Abstract
The Wiskott-Aldrich syndrome (WAS) is an X-linked recessive immune deficiency disorder characterized by thrombocytopenia, small platelet size, eczema, recurrent infections, and increased risk of autoimmune disorders and malignancies. X-linked thrombocytopenia (XLT) is an allelic variant of WAS which presents with a milder phenotype, generally limited to thrombocytopenia. WAS and XLT are caused by mutations of the Wiskott-Aldrich syndrome protein (WASP) gene which encodes a 502-amino acid protein, named WASP. WASP is thought to play a role in actin cytoskeleton organization and cell signaling. Here, we report the identification of 141 unique mutations, 71 not previously reported, from 227 WAS/XLT families with a total of 262 affected members. When possible we studied the effects of these mutations on transcription, RNA splicing, and protein expression. By analyzing a large number of patients with WAS/XLT at the molecular level we identified 5 mutational hotspots in the WASP gene and have been able to establish a strong association between genotype and phenotype. (Blood. 2004;104:4010-4019)
Introduction
The Wiskott-Aldrich syndrome (WAS) (Online Mendelian Inheritance in Man [OMIM] 301000) is a rare X-linked recessive immunodeficiency disorder characterized by thrombocytopenia and small-sized platelets, eczema, recurrent bacterial and viral infections, a high incidence of autoimmunity, and an increased risk of malignancies.1,2 Immunologic abnormalities characteristic for patients with WAS involve both B- and T-cell function3 and include defective monocyte chemotaxis4 as well as abnormal morphology of stimulated dendritic cells.5 Most affected infants have normal numbers of circulating lymphocytes but often develop lymphopenia by age 6 to 8 years or earlier3 possibly because of increased apoptosis.6,7 Patients with the classic WAS phenotype fail to respond to polysaccharide and to selected protein antigens.3 X-linked thrombocytopenia (XLT) (OMIM 313900) is a relatively benign disorder characterized by congenital thrombocytopenia and small platelets, easy bruising, sometimes associated with transient eczema. The data presented here indicate that the prevalence of XLT equals that of WAS. The incidence of WAS/XLT, estimated to be 10 in 1 million live births,8 has to be updated when new data from the US and European registries become available. The gene responsible for WAS, the Wiskott-Aldrich Syndrome protein gene (WASP), was cloned and sequenced in l994.9 The WASP gene has 12 exons containing 1823 base pairs and encodes a 502-amino acid protein which is predominantly expressed in hematopoietic cells.9 It was subsequently shown that mutations of the WASP gene can cause X-linked thrombocytopenia, indicating that XLT is a mild allelic variant of WAS.10,11 More recently, it was recognized that mutations within the Cdc42 binding site cause congenital X-linked neutropenia, without the platelet defect or any of the other findings characteristic for WAS/XLT.12
WASP contains several unique domains, including a pleckstrin homology (PH) domain, a guanosine triphosphatase (GTPase) binding domain (GBD), a proline-rich region with SH3 binding motifs, a verprolin and a cofilin homology domain, and the C-terminal acidic region which is involved in the reorganization of the actin cytoskeleton by activating Arp2/3 complex-mediated actin polymerization.13,14 Considering the unique functions of these domains, WASP appears to play critical roles in signal transduction and in the regulation of cytoskeletal reorganization.
Because of the complex structure and multifunctional design of WASP, and the variations in the clinical phenotype resulting from mutations of the WASP gene, a correlation between phenotype and genotype was examined by a number of investigators. A strong association, reported by some15-19 but not by others,2,20,21 may have implications on long-term prognosis and on the selection of therapies with variable risks.
In this analysis, we have examined the molecular defect in 227 unrelated WAS/XLT families studied in 2 centers, Seattle, Washington, and Brescia, Italy. We identified 141 unique mutations, of which 71 had not been previously reported. WASP mutations consisted of single nucleotide substitutions, splice site mutations, insertions, deletions, and complex mutations. A scoring system was designed to identify the clinical phenotype, and the effect of the mutations on transcription and protein expression was determined. Five hotspots, 2 preferentially associated with WAS and 3 with XLT, were identified, and a strong relationship between clinical phenotype and the expression of WASP was established.
Patients, materials, and methods
Patients
Of the 262 patients from 227 families included in this report, 184 (from 161 families) were studied molecularly at the University of Washington, Seattle, and included patients from the United States and Canada (n = 163), Central and South America (n = 5), Europe (n = 4), New Zealand (1), and South East Asia (n = 11), and the remaining 78 patients (from 68 families) were analyzed at the University of Brescia, Italy, and included patients from Europe (n = 68), the Middle East (n = 8), and New Zealand (n = 2) (Table 1). A total of 59 families listed in Table 1 were reported previously10,15,18,22,23 (see footnotes of Table 1). The clinical diagnoses of WAS or XLT were based on typical clinical and laboratory findings, including thrombocytopenia and small platelets, eczema, recurrent frequent or severe infections, and abnormal immune function. When possible, patients were assigned a score based on the clinical severity of their disease, using a previously described scoring system.11,24 Briefly, a score of 1 was assigned to patients with isolated thrombocytopenia and small platelets; a score of 0 to 1 (0.5) if thrombocytopenia was intermittent.23 A score of 2 was assigned to patients with microthrombocytopenia who had a history of localized eczema that responded promptly to standard therapy and/or occasionally suffered from uncomplicated upper respiratory infections. A score of 2 to 3 (2.5) was assigned to patients with microthrombocytopenia, persistent but therapy-responsive eczema, or frequent infections that may be severe enough to require intermittent antibiotic therapy; a score of 3 was given when both criteria, therapy-responsive eczema and frequent infections requiring intermittent antibiotics, were present. A score of 4 was assigned to patients with microthrombocytopenia, persistent and difficult-to-treat eczema requiring continuous treatment with steroid ointment and occasionally oral antibiotics for superinfection of the eczema, and/or severe life-threatening infections that may include abscesses, pneumonia (including Pneumocystis carinii pneumonia), meningitis, sepsis, and recurrent herpes simplex infection. A score of 5 was assigned to patients with WAS/XLT who developed autoimmunity or malignancy (most patients progressing to a score of 5 had classic WAS, some were originally diagnosed as XLT).
WASP mutations in 262 patients from 227 families
Patient . | Origin . | Age, y . | Score . | Exon . | Mutation type . | gDNA mutation . | cDNA mutation (if different from gDNA) . | Predicted protein change . | Western blot . |
|---|---|---|---|---|---|---|---|---|---|
| 1-S | US | 18 | 3 | 1 | Insertion | 41 insA | —†† | fs stop aa 37 | Absent |
| 2-B | Sweden | 6 (dec.) | 4 | 1 | Insertion | 41 insG | — | fs stop aa 37 | ND |
| 3-S | US | 4 | 5 | 1 | Deletion | 56-57delG | — | fs stop aa 44 | Absent |
| 4*-S | US | ? | 3 | 1 | Insertion | 62-64insC | — | fs stop aa 37 | Absent |
| 5-S | US | 0.2 | 3 | 1 | Insertion | 65-69insG | — | fs stop aa 37 | ND |
| 6*-S | US | 5.0 (dec.) | 5 | 1 | Nonsense | 71C>T | — | R13X | Absent |
| 7a*/b-S | US | 1.9/<2 | 4/3** | 1 | Nonsense | 71C>T | — | R13X | Absent |
| 8-B | Italy | 1.6 (dec.) | 4 | 1 | Nonsense | 71C>T | — | R13X | ND |
| 9-B | Italy | 8 | 4 | 1 | Nonsense | 74G>T | — | G14X | ND |
| 10-S | US | 5 | 3-4 | 1 | Missense | 104T>C | — | S24P | Reduced |
| 11-S | Thailand | 12 | 2 | 1 | Missense | 105C>T | ND | S24F | ND |
| 12§-B | Italy | 1.9 (dec.) | 5 | 1 | Deletion | 115-116delC | — | fs stop aa 44 | Absent |
| 13*-S | US | 12 | 2 | 1 | Missense | 113C>T | — | L27F | Reduced |
| 14-S | US | 7.5 | 3-4 | 1 | Deletion | 122-124del | — | Inframe | ND |
| 15-S | US | 1 | 1 | 1 | Missense | 125G>A | — | E31K | Absent |
| 16-S | Canada | 2 | 2 | 1 | Missense | 125G>A | — | E31K | Reduced |
| 17-B | Italy | 7 | 2-3 | 1 | Missense | 125G>A | — | E31K | Absent |
| 18-S | Chile | 0.3 | 1 | 1 | Missense | 125G>A | — | E31K | Reduced |
| 19-S | US | 0.4 | ND | 1 | Missense | 125G>A | — | E31K | Absent |
| 20-S | US | 0.5 | 1 | 1 | Nonsense | 134C>T | — | R34X | ND |
| 21-S | US | 1.1 | 3 | 1 | Nonsense | ND | 134C>T | R34X | Absent |
| 22—S | US | 0.2 | 5 | 1 | Nonsense | 134C>T | — | R34X | Absent |
| 23-B | Turkey | 5 | 2 | 1 | Missense | 138T>A | — | L35H | ND |
| 24*-S | US | 2.1 | 5 | 1 | Missense | 138,139TC>AT | — | L35H | Reduced |
| 25-S | Greece | 0.3 | 1 | 1 | Deletion | 140-142delTT | — | fs stop aa 36 | Absent |
| 26*-S | US | 4 | 4 | 1 | Deletion | 140-142delTT | — | fs stop aa 36 | Absent |
| 27-B | Italy | 2.3 (dec) | 5 | 1 | Deletion | 140-142delTT | — | fs stop aa 36 | ND |
| 28-S | US | 0.4 | ND | 1 | Insertion | 140-142insT | — | fs stop aa 37 | Absent |
| 29*-S | US | 8 | 2 | 1 | Missense | 150T>C | — | L39P | Reduced |
| 30a*/b*-S | US | 16/10 | 2/2 | 1 | Missense | 150T>C | — | L39P | Reduced |
| 31 a/b-B | Italy | 14/11 | 2/2-3 | 1 | Missense | 150T>C | — | L39P | Reduced |
| 32§-B | Italy | 6 (dec.) | 5 | 1 | Nonsense | 155C>T | — | R41X | Absent |
| 33§-B | Italy | 0.7 (dec.) | 5 | 1 | Nonsense | 155C>T | — | R41X | ND |
| 34-B | Italy | 9 | 4 | 1 | Nonsense | 155C>T | — | R41X | ND |
| 35-S | US | 0.2 | 2 | 1 | Nonsense | 155C>T | — | R41X | Absent |
| 36-S | US | 8 | 2 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 37-S | US | 0.3 | 1-2 | 2 | Missense | 168C>T | — | T45M | Absent |
| 38a*/b*-S | Canada | 8/2 | 2/2 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 39*-S | US | 8 | 1 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 40-S | US | ? | ND | 2 | Missense | 168C>T | — | T45M | Reduced |
| 41-B | Croatia | 6 | 2 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 42-S | US | 2.5 | 1 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 43*-S | Polynesia | 8 | 2 | 2 | Missense | 174C>A | — | A47D | Reduced |
| 44*-S | US | 22 | 2 | 2 | Missense | 177C>T | — | T48I | Reduced |
| 45-S | US | 6 | 4 | 2 | Deletion | 186-196del | — | fs stop aa 58 | Absent |
| 46-S | US | 9 | 1 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 47-S | US | 2.7 | 1 | 2 | Missense | 201C>T | — | A56V | ND |
| 48*-S | US | ? | 1 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 49-B | Italy | 16 | 1 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 50a/b-S | US | 75/3 | 1/2 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 51-S | US | 1 | ND | 2 | Deletion | 201-213del | — | fs stop aa 71 | ND |
| 52-S | US | ? | 2 | 2 | Missense | 206C>G | — | P58A | Reduced |
| 53a/b/c∥-B | Italy | 9/7/32 | 0.5/0.5/0.5 | 2 | Missense | 207C>G | — | P58R | Normal |
| 54*-S | Canada | 5 (dec.) | 5 | 2 | Deletion | 206-210delC | — | fs stop aa 75 | ND |
| 55a*/b*-S | US | 5/20 | 5/5 | 2 | Deletion | 211 delT | — | fs stop aa 75 | Absent |
| 56-B | Nicaragua | 1.5 | 4 | 2 | Complex (2 populations) | 211 delT | — | fs stop aa 75 | ND |
| 211T>C | P59P | ||||||||
| 57-B | Italy | 12 (dec.) | 4 | 2 | Missense | 224T>C | — | W64R | ND |
| 58-S | US | 13 (dec.) | 5 | 2 | Deletion | del 237-46 | — | fs stop aa 72 | Absent |
| 59-S | US | 1.2 | 2-3 | 2 | Missense | 252G>A | — | C73Y | Reduced |
| 60-B | Italy | 8 | 3 | 2 | Missense | 252G>A | — | C73Y | ND |
| 61-B | Russia | 1.8 | 2-3 | 2 | Missense | 255T>C | — | F74S | ND |
| 62-S | India | 5 | 2 | 2 | Missense | 257G>A | — | V75M | Reduced |
| 63a*/b*/c*-S | US | 5/4/17 | 2/2/2 | 2 | Missense | 257G>A | — | V75M | Reduced |
| 64a/b-B | Italy | 15/24 | 1/2 | 2 | Missense | 257G>A | — | V75M | Reduced |
| 65-S | US | 3 | 1 | 2 | Missense | 257G>A | — | V75M | Normal |
| 68-S | US | 6 | 2 | 2 | Missense | 261A>C | — | K76T | ND |
| 69-B | Italy | 9 | 2 | 2 | Missense | 263G>C | — | D77H | Reduced |
| 70a/b-B | Italy | 7/3 | 2/1 | 2 | Missense | 264A>G | — | D77G | Reduced |
| 71-S | US | 1 | 2 | 2 | Missense | 278T>C | — | S82P | Reduced |
| 72-B | Serbia | 9 | 5 | 2 | Deletion | 279-280delC | — | fs stop aa 126 | ND |
| 73a/b-B | New Zealand | 14/9 | 2/2 | 2 | Missense | 290C>A | — | R86S | ND |
| 74-S | US | 11 | 2 | 2 | Missense | 290C>G | — | R86G | Reduced |
| 75-S | US, African American | 19 | 2 | 2 | Missense | 290C>T | — | R86C | Absent |
| 76-S | US | 9 | 2-3 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 77-S | US | 4 | 2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 78-S | US | 5 | 2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 79a*/b*-S | US | ? | 2/2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 80*-S | US | ? | 1 | 2 | Missense | 290C>T | — | R86C | ND |
| 81-B | Israel | 8 | 1 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 82-S | US | 2 | 2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 83-B | Turkey | 13 | 2 | 2 | Missense | 290C>T | — | R86C | ND |
| 84-S | Malaysia | 6 | 1 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 85-S | US | 15 | 1-2 | 2 | Missense | 291G>A | — | R86H | Absent |
| 86*-S | US | 10 | 2 | 2 | Missense | 291G>A | — | R86H | Reduced |
| 87-S | US | 0.5 | 2-3** | 2 | Missense | 291G>A | — | R86H | Reduced |
| 88-B | Russia | 17.5 | 3 | 2 | Missense | 291G>A | — | R86H | Reduced |
| 89-S | US | 5.5 | 2 | 2 | Missense | 291G>T | — | R86L | Absent |
| 90*-S | Hungary | 3.5 | 4 | 3 | Deletion | 312-313delGT | ND | ND | ND |
| 91-S | US | 5 | 5 | 3 | Nonsense | 325G>A | — | W97X | Absent |
| 92-S | US | 1.3 | 5 | 3 | Nonsense | 325G>A | — | W97X | Absent |
| 93-S | US | 1.5 | 4 | 3 | Nonsense | 329C>T | ND | Q99X | Absent |
| 94*-S | US | ? | 5 | 3 | Complex | 329C>T | 329C>T/exon3del | Q99X/inframe Exon3del | Absent |
| 95-B | Finland | 2.5 | 1-2 | 3 | Missense | 334G>T | — | E100D | ND |
| 96-B | Italy | 5 | 2 | 3 | Missense | 348T>C | — | L105P | Reduced |
| 97*-S | US | <2 | 2** | 3 | Missense | 354A>G | — | Y107C | Reduced |
| 98-S | US | <2 | 2** | 3 | Deletion | 360-364delC | — | fs stop aa 126 | Absent |
| 99-S | US | 0.5 | 4 | 3 | Deletion | 360-364delC | — | fs stop aa 126 | Absent |
| 100-B | Italy | 7 | 4 | 3 | Insertion | 360-364insC | — | fs stop aa 121 | Absent |
| 101-S | US | ? | 2 | 3 | Insertion | 360-364insC | — | fs stop aa 121 | ND |
| 102-S | US | 2.1 | 5 | 3 | Deletion | 366-370delC | — | fs stop aa 126 | Absent |
| 103-S | US | 0.3 | 5 | 3 | Deletion | 371-372delT | — | fs stop aa 126 | Absent |
| 104-S | US | 0.9 | 1-2 | 3 | Missense | 390G>A | — | G119E | Reduced |
| 105a/b-S | Malaysia | 4/0.2 | 4/1 | 4 | Nonsense | 400C>A | Multiple products | C122X | Absent |
| 106*-S | ? | 5 | 4 | Missense | 407G>A | — | G125R | Absent | |
| 107-S | Greece | 1 | 5 | 4 | Missense | 411T>C | — | L126P | ND |
| 108-S | US | 0.3 | 5 | 4 | Missense | 416T>C | — | F128L | Absent |
| 109-S | US | ? | 5 | 4 | Missense | 416T>C | — | F128L | ND |
| 110*-S | US | 9 | 4 | 4 | Missense | 417T>C | — | F128S | ND |
| 111*-S | US | 8 | 3 | 4 | Complex | 425G>A,431G>A | — | E131K, E133K | Absent |
| 112a*/b*-S | US | 14 (dec.)/3 | 5/4 | 4 | Complex | 425G>A,290C>T | — | E131K,R86C | Absent |
| 113-S | Chile | 2 | 3 | 4 | Missense | 431G>A | — | E133K | ND |
| 114-B | Italy | 3.5 (dec.) | 5 | 4 | Missense | 431G>A | — | E133K | ND |
| 115-S | US | 12 | 2 | 4 | Missense | 433G>T | — | E133D | Reduced |
| 116-S | US | 0.7 | 1 | 4 | Missense | 433G>T | — | E133D | Reduced |
| 117*-S | US | 15 | 4 | 4 | Missense | 435C>T | — | A134V | Reduced |
| 118-S | US | 17 | 2 | 4 | Insertion | C435 ins 6bp | — | 134 DE ins | ND |
| 119-S | US | 2 | 2 | 4 | Complex | 447G>A, IVS11-1g>a | 447G>A, exon 11del | R149Q fs stop aa 543 | Reduced |
| 120-B | Italy | 3.5 (dec.) | 4 | 4 | Deletion | 451-458del | — | fs stop aa 165 | ND |
| 121-B | Italy | 13 (dec.) | 4 | 4 | Insertion | 471-476insA | ND | fs stop aa 168 | ND |
| 122#-S | US | 13 | 2 | 4 | Insertion | 471-476insA | — | fs stop aa 168 | Absent |
| 123-B | Italy | 6 (dec.) | 5 | 4 | Deletion | 485-486delAG | — | fs stop aa 167 | Absent |
| 124a/b‡-B | Serbia | 17/21 | 5/5 | 5 | Insertion | 512-516insC | — | fs stop aa 168 | ND |
| 125-S | US | 4 | 5 | 5 | Insertion | 512-516insC | — | fs stop aa 168 | Absent |
| 126-B | Italy | 22 | 5 | 6 | Deletion | 570delT | — | fs stop aa 260 | ND |
| 127-S | US | ? | 2 | 6 | Deletion | 597-597delC | ND | fs stop aa 260 | ND |
| 128-B | Italy | 3 | 4 | 7 | Deletion | 644delA | — | fs stop aa 260 | Absent |
| 129-S | US | 0.5 (dec.) | 2-3** | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 130a/b-S | US | 1.5/0.8 | 4/3 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 131-S | US | 1 | 5 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 132-S | US | ? | 3-4 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 133-S | US Latino | <2 | 2** | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 134*-S | US | 14 | 4 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 135*-S | US | 11 | 4 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 136-B | Italy | 5 | 4 | 7 | Nonsense | 665C>T | — | R211X | ND |
| 137-B | Italy | 6 | 4 | 7 | Nonsense | 665C>T | — | R211X | ND |
| 138-B | Italy | 2.1 (dec.) | 4 | 7 | Nonsense | 665C>T | — | R211X | ND |
| 139*-S | US | 1.9 | 4 | 7 | Splice | 705A>G | 705-768 del | fs stop aa 239 | Absent |
| 140-S | US | >5 | 5 | 7 | Splice | 705A>G | 705-768 del | fs stop aa 239 | Absent |
| 141-B | Sweden | <2 | 2** | 7 | Nonsense | 717C>G | — | S228X | ND |
| 142-S | US | 4 | 3-4 | 7 | Deletion | 731delA | — | fs stop aa 260 | Absent |
| 143‡B | Italy | 15 | 1 | 7 | Missense | 741C>G | — | A236G | Reduced |
| 144-S | US | 0.7 | ND | 7 | Complex | Exon 7(−5 to IVS7 +22) (del 27 bp) | 764-768 del. IVS7 +22 to 71 (ins 49 bp) | fs stop aa 257 | Absent |
| 145§-B | England | 4 (dec.) | 5 | 9 | Nonsense | 887G>T | — | E285X | ND |
| 146-B | Italy | 10 | 2 | 9 | Deletion | 907delC | — | fs stop aa 291 | ND |
| 147*-S | US | 2.5 | 5 | 9 | Nonsense | 923C>T | — | Q297X | Absent |
| 148-S | Turkey | 1.2 | 3 | 9 | Nonsense | 950G>T | — | E306X | Reduced |
| 149a*/b*†-S | US | ?/1.0 | 2/2 | 9 | Missense | 953A>G | — | M307V | ND |
| 150a*/b*-S | India | 3/1 | 3/2** | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 151a*/b*-S | US | ? | 5/5 | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 152-B | Italy | 7 | 5 | 10 | Nonsense | 995C>T | — | R321X | ND |
| 153-S | US | ? | ND | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 154a/b-S | US Latino | 7.0/0.2 | ND | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 155-S | US | 1 | 2 | 10 | Deletion | 1017-1018delC | Multiple products | Multiple products | Absent |
| 156a*/b-S | US | 3/0.5 | 2/2 | 10 | Insertion | 1025insA | ND | fs stop aa 335 | Absent |
| 157-B | Italy | 3 (dec.) | 5 | 10 | Deletion | 1028delG | ND | fs stop aa 444 | Reduced |
| 158*-S | US | ? | 2 | 10 | Complex | 1029insT | del 966-1121 (50%) | inframe del 52aa | Red/trunc |
| del exon 10 (25%) | fs stop aa358 | ||||||||
| fs stop aa335 | |||||||||
| 1029 insT (25%) | |||||||||
| 159*-S | US | 10 | 4 | 10 | Deletion | 1030-1035delG | — | fs stop aa 444 | Absent |
| 160-S | US | ? | ND | 10 | Deletion | 1030-1035delG | — | fs stop aa 444 | Absent |
| 161-S | Brasil | 2.2 | ND | 10 | Deletion | 1030-1035delG | — | fs stop aa 444 | ND |
| 162a/b-S | US | 7.0/5 | 4/4 | 10 | Deletion | 1088-1092delC | Multiple products | Multiple products, fs stop aa 444 | Absent |
| 163-S | US | 4 | 5 | 10 | Deletion | 1088-1092delC | ND | Multiple products, fs stop aa 444 | ND |
| 164-S | New Zealand | 10 | 2 | 10 | Deletion | 1107-1108delGA | Multiple products | Multiple products, fs stop aa 493 | Red/trunc |
| 165*-S | US | 8 | 5 | 10 | Deletion | 1109-1113delC | — | fs stop aa 444 | Absent |
| 166*-S | US | 6 | 4 | 10 | Complex | 1109C>A, 1110-1113delC | — | P359T, fs stop aa 444 | Red/trunc |
| 167-B | Turkey | 5 | 4 | 10 | Missense | 1115C>A | — | P361T | ND |
| 168-S | Malaysia | 9 | 5 | 10 | Deletion | 1115-1119delC | — | fs stop aa 444 | ND |
| 169§-B | Italy | <2 | 2** | 10 | Complex | 1115-1119insC | — | fs stop aa 494 | ND |
| (2 populat.) | 1110-1182del | infr.del24aa | |||||||
| 170-B | Turkey | 12 | 5 | 10 | Nonsense | 1124C>T | — | R364X | ND |
| 171-S | US | 2 | 3 | 10 | Deletion | 1145-1149delC | Exon 10 del | fs stop aa 444 | Absent |
| 172-B | Italy | 10 | 5 | 10 | Deletion | 1184-1185delC | — | fs stop aa 444 | Absent |
| 173-S | Canada | 0.5 | 3 | 10 | Insertion | 1187-1191 insC | — | fs stop aa 494 | ND |
| 174-B | Bosnia | 4 | 5 | 10 | Deletion | 1192delT | — | fs stop aa 444 | Absent |
| 175-S | Puerto Rico | 1 | 4 | 10 | Insertion | 1286 ins4bp | — | fs stop aa 495 | Absent |
| 176*-S | US | 1.8 | 4 | 10 | Insertion | 1301-1305 insG | — | fs stop aa 494 | Red/trunc |
| 177*-S | US Latino | 8 | 5 | 10 | Insertion | 1301-1305 insG | — | fs stop aa 494 | Red/trunc |
| 178*-S | US | 14 | 5 | 10 | Deletion | 1301-1305 insG | — | fs stop aa 494 | Red/trunc |
| 179∥-B | Italy | 9 | 0.5 | 11 | Missense | 1476T>A | — | 1481N | normal |
| 180-B | Italy | 7 | 5 | 11 | Insertion | 1486-1487insTC | — | fs lack of term. codon | ND |
| 181-B | Russia | 6 | 5 | 12 | Deletion | 1519delT | — | fs lack of term. codon | Absent |
| 182-S | US | 2.5 | 1 | 12 | Missense | 1542G>C | — | X503S | Absent |
| 183-B | Turkey | 8.5 | 2 | 12 | Missense | 1542G>C | — | X503S | ND |
| 184-S | US | <2 | 3-4** | 5′UTR-Intron 2 | Deletion | del 5′UTR -719 to IVS2+1 | No product | No product | Absent |
| 185-B | Russia | 6 | 4 | Intron 1 | Splice | IVSI-2a>g | ND | ND | ND |
| 186-S | US | 2.1 | 5 | Intron 2 | Splice | IVS2+1g>a | ND | ND | Absent |
| 187-S | US | 13 | 2 | Intron 2 | Splice | IVS2+1g>t | ND | ND | Absent |
| 188-S | US | 0.5 | ND | Intron 2 | Splice | IVS2-1 g>a | Exon 3 del | Inframe del 52 aa | ND |
| 189*-S | US | 4.0 | 3 | Intron 3 | Splice | IVS3-1 g>a | ins intron 3 | fs stop aa 201 | Absent |
| 190a/b-B | Russia | 5/7 | 3/3 | Intron 4 | Splice | IVS4-7 t>g | ND | ND | ND |
| 191-B | Croatia | 8 | 5 | Intron 6 | Splice | INS6+2t>g | ND | ND | ND |
| 192*-S | US | 7 | 1 | Intron 6 | Splice | IVS6+5 g>a | 70% ins38nt intron 6/30% normal | fs stop aa 190/normal | Reduced |
| 193-S | US | 14.0 | 2 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 194-S | US | 1.4 | 2 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 195a/b/c/d-S | US | 6/18/7/9 | 1/1/1/1 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 196a¶/b-S | US | 43 (dec.)/55 | 5/1 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 197-S | US African American | 12 | 2 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 198a/b-B | Italy | 4/1.8 (dec.) | 5/5 | Intron 7 | Splice | IVS7+1g>t | ND | ND | Absent |
| 199-B | Sweden | 10 (dec.) | 4 | Intron 7 | Splice | IVS7+1g>a | ND | ND | ND |
| 200-S | US | <3 | 2 | Intron 8 | Splice | IVS8+1g>a | Exon 8 del | fs stop aa 246 | Absent |
| 201-S | US | 1 | 4 | Intron 8 | Splice | IVS8+1g>a | Exon 8 del | fs stop aa 246 | Absent |
| 202-S | US American Indian | 10 | 3-4 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 203-S | US | <2 | 2 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 204-B | Italy | 7.9 | 2-3** | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | ND |
| 205-B | Italy | 1.5 (dec.) | 5 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 206-B | Sweden | 6 | 2 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | ND |
| 207-S | US | 0.1 | ND | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 208a/b/c/d-S | US Latino | 12/5/1.5/0.5 | 4**/3**/3**/2 | Intron 8 | Splice | IVS8+1g>c | ND | fs stop aa 246 | Absent |
| 209-S | Nepal | 2 | ND | Intron 8 | Splice | IVS8+1g>1 | Exon 8 del | ND | ND |
| 210-S | US | 0.8 | 3 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Absent |
| 211-S | Thailand | 9 | 4 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Absent |
| 212-S | US | 0.5 | 2 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Absent |
| 213-S | US | 1.0 | 3 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Reduced, normal size |
| 214-S | US | 2.0 | ND | Intron 8 | Splice | IVS8+2 to3 del tg | Exon 8 del | fs stop aa 246 | Absent |
| 215a§/b-B | Italy | 11/2.1 | 3/2 | Intron 8 | Splice | IVS8+3insT | ND | ND | ND |
| 216-S | US | 8 | 4 | Intron 8 | Splice | IVS8+5g>a | Exon 8 del | fs stop aa 246 | Absent |
| 217-S | US | 3.0 | 2 | Intron 8 | Splice | IVS8-2a>g | Intron 8ins | fs stop aa 328 | Absent |
| 218-S | US | 1 | 3 | Intron 9 | Splice | IVS9+1g>a | ND | ND | Absent |
| 219-B | Italy | 22 | 4 | Intron 9 | Splice | IVS9+2del tgag | ND | ND | ND |
| 220a*/b*-S | US | 2/<2 | 4/2** | Intron 9 | Splice | IVS9+2t>c | 80% ins 114nt of Intron 9 20% Intron9ins | fs stop aa 326 | Absent |
| 221-B | Sweden | 22 | 5 | Intron 9 | Splice | IVS9+2t>g | ND | ND | ND |
| 222*-S | US Greek | 1 | 4 | Intron 10 | Complex | IVS10del 9nt, ins 11 nt | 1372 ins 13nt | fs stop aa 498 | Red/trunc |
| 223§-B | Italy | <2 | 2-3** | Intron 10 | Splice | IVS10-3delC | ND | ND | ND |
| 224§-B | Italy | <2 | 1** | Intron 10 | Splice | IVS10-2a>t | ND | ND | Reduced |
| 225-B | Turkey | 7 | 4 | Intron 10 | Splice | IVS10-2a>c | ND | ND | ND |
| 226*-S | Canada | 4 | 4 | Intron 11 | Splice | IVS11+2t>c | Exon 11 del | fs stop aa 543 | Absent |
| 227*-S | US | ? | 5 | Intron 11 | Splice | IVS11+2t>g | Exon 11 del | fs stop aa 543 | Absent |
Patient . | Origin . | Age, y . | Score . | Exon . | Mutation type . | gDNA mutation . | cDNA mutation (if different from gDNA) . | Predicted protein change . | Western blot . |
|---|---|---|---|---|---|---|---|---|---|
| 1-S | US | 18 | 3 | 1 | Insertion | 41 insA | —†† | fs stop aa 37 | Absent |
| 2-B | Sweden | 6 (dec.) | 4 | 1 | Insertion | 41 insG | — | fs stop aa 37 | ND |
| 3-S | US | 4 | 5 | 1 | Deletion | 56-57delG | — | fs stop aa 44 | Absent |
| 4*-S | US | ? | 3 | 1 | Insertion | 62-64insC | — | fs stop aa 37 | Absent |
| 5-S | US | 0.2 | 3 | 1 | Insertion | 65-69insG | — | fs stop aa 37 | ND |
| 6*-S | US | 5.0 (dec.) | 5 | 1 | Nonsense | 71C>T | — | R13X | Absent |
| 7a*/b-S | US | 1.9/<2 | 4/3** | 1 | Nonsense | 71C>T | — | R13X | Absent |
| 8-B | Italy | 1.6 (dec.) | 4 | 1 | Nonsense | 71C>T | — | R13X | ND |
| 9-B | Italy | 8 | 4 | 1 | Nonsense | 74G>T | — | G14X | ND |
| 10-S | US | 5 | 3-4 | 1 | Missense | 104T>C | — | S24P | Reduced |
| 11-S | Thailand | 12 | 2 | 1 | Missense | 105C>T | ND | S24F | ND |
| 12§-B | Italy | 1.9 (dec.) | 5 | 1 | Deletion | 115-116delC | — | fs stop aa 44 | Absent |
| 13*-S | US | 12 | 2 | 1 | Missense | 113C>T | — | L27F | Reduced |
| 14-S | US | 7.5 | 3-4 | 1 | Deletion | 122-124del | — | Inframe | ND |
| 15-S | US | 1 | 1 | 1 | Missense | 125G>A | — | E31K | Absent |
| 16-S | Canada | 2 | 2 | 1 | Missense | 125G>A | — | E31K | Reduced |
| 17-B | Italy | 7 | 2-3 | 1 | Missense | 125G>A | — | E31K | Absent |
| 18-S | Chile | 0.3 | 1 | 1 | Missense | 125G>A | — | E31K | Reduced |
| 19-S | US | 0.4 | ND | 1 | Missense | 125G>A | — | E31K | Absent |
| 20-S | US | 0.5 | 1 | 1 | Nonsense | 134C>T | — | R34X | ND |
| 21-S | US | 1.1 | 3 | 1 | Nonsense | ND | 134C>T | R34X | Absent |
| 22—S | US | 0.2 | 5 | 1 | Nonsense | 134C>T | — | R34X | Absent |
| 23-B | Turkey | 5 | 2 | 1 | Missense | 138T>A | — | L35H | ND |
| 24*-S | US | 2.1 | 5 | 1 | Missense | 138,139TC>AT | — | L35H | Reduced |
| 25-S | Greece | 0.3 | 1 | 1 | Deletion | 140-142delTT | — | fs stop aa 36 | Absent |
| 26*-S | US | 4 | 4 | 1 | Deletion | 140-142delTT | — | fs stop aa 36 | Absent |
| 27-B | Italy | 2.3 (dec) | 5 | 1 | Deletion | 140-142delTT | — | fs stop aa 36 | ND |
| 28-S | US | 0.4 | ND | 1 | Insertion | 140-142insT | — | fs stop aa 37 | Absent |
| 29*-S | US | 8 | 2 | 1 | Missense | 150T>C | — | L39P | Reduced |
| 30a*/b*-S | US | 16/10 | 2/2 | 1 | Missense | 150T>C | — | L39P | Reduced |
| 31 a/b-B | Italy | 14/11 | 2/2-3 | 1 | Missense | 150T>C | — | L39P | Reduced |
| 32§-B | Italy | 6 (dec.) | 5 | 1 | Nonsense | 155C>T | — | R41X | Absent |
| 33§-B | Italy | 0.7 (dec.) | 5 | 1 | Nonsense | 155C>T | — | R41X | ND |
| 34-B | Italy | 9 | 4 | 1 | Nonsense | 155C>T | — | R41X | ND |
| 35-S | US | 0.2 | 2 | 1 | Nonsense | 155C>T | — | R41X | Absent |
| 36-S | US | 8 | 2 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 37-S | US | 0.3 | 1-2 | 2 | Missense | 168C>T | — | T45M | Absent |
| 38a*/b*-S | Canada | 8/2 | 2/2 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 39*-S | US | 8 | 1 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 40-S | US | ? | ND | 2 | Missense | 168C>T | — | T45M | Reduced |
| 41-B | Croatia | 6 | 2 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 42-S | US | 2.5 | 1 | 2 | Missense | 168C>T | — | T45M | Reduced |
| 43*-S | Polynesia | 8 | 2 | 2 | Missense | 174C>A | — | A47D | Reduced |
| 44*-S | US | 22 | 2 | 2 | Missense | 177C>T | — | T48I | Reduced |
| 45-S | US | 6 | 4 | 2 | Deletion | 186-196del | — | fs stop aa 58 | Absent |
| 46-S | US | 9 | 1 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 47-S | US | 2.7 | 1 | 2 | Missense | 201C>T | — | A56V | ND |
| 48*-S | US | ? | 1 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 49-B | Italy | 16 | 1 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 50a/b-S | US | 75/3 | 1/2 | 2 | Missense | 201C>T | — | A56V | Reduced |
| 51-S | US | 1 | ND | 2 | Deletion | 201-213del | — | fs stop aa 71 | ND |
| 52-S | US | ? | 2 | 2 | Missense | 206C>G | — | P58A | Reduced |
| 53a/b/c∥-B | Italy | 9/7/32 | 0.5/0.5/0.5 | 2 | Missense | 207C>G | — | P58R | Normal |
| 54*-S | Canada | 5 (dec.) | 5 | 2 | Deletion | 206-210delC | — | fs stop aa 75 | ND |
| 55a*/b*-S | US | 5/20 | 5/5 | 2 | Deletion | 211 delT | — | fs stop aa 75 | Absent |
| 56-B | Nicaragua | 1.5 | 4 | 2 | Complex (2 populations) | 211 delT | — | fs stop aa 75 | ND |
| 211T>C | P59P | ||||||||
| 57-B | Italy | 12 (dec.) | 4 | 2 | Missense | 224T>C | — | W64R | ND |
| 58-S | US | 13 (dec.) | 5 | 2 | Deletion | del 237-46 | — | fs stop aa 72 | Absent |
| 59-S | US | 1.2 | 2-3 | 2 | Missense | 252G>A | — | C73Y | Reduced |
| 60-B | Italy | 8 | 3 | 2 | Missense | 252G>A | — | C73Y | ND |
| 61-B | Russia | 1.8 | 2-3 | 2 | Missense | 255T>C | — | F74S | ND |
| 62-S | India | 5 | 2 | 2 | Missense | 257G>A | — | V75M | Reduced |
| 63a*/b*/c*-S | US | 5/4/17 | 2/2/2 | 2 | Missense | 257G>A | — | V75M | Reduced |
| 64a/b-B | Italy | 15/24 | 1/2 | 2 | Missense | 257G>A | — | V75M | Reduced |
| 65-S | US | 3 | 1 | 2 | Missense | 257G>A | — | V75M | Normal |
| 68-S | US | 6 | 2 | 2 | Missense | 261A>C | — | K76T | ND |
| 69-B | Italy | 9 | 2 | 2 | Missense | 263G>C | — | D77H | Reduced |
| 70a/b-B | Italy | 7/3 | 2/1 | 2 | Missense | 264A>G | — | D77G | Reduced |
| 71-S | US | 1 | 2 | 2 | Missense | 278T>C | — | S82P | Reduced |
| 72-B | Serbia | 9 | 5 | 2 | Deletion | 279-280delC | — | fs stop aa 126 | ND |
| 73a/b-B | New Zealand | 14/9 | 2/2 | 2 | Missense | 290C>A | — | R86S | ND |
| 74-S | US | 11 | 2 | 2 | Missense | 290C>G | — | R86G | Reduced |
| 75-S | US, African American | 19 | 2 | 2 | Missense | 290C>T | — | R86C | Absent |
| 76-S | US | 9 | 2-3 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 77-S | US | 4 | 2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 78-S | US | 5 | 2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 79a*/b*-S | US | ? | 2/2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 80*-S | US | ? | 1 | 2 | Missense | 290C>T | — | R86C | ND |
| 81-B | Israel | 8 | 1 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 82-S | US | 2 | 2 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 83-B | Turkey | 13 | 2 | 2 | Missense | 290C>T | — | R86C | ND |
| 84-S | Malaysia | 6 | 1 | 2 | Missense | 290C>T | — | R86C | Reduced |
| 85-S | US | 15 | 1-2 | 2 | Missense | 291G>A | — | R86H | Absent |
| 86*-S | US | 10 | 2 | 2 | Missense | 291G>A | — | R86H | Reduced |
| 87-S | US | 0.5 | 2-3** | 2 | Missense | 291G>A | — | R86H | Reduced |
| 88-B | Russia | 17.5 | 3 | 2 | Missense | 291G>A | — | R86H | Reduced |
| 89-S | US | 5.5 | 2 | 2 | Missense | 291G>T | — | R86L | Absent |
| 90*-S | Hungary | 3.5 | 4 | 3 | Deletion | 312-313delGT | ND | ND | ND |
| 91-S | US | 5 | 5 | 3 | Nonsense | 325G>A | — | W97X | Absent |
| 92-S | US | 1.3 | 5 | 3 | Nonsense | 325G>A | — | W97X | Absent |
| 93-S | US | 1.5 | 4 | 3 | Nonsense | 329C>T | ND | Q99X | Absent |
| 94*-S | US | ? | 5 | 3 | Complex | 329C>T | 329C>T/exon3del | Q99X/inframe Exon3del | Absent |
| 95-B | Finland | 2.5 | 1-2 | 3 | Missense | 334G>T | — | E100D | ND |
| 96-B | Italy | 5 | 2 | 3 | Missense | 348T>C | — | L105P | Reduced |
| 97*-S | US | <2 | 2** | 3 | Missense | 354A>G | — | Y107C | Reduced |
| 98-S | US | <2 | 2** | 3 | Deletion | 360-364delC | — | fs stop aa 126 | Absent |
| 99-S | US | 0.5 | 4 | 3 | Deletion | 360-364delC | — | fs stop aa 126 | Absent |
| 100-B | Italy | 7 | 4 | 3 | Insertion | 360-364insC | — | fs stop aa 121 | Absent |
| 101-S | US | ? | 2 | 3 | Insertion | 360-364insC | — | fs stop aa 121 | ND |
| 102-S | US | 2.1 | 5 | 3 | Deletion | 366-370delC | — | fs stop aa 126 | Absent |
| 103-S | US | 0.3 | 5 | 3 | Deletion | 371-372delT | — | fs stop aa 126 | Absent |
| 104-S | US | 0.9 | 1-2 | 3 | Missense | 390G>A | — | G119E | Reduced |
| 105a/b-S | Malaysia | 4/0.2 | 4/1 | 4 | Nonsense | 400C>A | Multiple products | C122X | Absent |
| 106*-S | ? | 5 | 4 | Missense | 407G>A | — | G125R | Absent | |
| 107-S | Greece | 1 | 5 | 4 | Missense | 411T>C | — | L126P | ND |
| 108-S | US | 0.3 | 5 | 4 | Missense | 416T>C | — | F128L | Absent |
| 109-S | US | ? | 5 | 4 | Missense | 416T>C | — | F128L | ND |
| 110*-S | US | 9 | 4 | 4 | Missense | 417T>C | — | F128S | ND |
| 111*-S | US | 8 | 3 | 4 | Complex | 425G>A,431G>A | — | E131K, E133K | Absent |
| 112a*/b*-S | US | 14 (dec.)/3 | 5/4 | 4 | Complex | 425G>A,290C>T | — | E131K,R86C | Absent |
| 113-S | Chile | 2 | 3 | 4 | Missense | 431G>A | — | E133K | ND |
| 114-B | Italy | 3.5 (dec.) | 5 | 4 | Missense | 431G>A | — | E133K | ND |
| 115-S | US | 12 | 2 | 4 | Missense | 433G>T | — | E133D | Reduced |
| 116-S | US | 0.7 | 1 | 4 | Missense | 433G>T | — | E133D | Reduced |
| 117*-S | US | 15 | 4 | 4 | Missense | 435C>T | — | A134V | Reduced |
| 118-S | US | 17 | 2 | 4 | Insertion | C435 ins 6bp | — | 134 DE ins | ND |
| 119-S | US | 2 | 2 | 4 | Complex | 447G>A, IVS11-1g>a | 447G>A, exon 11del | R149Q fs stop aa 543 | Reduced |
| 120-B | Italy | 3.5 (dec.) | 4 | 4 | Deletion | 451-458del | — | fs stop aa 165 | ND |
| 121-B | Italy | 13 (dec.) | 4 | 4 | Insertion | 471-476insA | ND | fs stop aa 168 | ND |
| 122#-S | US | 13 | 2 | 4 | Insertion | 471-476insA | — | fs stop aa 168 | Absent |
| 123-B | Italy | 6 (dec.) | 5 | 4 | Deletion | 485-486delAG | — | fs stop aa 167 | Absent |
| 124a/b‡-B | Serbia | 17/21 | 5/5 | 5 | Insertion | 512-516insC | — | fs stop aa 168 | ND |
| 125-S | US | 4 | 5 | 5 | Insertion | 512-516insC | — | fs stop aa 168 | Absent |
| 126-B | Italy | 22 | 5 | 6 | Deletion | 570delT | — | fs stop aa 260 | ND |
| 127-S | US | ? | 2 | 6 | Deletion | 597-597delC | ND | fs stop aa 260 | ND |
| 128-B | Italy | 3 | 4 | 7 | Deletion | 644delA | — | fs stop aa 260 | Absent |
| 129-S | US | 0.5 (dec.) | 2-3** | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 130a/b-S | US | 1.5/0.8 | 4/3 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 131-S | US | 1 | 5 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 132-S | US | ? | 3-4 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 133-S | US Latino | <2 | 2** | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 134*-S | US | 14 | 4 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 135*-S | US | 11 | 4 | 7 | Nonsense | 665C>T | — | R211X | Absent |
| 136-B | Italy | 5 | 4 | 7 | Nonsense | 665C>T | — | R211X | ND |
| 137-B | Italy | 6 | 4 | 7 | Nonsense | 665C>T | — | R211X | ND |
| 138-B | Italy | 2.1 (dec.) | 4 | 7 | Nonsense | 665C>T | — | R211X | ND |
| 139*-S | US | 1.9 | 4 | 7 | Splice | 705A>G | 705-768 del | fs stop aa 239 | Absent |
| 140-S | US | >5 | 5 | 7 | Splice | 705A>G | 705-768 del | fs stop aa 239 | Absent |
| 141-B | Sweden | <2 | 2** | 7 | Nonsense | 717C>G | — | S228X | ND |
| 142-S | US | 4 | 3-4 | 7 | Deletion | 731delA | — | fs stop aa 260 | Absent |
| 143‡B | Italy | 15 | 1 | 7 | Missense | 741C>G | — | A236G | Reduced |
| 144-S | US | 0.7 | ND | 7 | Complex | Exon 7(−5 to IVS7 +22) (del 27 bp) | 764-768 del. IVS7 +22 to 71 (ins 49 bp) | fs stop aa 257 | Absent |
| 145§-B | England | 4 (dec.) | 5 | 9 | Nonsense | 887G>T | — | E285X | ND |
| 146-B | Italy | 10 | 2 | 9 | Deletion | 907delC | — | fs stop aa 291 | ND |
| 147*-S | US | 2.5 | 5 | 9 | Nonsense | 923C>T | — | Q297X | Absent |
| 148-S | Turkey | 1.2 | 3 | 9 | Nonsense | 950G>T | — | E306X | Reduced |
| 149a*/b*†-S | US | ?/1.0 | 2/2 | 9 | Missense | 953A>G | — | M307V | ND |
| 150a*/b*-S | India | 3/1 | 3/2** | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 151a*/b*-S | US | ? | 5/5 | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 152-B | Italy | 7 | 5 | 10 | Nonsense | 995C>T | — | R321X | ND |
| 153-S | US | ? | ND | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 154a/b-S | US Latino | 7.0/0.2 | ND | 10 | Nonsense | 995C>T | — | R321X | Absent |
| 155-S | US | 1 | 2 | 10 | Deletion | 1017-1018delC | Multiple products | Multiple products | Absent |
| 156a*/b-S | US | 3/0.5 | 2/2 | 10 | Insertion | 1025insA | ND | fs stop aa 335 | Absent |
| 157-B | Italy | 3 (dec.) | 5 | 10 | Deletion | 1028delG | ND | fs stop aa 444 | Reduced |
| 158*-S | US | ? | 2 | 10 | Complex | 1029insT | del 966-1121 (50%) | inframe del 52aa | Red/trunc |
| del exon 10 (25%) | fs stop aa358 | ||||||||
| fs stop aa335 | |||||||||
| 1029 insT (25%) | |||||||||
| 159*-S | US | 10 | 4 | 10 | Deletion | 1030-1035delG | — | fs stop aa 444 | Absent |
| 160-S | US | ? | ND | 10 | Deletion | 1030-1035delG | — | fs stop aa 444 | Absent |
| 161-S | Brasil | 2.2 | ND | 10 | Deletion | 1030-1035delG | — | fs stop aa 444 | ND |
| 162a/b-S | US | 7.0/5 | 4/4 | 10 | Deletion | 1088-1092delC | Multiple products | Multiple products, fs stop aa 444 | Absent |
| 163-S | US | 4 | 5 | 10 | Deletion | 1088-1092delC | ND | Multiple products, fs stop aa 444 | ND |
| 164-S | New Zealand | 10 | 2 | 10 | Deletion | 1107-1108delGA | Multiple products | Multiple products, fs stop aa 493 | Red/trunc |
| 165*-S | US | 8 | 5 | 10 | Deletion | 1109-1113delC | — | fs stop aa 444 | Absent |
| 166*-S | US | 6 | 4 | 10 | Complex | 1109C>A, 1110-1113delC | — | P359T, fs stop aa 444 | Red/trunc |
| 167-B | Turkey | 5 | 4 | 10 | Missense | 1115C>A | — | P361T | ND |
| 168-S | Malaysia | 9 | 5 | 10 | Deletion | 1115-1119delC | — | fs stop aa 444 | ND |
| 169§-B | Italy | <2 | 2** | 10 | Complex | 1115-1119insC | — | fs stop aa 494 | ND |
| (2 populat.) | 1110-1182del | infr.del24aa | |||||||
| 170-B | Turkey | 12 | 5 | 10 | Nonsense | 1124C>T | — | R364X | ND |
| 171-S | US | 2 | 3 | 10 | Deletion | 1145-1149delC | Exon 10 del | fs stop aa 444 | Absent |
| 172-B | Italy | 10 | 5 | 10 | Deletion | 1184-1185delC | — | fs stop aa 444 | Absent |
| 173-S | Canada | 0.5 | 3 | 10 | Insertion | 1187-1191 insC | — | fs stop aa 494 | ND |
| 174-B | Bosnia | 4 | 5 | 10 | Deletion | 1192delT | — | fs stop aa 444 | Absent |
| 175-S | Puerto Rico | 1 | 4 | 10 | Insertion | 1286 ins4bp | — | fs stop aa 495 | Absent |
| 176*-S | US | 1.8 | 4 | 10 | Insertion | 1301-1305 insG | — | fs stop aa 494 | Red/trunc |
| 177*-S | US Latino | 8 | 5 | 10 | Insertion | 1301-1305 insG | — | fs stop aa 494 | Red/trunc |
| 178*-S | US | 14 | 5 | 10 | Deletion | 1301-1305 insG | — | fs stop aa 494 | Red/trunc |
| 179∥-B | Italy | 9 | 0.5 | 11 | Missense | 1476T>A | — | 1481N | normal |
| 180-B | Italy | 7 | 5 | 11 | Insertion | 1486-1487insTC | — | fs lack of term. codon | ND |
| 181-B | Russia | 6 | 5 | 12 | Deletion | 1519delT | — | fs lack of term. codon | Absent |
| 182-S | US | 2.5 | 1 | 12 | Missense | 1542G>C | — | X503S | Absent |
| 183-B | Turkey | 8.5 | 2 | 12 | Missense | 1542G>C | — | X503S | ND |
| 184-S | US | <2 | 3-4** | 5′UTR-Intron 2 | Deletion | del 5′UTR -719 to IVS2+1 | No product | No product | Absent |
| 185-B | Russia | 6 | 4 | Intron 1 | Splice | IVSI-2a>g | ND | ND | ND |
| 186-S | US | 2.1 | 5 | Intron 2 | Splice | IVS2+1g>a | ND | ND | Absent |
| 187-S | US | 13 | 2 | Intron 2 | Splice | IVS2+1g>t | ND | ND | Absent |
| 188-S | US | 0.5 | ND | Intron 2 | Splice | IVS2-1 g>a | Exon 3 del | Inframe del 52 aa | ND |
| 189*-S | US | 4.0 | 3 | Intron 3 | Splice | IVS3-1 g>a | ins intron 3 | fs stop aa 201 | Absent |
| 190a/b-B | Russia | 5/7 | 3/3 | Intron 4 | Splice | IVS4-7 t>g | ND | ND | ND |
| 191-B | Croatia | 8 | 5 | Intron 6 | Splice | INS6+2t>g | ND | ND | ND |
| 192*-S | US | 7 | 1 | Intron 6 | Splice | IVS6+5 g>a | 70% ins38nt intron 6/30% normal | fs stop aa 190/normal | Reduced |
| 193-S | US | 14.0 | 2 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 194-S | US | 1.4 | 2 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 195a/b/c/d-S | US | 6/18/7/9 | 1/1/1/1 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 196a¶/b-S | US | 43 (dec.)/55 | 5/1 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 197-S | US African American | 12 | 2 | Intron 6 | Splice | IVS6+5 g>a | ins 38nt intron 6/normal | fs stop aa 190/normal | Reduced |
| 198a/b-B | Italy | 4/1.8 (dec.) | 5/5 | Intron 7 | Splice | IVS7+1g>t | ND | ND | Absent |
| 199-B | Sweden | 10 (dec.) | 4 | Intron 7 | Splice | IVS7+1g>a | ND | ND | ND |
| 200-S | US | <3 | 2 | Intron 8 | Splice | IVS8+1g>a | Exon 8 del | fs stop aa 246 | Absent |
| 201-S | US | 1 | 4 | Intron 8 | Splice | IVS8+1g>a | Exon 8 del | fs stop aa 246 | Absent |
| 202-S | US American Indian | 10 | 3-4 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 203-S | US | <2 | 2 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 204-B | Italy | 7.9 | 2-3** | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | ND |
| 205-B | Italy | 1.5 (dec.) | 5 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 206-B | Sweden | 6 | 2 | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | ND |
| 207-S | US | 0.1 | ND | Intron 8 | Splice | IVS8+1g>a | ND | fs stop aa 246 | Absent |
| 208a/b/c/d-S | US Latino | 12/5/1.5/0.5 | 4**/3**/3**/2 | Intron 8 | Splice | IVS8+1g>c | ND | fs stop aa 246 | Absent |
| 209-S | Nepal | 2 | ND | Intron 8 | Splice | IVS8+1g>1 | Exon 8 del | ND | ND |
| 210-S | US | 0.8 | 3 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Absent |
| 211-S | Thailand | 9 | 4 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Absent |
| 212-S | US | 0.5 | 2 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Absent |
| 213-S | US | 1.0 | 3 | Intron 8 | Splice | IVS8+1 to+6 del gtga | Exon 8 del | fs stop aa 246 | Reduced, normal size |
| 214-S | US | 2.0 | ND | Intron 8 | Splice | IVS8+2 to3 del tg | Exon 8 del | fs stop aa 246 | Absent |
| 215a§/b-B | Italy | 11/2.1 | 3/2 | Intron 8 | Splice | IVS8+3insT | ND | ND | ND |
| 216-S | US | 8 | 4 | Intron 8 | Splice | IVS8+5g>a | Exon 8 del | fs stop aa 246 | Absent |
| 217-S | US | 3.0 | 2 | Intron 8 | Splice | IVS8-2a>g | Intron 8ins | fs stop aa 328 | Absent |
| 218-S | US | 1 | 3 | Intron 9 | Splice | IVS9+1g>a | ND | ND | Absent |
| 219-B | Italy | 22 | 4 | Intron 9 | Splice | IVS9+2del tgag | ND | ND | ND |
| 220a*/b*-S | US | 2/<2 | 4/2** | Intron 9 | Splice | IVS9+2t>c | 80% ins 114nt of Intron 9 20% Intron9ins | fs stop aa 326 | Absent |
| 221-B | Sweden | 22 | 5 | Intron 9 | Splice | IVS9+2t>g | ND | ND | ND |
| 222*-S | US Greek | 1 | 4 | Intron 10 | Complex | IVS10del 9nt, ins 11 nt | 1372 ins 13nt | fs stop aa 498 | Red/trunc |
| 223§-B | Italy | <2 | 2-3** | Intron 10 | Splice | IVS10-3delC | ND | ND | ND |
| 224§-B | Italy | <2 | 1** | Intron 10 | Splice | IVS10-2a>t | ND | ND | Reduced |
| 225-B | Turkey | 7 | 4 | Intron 10 | Splice | IVS10-2a>c | ND | ND | ND |
| 226*-S | Canada | 4 | 4 | Intron 11 | Splice | IVS11+2t>c | Exon 11 del | fs stop aa 543 | Absent |
| 227*-S | US | ? | 5 | Intron 11 | Splice | IVS11+2t>g | Exon 11 del | fs stop aa 543 | Absent |
The “B” associated with the patient number identifies Brescia, the “S” identifies Seattle as the study center. a/b/c/d indicates individuals from the same family; ND, not determined; Red/trunc, reduced in quantity and truncated (small protein size); and dec., deceased.
Reported in Zhu et al,15 1997.
Reported by Kwan et al,22 1995.
Reported by Villa et al,10 1995.
Reported by Wengler et al,18 1995.
Reported by Notarangelo et al,23 2002.
This patient had a score of 1, at age 43 developed fatal lymphoma (score 5).
Remutation occurred in an early lymphocyte population.
This patient underwent BMT before 2 years of age.
The cDNA mutation was the same as the gDNA mutation.
PBMC isolation and cell lines
After informed consent was obtained to sequence DNA and set up cell lines for biochemical and immunologic analysis, heparinized blood was collected by venipuncture and shipped overnight or transported on the same day at room temperature to 1 of the 2 centers. Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque (Amersham Pharmacia Biotech, Piscataway, NJ) density gradient centrifugation and washed twice with phosphate-buffered saline (PBS). B-lymphoblastoid cell lines (B-LCLs) were established by infecting PBMCs with Epstein-Barr virus (EBV) containing supernatant as previously described.25 T-cell lines were established as follows: isolated PBMCs were cultured at a concentration of 2 × 106/mL in the presence of phytohemagglutinin (PHA; 3-5 μg added at weekly intervals). During the 2nd and the 3rd week, interleukin 2 (IL-2) at 500 U/mL was added. After 2 to 4 weeks in culture the cells were lysed and studied for the presence of WASP by Western blot analysis.
Nucleic acid isolation and cDNA synthesis
Genomic DNA (gDNA) was extracted from whole blood or buffy coat using the QIAamp DNA Blood mini kit (Qiagen, Valencia, CA). Total RNA was isolated from PBMCs or from established lymphoblastoid cell lines with Trizol (Life Technologies, Grand Island, NY) and submitted to reverse transcriptase-polymerase chain reaction (RT-PCR) using the Thermoscript RT-PCR kit (Life Technologies) according to manufacturers' instructions.
PCR reaction and sequence analysis
cDNA (1 μg) was amplified in 2 overlapping fragments using primers and conditions previously reported.26 Direct sequencing was performed using the ABI Prism Big Dye Terminator Cycle Sequencing System (Perkin Elmer, Chatsworth, CA), and chromatograms were generated on an Applied Biosystems Model 373 or 310 DNA sequencer according to manufacturers' instructions. gDNA (0.2-1 μg) was amplified using intronic primer pairs that flanked 1 to 4 exons, including all intron/exon junctions. For the patients analyzed at the University of Washington, we selected primer pairs reported previously by Derry et al27 ; for the patients analyzed at the University of Brescia, the primers were previously reported by Giliani et al.28
Western blot analysis
B-LCLs or IL-2-dependent T-cell lines from patients and from a WASP-positive (healthy individual) and a WAS-negative (WAS patient with a nonsense mutation in exon 1) control subject were lysed and submitted to Western blot analysis as previously described.15,23 From each sample, 20 μg total protein was electrophoresed through a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a polyvinylidene diflouride (PVDF) Immobilon-P membrane (Millipore, Bedford, MA). For the experiments performed in Seattle, the membrane was incubated with the polyclonal anti-WASP antibody 503, and for those carried out in Brescia, the anti-WASP monoclonal antibody 3F3-A529 was used. The membrane was then washed 4 times, and results were visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).15
Results
Clinical scores
Sufficient information was available to clinically score 248 patients with WAS/XLT (Table 1). A score of 0 to 1 (0.5 in Table 1) was given to 4 patients with intermittent XLT,23 a score of 1 (including 4 patients with a score of 1-2) was given to 33 patients, a score of 2 was given to 73 patients (including 9 patients with a score of 2-3), a score of 3 was given to 29 patients (including 5 patients with a score of 3-4), and a score of 4 was given to 47 patients; 53 patients progressed to a score of 5.
Mutation analysis
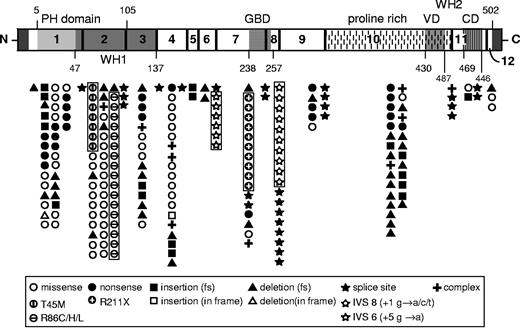
We identified and characterized at total of 141 unique WASP mutations in 262 patients with WAS/XLT from 227 unrelated families on the gDNA, mRNA, and/or protein level (Figure 1; Table 1).
A schematic illustration of WASP representing the 12 exons and the major functional domains. The mutations of WASP listed in Table 1 are visualized according to their location in the exons and the exon/intron junctions. Each symbol represents a single family with a WASP mutation. Missense mutations are located mostly in exons 1 to 4; deletions and insertions are distributed throughout the WASP gene, and splice site mutations are found predominantly in introns 6, 8, 9, and 10. PH indicates pleckstrin hemology; WH1, WAS homology 1; GBD, GTPase binding domain; VD, verprolin homology domain; and CD, cofilin homology domain.
A schematic illustration of WASP representing the 12 exons and the major functional domains. The mutations of WASP listed in Table 1 are visualized according to their location in the exons and the exon/intron junctions. Each symbol represents a single family with a WASP mutation. Missense mutations are located mostly in exons 1 to 4; deletions and insertions are distributed throughout the WASP gene, and splice site mutations are found predominantly in introns 6, 8, 9, and 10. PH indicates pleckstrin hemology; WH1, WAS homology 1; GBD, GTPase binding domain; VD, verprolin homology domain; and CD, cofilin homology domain.
As shown in Table 2, the most common mutations were missense mutations (n = 80 families) followed by splice site mutations (n = 44), deletions (n = 40), and nonsense mutations (n = 35). Insertions and complex mutations made up less than 13% of the mutations identified.
Distribution of mutations in 227 WAS/XLT families
Mutation type . | Families affected (%) . |
|---|---|
| Missense | 80 (35.3) |
| Splice | 44 (19.4) |
| Deletion | 40 (17.6) |
| Nonsense | 35 (15.4) |
| Insertion | 18 (7.9) |
| Complex | 10 (4.4) |
| Total | 227 (100) |
Mutation type . | Families affected (%) . |
|---|---|
| Missense | 80 (35.3) |
| Splice | 44 (19.4) |
| Deletion | 40 (17.6) |
| Nonsense | 35 (15.4) |
| Insertion | 18 (7.9) |
| Complex | 10 (4.4) |
| Total | 227 (100) |
Protein expression
We were able to directly determine the quantity and size of WASP in cell lysates generated from B-LCL or T-cell lines established from 187 individuals belonging to 160 unrelated families. The 2 anti-WASP antibodies used gave identical results. Mutated WASP of normal size was present, although often at reduced amounts, in affected males from 69 (43%) families which were classified as “WASP positive” (Table 3). In contrast, WASP was absent, reduced in quantity and size (truncated) in affected members of the remaining 91 families (57%), which were classified as “WASP negative.”
Types of mutations correlate with the expression of WASP
Mutation type . | WASP+ families affected (%) . | WASP− families affected (%) . |
|---|---|---|
| Missense | 51 (73.9) | 10 (11) |
| Splice | 8 (l* = 1, V* = 7) (11.6) | 23 (l* = 19, V* = 4) (25.3) |
| Complex | 4 (5.8) | 4 (4.4) |
| Deletion/insertion | 5 (7.2) | 32 (35.1) |
| Nonsense | 1 (1.5) | 22 (24.2) |
| Totals | 69 (100) | 91 (100) |
Mutation type . | WASP+ families affected (%) . | WASP− families affected (%) . |
|---|---|---|
| Missense | 51 (73.9) | 10 (11) |
| Splice | 8 (l* = 1, V* = 7) (11.6) | 23 (l* = 19, V* = 4) (25.3) |
| Complex | 4 (5.8) | 4 (4.4) |
| Deletion/insertion | 5 (7.2) | 32 (35.1) |
| Nonsense | 1 (1.5) | 22 (24.2) |
| Totals | 69 (100) | 91 (100) |
The distribution of mutations observed in WASP+ and WASP− families is significantly different (ϰ2 = 72.3, 4 df, P < .001). The association was also highly significant, using the r × c version of the Fisher exact test (P < .001). l* indicates invariant sites; V*, variant sites.
Most WASP+ patients had scores of 1 of 2.5 (2-3), characteristic of the XLT phenotype (Table 1). The majority of the WASP+ families had missense mutations (n = 51, 73.9%) followed by a much smaller percentage who had splice site mutations (n = 8, 11.6%) (Table 3). Of the 8 WASP+ families with splice site mutations, 7 had mutations affecting a variant splice site and only 1 affected an invariant splice site. The opposite is true for WASP- families with splice site mutations: 19 had mutations affecting an invariant splice site and only 4 had a variant splice site mutation.
Most patients with deletions or insertions affecting exon 10 that resulted in frameshift and stop at either amino acid (aa) 444 (n = 13) or at aa 493/494 (n = 9) failed to express WASP; those whose lymphocytes expressed WASP (n = 7) had a reduced quantity of truncated, presumably nonfunctional, WASP. Of the 20 patients that could be scored, 16 had a WAS phenotype; of the 4 patients with the XLT phenotype, 2 were 2 years of age or younger (Table 1).
The majority of the WASP-negative patients had scores of 3 or greater (Table 1). Mutations associated with absence of WASP were predominantly deletions/insertions, nonsense mutations, or splice site mutations accounting for 85% of the WASP- families (Table 3). The rest were missense and complex mutations. An exception was WASP-negative infants who were scored when aged 2 years or younger and who had mutations that were expected to result in a severe WAS phenotype (eg, nonsense mutations, deletions, insertions, splice site, complex mutations, and missense mutations in exon 4 that resulted in failure to express WASP). Of 49 infants with these characteristics, 32 (65%) had scores of 3 to 5, whereas 17 (35%) had scores of 1 to 2.5. In contrast, of 69 WASP- patients older than 2 years when the scores were obtained, and with similar mutations, 64 (93%) had scores of 3 to 5 and only 5 (7%) had scores of 1 to 2.5 (χ2 = 14.23, 1 degree of freedom [df], P = .0002), suggesting that it may take up to several years for a WASP- infant to present with the severe WAS phenotype expected from mutations that result in lack of WASP. It may, therefore, be prudent to consider scores of 1 to 2.5 as indeterminate when observed in infants aged 2 years or younger with mutations that predict absence of WASP and a severe WAS phenotype.
Mutational hotspots
In the 227 unrelated families studied we found 5 mutational “hotspots” which were defined as occurring in 6 or more unrelated families (> 2.5%) (Table 4).
WASP hotspot mutations and clinical phenotype
Mutation . | Affected families (no. patients) . | % of total families . | Score 1-2.5*, no. patients . | Score 3-5, no. patients . |
|---|---|---|---|---|
| 168C > T (T45M) | 7 (8†) | (3.1) | 7 | 0 |
| 290C > N/291G > N (R86S/G/C/H/L) | 17 (19) | (7.5) | 19 | 0 |
| IVS6 + 5g > a fs stop aa 190/normal | 6 (10) | (2.6) | 9 | 1 |
| 665C > T (R211X) | 10 (11) | (4.4) | 1 | 10 |
| IVS8 + 1g > a/c/t, fs stop aa246 | 10 (13†) | (4.4) | 5 | 6 |
| Totals | 50 (61) | (22) | 41 | 17 |
Mutation . | Affected families (no. patients) . | % of total families . | Score 1-2.5*, no. patients . | Score 3-5, no. patients . |
|---|---|---|---|---|
| 168C > T (T45M) | 7 (8†) | (3.1) | 7 | 0 |
| 290C > N/291G > N (R86S/G/C/H/L) | 17 (19) | (7.5) | 19 | 0 |
| IVS6 + 5g > a fs stop aa 190/normal | 6 (10) | (2.6) | 9 | 1 |
| 665C > T (R211X) | 10 (11) | (4.4) | 1 | 10 |
| IVS8 + 1g > a/c/t, fs stop aa246 | 10 (13†) | (4.4) | 5 | 6 |
| Totals | 50 (61) | (22) | 41 | 17 |
Three of the hotspot mutations (T45M, R86N, IVS6 + 5g > a) are associated with low scores, and 2 hotspot mutations (R211X, IVS8 + 1g > a) are associated with high scores (ϰ2 = 36.11, 4 df, P < .001). Fisher exact test for r × c tables, P < .001.
A score of 2 to 3 is listed as a score of 2.5; scores 1 to 2.5 are considered XLT; scores of 3 to 5 represent the WAS phenotype.
One patient with the 168C > T missense mutation and 2 patients with the IVS8 + 1g > a spalice site mutation could not be scored because of insufficient clinical data.
The 168C>T mutation, found in 7 families, results in the substitution of threonine with methionine at position 45 and the 290C>N/291G>N mutations, observed in 17 unrelated families, result in the substitution of arginine at position 86 with either a serine, glycine, cysteine, histidine, or leucine. The IVS6+5g>a mutation, present in 6 unrelated families, results in both abnormal and normal splicing products. The 665C>T mutation, which converts arginine at codon 211 to a stop codon, was found in 10 families, and the IVS8+1g>n mutation, identified in 10 families, results in the deletion of exon 8, leading to frameshift and premature stop of translation. These 5 mutations account for 22% of all families included in this study. Three of these 5 mutations (168C>T, 290C>N/291G>N, and IVS6+5g>a) were consistently found in WASP+ patients with a mild phenotype (XLT) who had a low score, whereas the other 2 (665C>T and IVS8+1g>n) were predominantly WASP- and had a high score (P < .001).
Remutation
Patient 122-S was found to have gDNA with a single nucleotide insertion (471-476insA) which results in frameshift and premature stop at aa 168. This mutation is expected to cause severe WAS. However, at 12 years of age, patient 122-S has mild disease with a score of 2. His cDNA, generated from mRNA isolated from PBMCs, consists of 2 populations: the majority has the 471-476insA mutation, and a small fraction has a normal sequence. Western blot analysis revealed a small amount of WASP in extracts from both EBV-induced B lymphoblasts and IL-2-dependent T lymphoblasts, suggesting that the remutation occurred in an early lymphoid precursor population. A remutation (211T>C, resulting in the silent mutation P59P) was also observed in patient 56-B.
Discussion
Mutations of the WASP gene result in 3 distinct phenotypes: the classic WAS triad of thrombocytopenia/small platelets, recurrent infections as a result of immunodeficiency, and eczema1,2 ; the milder XLT variant, characterized predominantly by thrombocytopenia/small platelets10,11 ; and congenital neutropenia without the clinical findings characteristic for WAS/XLT.12
To investigate a possible phenotype/genotype association we (1) developed a scoring system that allowed the differentiation of WAS and XLT, (2) analyzed genomic DNA and cDNA from each subject for mutations of WASP, and (3) determined the effect of the mutation on WASP expression by cultured lymphoblasts.
Of the 141 unique WASP mutations identified in our series of 227 unrelated families, 71 had not been previously reported.9-11,15-23,27-44 Most mutations consisted of single nucleotide substitutions, small insertions and deletions, and splice site mutations and were distributed throughout the coding region and the intron-exon junctions. Except for a large deletion involving the 5′ untranslated region (UTR) and exons 1 and 2 (del5′UTR [-719]-intron2+1), mutations were not found outside the coding region, and none of the WAS/XLT patients were found to lack WASP expression without having a demonstrable mutation affecting either the exons or the exon-intron junctions.
As was observed in smaller series reported previously,9-11,15-22,30,35,39,43 the predominant mutations of WASP were missense mutations which were typically located in exons 1, 2, 3, and 4. Only 4 missense mutations, 1 each in exon 7, 9, 10, and 11, were observed downstream of exon 4. One of those, P361T in exon 10, is the only missense mutation identified to date affecting the polyproline region of WASP. In addition, 2 unrelated families, both with the XLT phenotype, had a point mutation affecting the termination codon of exon 12 (codon 503X>S), resulting in the absence of WASP.
The second most common WASP mutation affected splice sites and occurred predominantly in the downstream half (introns 6-11) of the WASP gene, similar to previous reports.15,17-19,30 Mutations affecting variant splice sites resulted in multiple splicing products, and often included small amounts of normal WASP. Insertions and deletions, with 2 exceptions, resulted in frameshift and premature stop of translation. Deletions were more common in areas with a series of cytosine or guanosine nucleotides and typically involved less than 10 nucleotides. Complex mutations were rare and involved double missense mutations, point mutations followed by a deletion, or a combination of deletions and insertions.
Of the 5 “hotspots,” 3 were associated with mild diseases: the substitution of arginine at position 86 with 1 of 5 different amino acids, was observed most frequently (17 families), followed by substitution of threonine with methionine at position 45 (7 families), and the variant splice site mutation IVS6+5g>a that generates simultaneously abnormal and reduced amounts of normal WASP (6 families). The other 2 hotspot mutations, the nonsense mutation Arg211X and the splice site mutation (IVS8+1g>n), both present in 10 families, were preferentially associated with the WAS phenotype. There was no association with ethnicity or geographic origin, and it is unclear why these 5 hotspot mutations occur in more than 20% of WAS/XLT families reported here. It is possible that these sequences are prone to mistakes during DNA replication. The alternative explanation that these hotspots represent naturally occurring polymorphisms is highly unlikely because no other mutations within the WASP gene could be identified in any of these patients with WAS/XLT, and none of these hotspot mutations was observed in more than 300 normal X chromosomes in which WASP was sequenced.
Thrombocytopenia and small-sized platelets were present in all patients with WASP mutations, including the members of 2 families with missense mutations in exon 2 (P58A) and exon 11 (I481N),23 respectively, who had intermittent petechiae and thrombocytopenia, with platelet counts as low as 30 × 109/L alternating with normal platelet counts (100-250 × 109/L) when asymptomatic. No patients with congenital neutropenia were identified, possibly reflecting a selection bias.
To explore a phenotype/genotype correlation, we divided the patients into 2 categories: WASP positive if the mutated protein was of normal size and WASP negative if protein was absent or truncated. Patients with mutations that allowed the expression of normal-sized mutated protein, often in reduced quantity, developed the XLT phenotype, whereas those patients whose lymphocytes could not express WASP or expressed only truncated WASP were more likely to present with the WAS phenotype (P < .001). Progression to a score of 5 because of either autoimmune disease or malignancy was observed in both groups, but far more frequently in WASP-negative patients. Exceptions to this rule exist as shown in Tables 1 and 3 and may make it difficult in individual cases to accurately predict the clinical course based solely on the type of mutation of the WASP gene. There are several explanations for these occasional discrepancies. Splice site mutations, especially if affecting variant intronic nucleotide positions often allow the generation of multiple splicing products, including normally spliced mRNA, resulting in a small quantity of normal WASP. For example, the hotspot mutation IVS6+5g>a, causes the insertion of 38 nucleotides from the proximal end of intron 6, which results in a frameshift and early termination of transcription, but also in the production of a small amount of normal WASP mRNA. Another mechanism that can modify the clinical phenotype is “remutation” because of in vivo (somatic) reversion of the inherited mutation. This mechanism was observed in patient 122-S who was found to have 2 populations of WASP cDNA and protein, mutated and normal. In contrast to the XLT phenotype (score 2) observed in patient122-S, a severe WAS phenotype (score of 4) was present in a nonrelated patient (121-B) with the same mutation. Wada et al31 reported a patient with WAS/XLT with remarkable clinical improvement after puberty when he was found to have undergone remutation. There may be other genes that are capable of modulating the clinical phenotype of patients with WAS/XLT. Disease-modifying genes have been demonstrated in patients with chronic granulomatous disease (CGD) whose clinical phenotypes were strongly influenced by polymorphisms of myeloperoxidase and FcγRIIIb.45 Genes that determine an individual's probability to develop atopic diseases, such as allergies and eczema, or genes that control the effectiveness of host defense against infections may modify the WAS/XLT phenotype. Finally, environmental factors, such as exposure to common or uncommon infectious agents, failure to establish the diagnosis at an early age, or less than optimal care, may affect the severity of the clinical presentation. Scoring before the age of 2 years is unreliable because it often suggests a phenotype that is milder than expected from the type of WASP mutation identified. For this reason, scores of 1 to 2.5 observed in infants aged 2 years should be considered as indeterminate.
Of the 248 patients with clinical information that allowed a score to be established, 53 (mostly nonsense or splice site mutations resulting in the absence of WASP) eventually reached a score of 5, indicating the progression to autoimmune disease or malignancy. Only 6 of the 53 patients with a score of 5 were found to have missense mutations, and all but 1 patient had exon 4 mutations and failed to express WASP. We observed a high rate of autoimmunity or malignancies in patients with splice site mutations that resulted in premature termination at codon 444 or 493. A strikingly higher incidence of lymphoma has recently been reported in patients with WAS/XLT with splice site mutations, nonsense mutations, and mutations resulting in frameshift than in patients with missense mutations.46
We conclude from this in-depth analysis of 2 large cohorts of families with WASP mutations, one predominantly from North America and the other from Europe, that the clinical phenotype of WAS or XLT, severe or mild, is strongly influenced by the effect of the mutation on protein expression. The frequency and severity of infections, the extent of eczema, and the progression to autoimmune diseases and malignancies correlated, although not always invariably, with the absence of WASP in patient lymphocytes. However, patients with missense mutations allowing expression of mutated WASP and those with splice site mutations resulting in multiple products, including small amounts of normal WASP, present overwhelmingly with the milder XLT phenotype. As expected, there is some overlap, especially in infants aged 2 years or younger, which makes it more difficult for the treating physician to predict the final outcome in a patient with a new diagnosis of WASP mutation. The dismal prognosis for patients with the WAS phenotype justifies the use of procedures such as stem cell transplantation that, by themselves, are associated with considerable morbidity and potential mortality.
Prepublished online as Blood First Edition Paper, July 29, 2004; DOI 10.1182/blood-2003-05-1592.
Supported in part by grants from the National Institutes of Health (HD17427-33), the March of Dimes Birth Defects Foundation (96-0330), the Immunodeficiency Foundation, the Jeffrey Modell Foundation, the DeJoria Wiskott-Aldrich Research Fund (H.D.O.); the MIUR-FIRB (project RBNE01Y3N3_004), MURST (Cofin 2002), the European Union “Quality of Life and Management of Living Resources” (contract QLG1-CT-99-01 090), and the European Union (grant QLGI-CT-2001-01 395); and the Berlucchi Foundation (L.D.N.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Lianne Sheppard for assistance with the statistical data analysis. The following individuals allowed the Seattle and Brescia laboratories to study their patients and provided material that was invaluable for this investigation: O. Alvarez, N. Amaurilio, J. Anžič, R. Ashok, U. Aysegul, D. Beardsley, V. Berdovicas, L. Bird, M. Blaese, V. Bonagura, W. G. Borges, C. Bottom, L.A. Boxer, L. Bruckner, R. Buckley, J. Bussell, M. Caniglia, F. Candotti, L. Cardinali, J. Casella, B. Casey, H. Chapel, M. Clayton, G. Colon, M.E. Conley, V. Correra, M. Cowan, C. Cunningham-Rundles, C. Curry, N. Day, G. del Toro, G. Eames, M. Elder, G. Elinder, A. Etzioni, A. Fasth, C. Fernandes, L. Filipovitch, C. Frantz, T. Fuengoer, R. Fuleihan, R. Galanello, E. Gelfand, V. Gentile, C. Gibbons, E. Gillan, W. Greg, M. Grimley, P. Harper, T. Hays, W. Hicks, H. Hill, R. Hostoffer, J. Hutter, R. Insel, A.-M. Irani, J. Jazbec, A. Junker, W. Kamchaisatian, M. Kanariou, Z. Karakas, G. Kato, H. Kayserili, B. Kerlin, A. Kheradpour, G. Kleiner, L. Kobrynski, A. Laszlo, C. L. Lee, E. Lichtenberg, J. Lipton, J. Litzman, A. Malta Corea, P. Martin, S. Martino, M. Masi, D. De Mattia, B. Mazer, C. Mendez, C. Mercer, C. Mullen, D. Murphey, D. Nugent, M. Oblender, J. Oleske, K. O'Neill, W. Owen, L. Pachman, S. Pasic, M. Pelidis, M. J. Petruzzi, A. Plebani, V. Poggi, K. Provo, M. Pulsipher, M. Rabusin, U. Ramenghi, I. Reznick, F. Rosen, H. Rosenblatt, C. Rossbach, E. Salman, O. Sanal, J. Sande, R. Schiff, M. Schumacher, S. Scommegna, R. Shapiro, A. Shigeoka, B. Sibly, J. W. Sleasman, D. Steele, C. Stotts, K. Sullivan, F. Tancredi, W. Tcheng, D. Toro, M. T. Trakultivakorn, A. Tsang, S. Turk, E. Tyler, W. Vasconcelos, K. Vettenranta, A. Ventura, M. Wang, K. Weinberg, G. Wilson, J. Winkelstein, L. Wolff, D. Wright, and J. Wu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal