF1.5) pEGFP-CF-PKCδ containing the catalytic fragment of PKCδ. At 48 hours after transfection, semiquantitative RT-PCR (top and middle) and Western blot (bottom) for PLSCR1 were performed.
PLSCR1 is not phosphorylated by PKCδ
The observation that PKCδ could up-regulate PLSCR1 (Figure 5C) raised the question of whether PLSCR1 was being phosphorylated by PKCδ, as had been reported for in vitro phosphorylation of PLSCR1 immunoprecipitated from cells, and for in vivo phosphorylation in apoptotic Jurkat cells.14 For these experiments, PLSCR1 purified from human erythrocytes was used as substrate in an in vitro phosphorylation assay with recombinant PKCδ. As shown in Figure 6A, phosphorylation of PLSCR1 was not observed under these conditions, even though autophosphorylation of PKCδ and phosphorylation of histone were readily detected. Intracellular phosphorylation of PLSCR1 was also not observed when apoptosis was induced in Jurkat cells by Fas ligation (Figure 6B), in contrast to the report by Frasch et al.14 Taken together, our results suggest that PLSCR1 is not a substrate for phosphorylation by PKCδ.
PLSCR1 is not phosphorylated by PKCδ in vitro and is not phosphorylated in apoptotic Jurkat cells. (A) PLSCR1 was incubated with recombinant PKCδ in an in vitro phosphorylation assay as described in “Materials and methods.” Proteins were resolved by SDS-PAGE and visualized by autoradiography (top panel), or immunoblotting for PLSCR1 with monoclonal antibody 4D2 (bottom panel). Histone served as positive control as a substrate for PKCδ (top, right). Note also autophosphorylation of PKCδ. (B) Jurkat T cells were incubated in medium containing [γ-32P] ATP for 2 hours at 37°C in absence (lane 1) or presence (lanes 2-3) of anti-Fas antibody to induce apoptosis. Samples were immunoprecipitated with anti-PLSCR1 antibody 4D2 (lanes 1,3) or normal mouse IgG (lane 2). Immunoprecipitates and an aliquot of cell lysate were separated by SDS-PAGE, transferred to PVDF membranes, and either immunoblotted for PLSCR1 (left top) or visualized by autoradiography (left bottom). Extent of apoptosis was assessed by quantifying PtdSer exposure on Jurkat cells following incubation for 2 hours in presence (bold line) or absence (thin line) of anti-Fas, as measured by the binding of factor Va light chain (right panel). Histogram of mean fluorescence is shown. See “Materials and methods” for details.
PLSCR1 is not phosphorylated by PKCδ in vitro and is not phosphorylated in apoptotic Jurkat cells. (A) PLSCR1 was incubated with recombinant PKCδ in an in vitro phosphorylation assay as described in “Materials and methods.” Proteins were resolved by SDS-PAGE and visualized by autoradiography (top panel), or immunoblotting for PLSCR1 with monoclonal antibody 4D2 (bottom panel). Histone served as positive control as a substrate for PKCδ (top, right). Note also autophosphorylation of PKCδ. (B) Jurkat T cells were incubated in medium containing [γ-32P] ATP for 2 hours at 37°C in absence (lane 1) or presence (lanes 2-3) of anti-Fas antibody to induce apoptosis. Samples were immunoprecipitated with anti-PLSCR1 antibody 4D2 (lanes 1,3) or normal mouse IgG (lane 2). Immunoprecipitates and an aliquot of cell lysate were separated by SDS-PAGE, transferred to PVDF membranes, and either immunoblotted for PLSCR1 (left top) or visualized by autoradiography (left bottom). Extent of apoptosis was assessed by quantifying PtdSer exposure on Jurkat cells following incubation for 2 hours in presence (bold line) or absence (thin line) of anti-Fas, as measured by the binding of factor Va light chain (right panel). Histogram of mean fluorescence is shown. See “Materials and methods” for details.
Inhibition of PLSCR1 expression by siRNA partially blocks ATRA- and PMA-induced differentiation
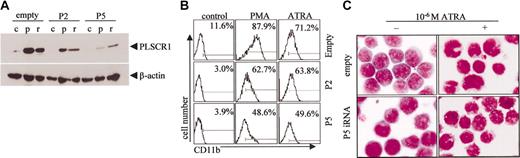
We investigated a possible role for PLSCR1 in leukemic cell differentiation by blocking PLSCR1 expression with siRNA. Of the 5 target sequences we selected to silence PLSCR1 expression, stable transfection of U937 cells with P2 and P5 reduced basal, and significantly inhibited ATRA- and PMA-induced PLSCR1 expression (Figure 7A and data not shown). Of note, P5 siRNA was more effective in suppressing PLSCR1 expression than P2 siRNA. Interestingly, stable transfection with P2 and particularly P5 siRNA also significantly inhibited ATRA- and PMA-induced cell differentiation, as evidenced by CD11b expression (Figure 7B) and morphologic features (Figure 7C), while differentiation induced by DMSO, VD3, and SB was not affected (data not shown), strongly indicating a role for PLSCR1 in ATRA/PMA-induced leukemic cell differentiation.
Effects of silencing PLSCR1 expression by siRNA on ATRA/PMA-induced leukemic cell differentiation. U937 cells stably transfected with empty vector or P2/P5 siRNA-carrying vectors were treated without (c) or with 10-6 M ATRA (r) or 100 nM PMA (p) for 3 days. (A) PLSCR1 was detected by Western blot with β-actin as loading control. (B) CD11b+ cells were measured by flow cytometry. In the histograms, each value represents the mean from triplicate samples with a variance of less than 15%. (C) Cells were collected onto slides by cytospin, stained by Wright staining, and observed under microscope (100×/1.30 oil). PMA-induced morphologic differentiation was also inhibited by P5 iRNA (data not shown). All experiments were repeated at least 3 times with similar results.
Effects of silencing PLSCR1 expression by siRNA on ATRA/PMA-induced leukemic cell differentiation. U937 cells stably transfected with empty vector or P2/P5 siRNA-carrying vectors were treated without (c) or with 10-6 M ATRA (r) or 100 nM PMA (p) for 3 days. (A) PLSCR1 was detected by Western blot with β-actin as loading control. (B) CD11b+ cells were measured by flow cytometry. In the histograms, each value represents the mean from triplicate samples with a variance of less than 15%. (C) Cells were collected onto slides by cytospin, stained by Wright staining, and observed under microscope (100×/1.30 oil). PMA-induced morphologic differentiation was also inhibited by P5 iRNA (data not shown). All experiments were repeated at least 3 times with similar results.
Discussion
Since the successful introduction of ATRA for the treatment of APL 15 years ago, a potentially less toxic cancer therapeutic strategy known as “differentiation therapy” has been developed, which uses drugs to induce cancer cells to undergo terminal differentiation, thus preventing their further proliferation.36-38 Therefore, understanding the mechanisms by which ATRA and other agents induce leukemic cell differentiation has attracted significant attention. Because PLSCR1 has previously been implicated in the proliferation and terminal differentiation of myeloid precursor cells,10 and a truncated mutation of PLSCR1 has been reported to confer a leukemogenic phenotype,3 we sought to investigate the role of differentiation-inducing agents on cellular PLSCR1 expression. Nakamaki et al18 reported the specific induction of PLSCR1 mRNA upon granulocytic differentiation of the promyelocytic leukemia NB4 and HT93 cells by ATRA. By contrast, no increase in PLSCR1 mRNA was observed when the bipotential myeloid leukemia HL-60 cells were induced to differentiate toward monocytes/macrophages, during erythroid differentiation induced by hemin in erythroid leukemia K562 and HEL cells or during megakaryocytic differentiation induced by PMA in K562 cells. In the present study, we showed that pharmacologic concentrations of ATRA elevated PLSCR1 mRNA and protein levels in NB4 and HL-60 cells with the induction of differentiation toward granulocytes. However, by contrast to the report of Nakamaki et al,18 PLSCR1 was also up-regulated upon ATRA-induced monocytic differentiation of U937 cells. Additionally, PMA, which induced these cells (NB4, U937 and HL60) to differentiate toward the monocytic phenotype, also potently enhanced PLSCR1 expression. Moreover, only minimal or no elevation of PLSCR1 was observed upon treatment with other granulocytic or monocytic differentiation-inducing agents, including DMSO, SB, or VD3 in NB4 and U937 cells, although elevated expression of PLSCR1 was seen in DMSO-treated HL60 cells. These results indicated that a downstream signaling pathway common to both ATRA and PMA might contribute to the regulation of PLSCR1 expression, which was further supported by the fact that PMA and ATRA also up-regulated PLSCR1 expression in Cos-7 cells. Since PMA is a strong activator of many isoforms of PKC39 and ATRA has been shown to directly bind to PKC isozymes and modulate the activity of PKCδ,40,41 we hypothesized that PKCδ was mediating the response of PLSCR1 expression to these 2 differentiation-inducing agents. Indeed, the PKCδ-specific inhibitor rottlerin almost completely abrogated ATRA- and PMA-induced PLSCR1 up-regulation, and ectopic expression of an active form of PKCδ induced PLSCR1 expression, providing the first evidence for a role for PKCδ in PLSCR1 expression. The mechanism by which PKCδ up-regulates PLSCR1 expression remains to be investigated. We speculate that it is likely indirect, and not through phosphorylation of PLSCR1. Although PKCδ has been previously reported to phosphorylate PLSCR1 with consequent activation of PL scramblase activity when coexpressed in Chinese hamster ovary (CHO) cells, or in apoptotic cells,14 we have been unable to demonstrate PKCδ-mediated phosphorylation of purified PLSCR1 directly in vitro, and by contrast to the report by Frasch et al,14 we did not observe phosphorylation of PLSCR1 in apoptotic Jurkat cells. The discrepancy remains unresolved.
In a recent report, Kambhampati et al41 demonstrated activation of PKCδ upon ATRA-induced differentiation of NB4 and HL-60 cells, and inhibition of PKCδ activity abrogated ATRA-induced cell differentiation, suggesting a critical role for PKCδ in mediating the biologic effects of ATRA in malignant cells. Furthermore, they showed that PKCδ forms a complex with the retinoic acid receptor α (RARα) and binds to retinoic acid–responsive elements (RAREs), and that inhibition of PKCδ blocked ATRA-dependent gene transcription via RARE. Exactly how activation of PKCδ mediates leukemic cell differentiation is unknown. Here we demonstrate that in maturation-resistant, ATRA-responsive NB4-LR1 cells,22,28 up-regulation of PLSCR1 by ATRA is not observed, suggesting a possible role for PLSCR1 in ATRA-induced cell differentiation. Consistent with this, silencing of PLSCR1 expression with siRNA inhibits ATRA- and PMA-induced cell differentiation, as assessed by morphologic (Figure 7C) and functional (increased CD11b expression, Figure 7B) criteria. Of note, antisense PLSCR1 transfection was also shown to significantly suppress ATRA-induced differentiation of NB4 cells.18 Taken together, these results indicate that as a protein that is induced upon PKCδ activation, PLSCR1 is required for leukemic cell differentiation by these agents. Whether PLSCR1 affects cell differentiation through its activity at the plasma membrane or whether ATRA- and PMA-induced PLSCR1 exerts its effects following its translocation into the nucleus17 remains to be elucidated.
Prepublished online as Blood First Edition Paper, August 12, 2004; DOI 10.1182/blood-2004-04-1630.
Supported in part by the National Key Program (973) for Basic Research of China (NO2002CB512806 and NO2002CB512805; G.-Q.C. and Q.Z.), the Key Project for International Collaboration of Ministry of Science and Technology of China (2003DF000038; G.-Q.C.), 100-Talent Program of the Chinese Academy of Sciences (G.-Q.C.), grants (02DJ14008 and 03XD14016; G.-Q.C.) and Prestar program (Q.Z.) from Science and Technology Committee of Shanghai, and grants HL036946 and HL063819 (P.J.S.) from the National Institutes of Health.
K.-W.Z, X.L., and Q.Z. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This article is dedicated to Dr Zhen-Yi Wang, who first discovered clinical efficacy of ATRA in the treatment of APL by differentiation induction on the occasion of his 80th birthday. Ke-Wen Zhao is a PhD candidate at Shanghai Institutes for Biological Sciences, and this work is submitted in partial fulfillment of the requirement for the PhD.

![Figure 6. PLSCR1 is not phosphorylated by PKCδ in vitro and is not phosphorylated in apoptotic Jurkat cells. (A) PLSCR1 was incubated with recombinant PKCδ in an in vitro phosphorylation assay as described in “Materials and methods.” Proteins were resolved by SDS-PAGE and visualized by autoradiography (top panel), or immunoblotting for PLSCR1 with monoclonal antibody 4D2 (bottom panel). Histone served as positive control as a substrate for PKCδ (top, right). Note also autophosphorylation of PKCδ. (B) Jurkat T cells were incubated in medium containing [γ-32P] ATP for 2 hours at 37°C in absence (lane 1) or presence (lanes 2-3) of anti-Fas antibody to induce apoptosis. Samples were immunoprecipitated with anti-PLSCR1 antibody 4D2 (lanes 1,3) or normal mouse IgG (lane 2). Immunoprecipitates and an aliquot of cell lysate were separated by SDS-PAGE, transferred to PVDF membranes, and either immunoblotted for PLSCR1 (left top) or visualized by autoradiography (left bottom). Extent of apoptosis was assessed by quantifying PtdSer exposure on Jurkat cells following incubation for 2 hours in presence (bold line) or absence (thin line) of anti-Fas, as measured by the binding of factor Va light chain (right panel). Histogram of mean fluorescence is shown. See “Materials and methods” for details.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/12/10.1182_blood-2004-04-1630/5/m_zh80230470040006.jpeg?Expires=1769089271&Signature=BEDY5kywPZcL47mncSiUvHbRpJ4cJicTL9l~UT3PzebZY6U~mGyXkH1TsurUn0rKTt4CtI1xshJ8XAeu6-hT3J6dkV3bFd~sN92cyr9Dn9MIqmFRq6ZC40BKKE4M2fBXjjXCGEWkRi0zDxB~SeqxXoU57NKmj0mCb2~fgkzd3HC1J-1DYQzFfEbpYq7s0gzz~0hvGd2THlFo-ftGLLQNQf244UZvdhL-2wsSnbVG0JIN92MP5abN6lTmrF-MS48zssb9ym6PnYX4I0KmiBZNBbF125qknKq-sUxybrlQU5acF6GSfQFfX1MX0u7MLJw82HbCCU6Is9pcbZKQ3B9M5g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal