Two papers in this issue identify mutations in genes encoding proteins that transduce growth and survival signals in subsets of patients with acute lymphoblastic leukemia. These data provide new biologic insights and identify patients who might benefit from from targeted therapies.
Deregulated signaling pathways are a central theme in the pathogenesis of myeloid malignancies. Genetic lesions such as the BCR/ABL fusion, PTPN11 point mutations, and FLT3 activating loop mutations aberrantly activate kinase cascades that regulate proliferation and survival. The Ras family of guanosine 5′-triphosphatases (GTPases) has emerged as a potential nodal point in these deregulated signaling networks, and oncogenic mutations of NRAS and KRAS2 are found in many patients with myelodysplastic syndrome (MDS), myeloproliferative disorders (MPDs), and acute myeloid leukemia (AML). Altered signal transduction has received considerably less attention in acute lymphoblastic leukemia (ALL), although the blasts of approximately 25% of adult ALL patients express BCR/ABL fusions and RAS mutations occur in another 10% of cases. In this issue of Blood, articles by Tartaglia and colleagues (page 307) and Paietta and colleagues (page 558) identify mutations in PTPN11 and FLT3 in discrete subsets of ALL specimens.
The PTPN11 gene encodes SHP-2 (Src homology 2 domain containing tyrosine phosphatase), a nonreceptor tyrosine phosphatase that relays signals from activated growth factor receptors to Ras and other effectors in a variety of cell types.1 Germ line PTPN11 mutations cause Noonan syndrome,2 a congenital disorder that is associated with an increased risk of juvenile myelomonocytic leukemia (JMML). Indeed, somatic PTPN11 mutations occur in approximately 35% of de novo JMML specimens and in a small percentage of patients with other myeloid malignancies.3,4 Tartaglia and colleagues now report a significant incidence of PTPN11 mutations in pediatric patients with common and B-precursor ALL that do not coexist in clones harboring chimeric fusion genes such as TEL/AML1, BCR/ABL, E2A/PBX1, or MLL/AF4. These investigators also detected NRAS and KRAS2 mutations in the same subset of ALL patients but found only one case with mutations in both PTPN11 and RAS. These data are reminiscent of JMML where RAS, NF1, and PTPN11 mutations are largely mutually exclusive.3,4 Elsewhere in this issue, Paietta and colleagues report FLT3 mutations in 3 cases of adult T-cell ALL, each of which showed high expression of CD117/KIT. These patients also exhibited nonrandom chromosomal translocations. This observation implicates aberrant signal transduction via altered FLT3 in a small percentage of adults with T-cell ALL, a phenomenon not previously reported.5
Like all good research, the reports in this issue of Blood raise new questions. First, is hyperactive Ras the critical biochemical lesion that results from PTPN11, BCR/ABL, and FLT3 mutations? The finding that individual leukemias rarely demonstrate mutations in more than one of these genes provides indirect genetic evidence that this might be true. Whereas biochemical data infer that FLT3 mutations and BCR/ABL encode proteins that deregulate Ras,5 this issue is less certain for PTPN11, where overexpression of mutant SHP-2 proteins is associated with modest Ras activation in COS7 cells but not in similarly transduced Ba/F3 cells.4,6 SHP-2 has other important biochemical targets such as Src kinases,7 and it is conceivable that deregulating a parallel signaling pathway may have similar phenotypic effects on Ras activation.
Second, the absence of chimeric fusion genes and signaling gene mutations in ALL is in direct contrast to AML, where oncogenic RAS, FLT3, and KIT mutations frequently coexist with fusion genes such as AML1/ETO or PML/RARα.8 It will therefore be important to ascertain if there is cooperation between these distinct classes of leukemia-associated mutations in ALL, or if leukemias that express common fusions such as TEL/AML1 evolve along a distinct genetic pathway from clones with mutations in PTPN11 or RAS mutations. As suggested by Paietta et al, answering this question has therapeutic implications as additional inhibitors of signaling molecules enter clinical trials.FIG1
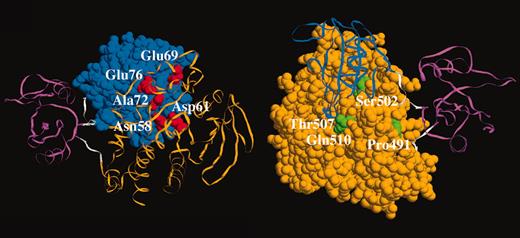
Somatic PTPN11 mutations in childhood acute leukemia. See the complete figure in the article beginning on page 307.
Somatic PTPN11 mutations in childhood acute leukemia. See the complete figure in the article beginning on page 307.
Finally, studies of myeloid malignancies have shown that the specific signaling molecule that is altered by mutation is nonrandom. For instance, RAS mutations appear to be more frequent in cases of AML associated with mutations that perturb the core-binding factor complex, whereas FLT3 mutations are strongly associated with PML/RARα. These data remind us that, as is true of leukemia-associated transcription factor fusions, there are certain to be differences with respect to how mutant signaling molecules undermine hematopoietic growth control and how these differences, in turn, will influence the context in which specific mutations are selected during leukemogenesis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal