Abstract
The glycoprotein Ib-V-IX (GPIb-V-IX) complex interacts with subendothelial von Willebrand factor (VWF) to ensure recruitment of platelets at sites of vascular injury, a process that culminates in integrin αIIbβ3-dependent stable adhesion and spreading. Interaction of the 14-3-3ζ adaptor protein with the C-terminal 606-610 phosphoserine motif of the GPIbα subunit has been implicated in the control of αIIbβ3 activation and cell spreading. In this study, we have examined potentially novel 14-3-3ζ binding sites by expressing mutant forms of GPIbα in Chinese-hamster-ovary (CHO) cells. Analysis of a series of neighboring 11-12 residue deletions identified a critical role for the 580-LVAGRRPSALS-590 sequence in promoting GPIbα-14-3-3ζ interaction. Development of a phosphospecific antibody demonstrated high levels of phosphorylation of the Ser587 and Ser590 residues in resting platelets (which became dephosphorylated during platelet spreading on VWF), and peptides containing these phosphorylated residues effectively displaced 14-3-3ζ from GPIbα. Analysis of single and double alanine substitutions of Ser587 and Ser590 demonstrated a major role for these residues in promoting GPIbα-14-3-3ζ binding. Moreover, these cell lines exhibited a defect in cell spreading on immobilized VWF. These studies demonstrate the existence of a second major 14-3-3ζ binding site within the cytoplasmic tail of GPIbα that has an important functional role in regulating integrin-dependent cell spreading. (Blood. 2004;104:420-427)
Introduction
The 14-3-3 protein family consists of ubiquitous homodimeric or heterodimeric intracellular adaptor proteins that take part in the signaling pathways of numerous biologic responses.1 There have been 5 isoforms identified in human platelets, the ϵ and η isoforms being weakly expressed relative to the more abundant γ, ζ, and β isoforms.2 The crystal structure of 14-3-3ζ revealed the association of 2 monomers, each composed of a bundle of 9 antiparallel helices, to form a large negatively charged groove that could be implicated in interactions with other proteins.3 The 14-3-3 proteins bind to specific phosphoserine-containing motifs,4 and their dimeric nature allows them to act as intramolecular and intermolecular phosphorylation-dependent bridges.5 The functions of the different isoforms in platelets are, however, still poorly understood.
In a seminal study, it was reported that 14-3-3ζ interacted with the platelet von Willebrand factor (VWF) receptor, the glycoprotein Ib-V-IX (GPIb-V-IX) complex.6 A binding site for 14-3-3ζ was found at the C-terminus of GPIbα within a serine-rich 606-SGHSL-610 sequence bearing similarities to phosphoserine containing 14-3-3ζ binding motifs.1,7 Further work showed that the serine at position 609 was predominantly phosphorylated in resting platelets and that phosphorylation was required for 14-3-3ζ binding.8 Glutathione S-transferase (GST)-14-3-3ζ pull-down and immunoprecipitation studies of GPIb-IX-transfected cells pointed to the existence of a separate binding or regulatory site in the GPIbα 570-590 domain.9 Experiments using synthetic GPIbβ and GPV intracellular peptides suggested that these subunits could also interact with 14-3-3ζ.10 Phosphorylation of the GPIbβ serine at position 166 by a protein kinase A (PKA)-dependent mechanism has been proposed to play a role in regulating 14-3-3ζ interactions.9-11
Recent studies have demonstrated that the GPIb-14-3-3ζ interaction is dynamically regulated, such that shear-induced activation of platelets leads to dissociation of 14-3-3ζ from GPIb.11 While the precise mechanisms regulating this have not been defined, changes in the phosphorylation status of Ser609 may play an important role. A functional correlation between 14-3-3ζ release from GPIb and integrin activation has been suggested from studies in Chinese-hamster-ovary (CHO) cells transfected with the GPIb-IX complex following deletion of the Glu591-Leu610 GPIbα C-terminal domain.8,9 This deletion prevented 14-3-3ζ-GPIb interactions, leading to defective integrin dependent spreading.12 However, 2 further studies, examining integrin activation using CHO cells transfected with the same constructs, have demonstrated either normal or increased cell spreading.13,14
To establish a precise functional map of the 14-3-3ζ binding regions in the GPIbα cytoplasmic tail, CHO cells were transfected with constructs of the GPIb-IX complex containing short 11-12 residue deletions and single mutations of the GPIbα intracellular domain. The effects of these mutations on GPIb-14-3-3ζ association and cell spreading on VWF were examined. In addition to the known 605-610 region, a new binding site was identified between residues 580 and 590 that resembled other 14-3-3ζ ligands and required Ser residues at positions 587 and 590. Phosphorylation of the diSer587/590 motif was demonstrated in resting platelets using a specific antibody, while its dephosphorylation and that of Ser609 were observed during platelet adhesion and activation on a VWF matrix. Following phosphatase treatment, dephosphorylation led to 14-3-3ζ release from GPIb-IX. Furthermore, lack of 14-3-3ζ association with GPIbα in cells expressing the GPIbα 580-590 deletion mutant, Ser587/590 to Ala substitution, or deletion of the 605-610 sequence resulted in a reduced ability of the VWF-GPIbα interaction to activate endogenous integrins and induce cell spreading.
Materials and methods
Materials
Human VWF (HVWF) was purified according to a published procedure.15 Botrocetin and monoclonal antibodies (mAbs) ALMA.12 against GPIbα and RAM.1 against GPIbβ were developed in our laboratories. The polyclonal antibody C-16 against 14-3-3ζ was from Santa Cruz Biotechnology (Santa Cruz, CA), while the polyclonal antibody K3584 against the GPIb-IX complex was a gift from Dr B. Steiner (Hoffmann-La Roche, Basel, Switzerland). There were 4 peptides representing different phosphorylation states of the 580-590 sequence of human GPIbα (LVAGRRPSALS, LVAGRRPpSALS, LVAGRRPSALpS, and LVAGRRPpSALpS) synthesized by Eurogentec (Seraing, Belgium), who also raised rabbit polyclonal antiserum pSGPIb1 against peptide LVAGRRPpSALpS. GPIbα peptides YSGHpSL and YSGHSL containing an N-terminal cysteine were synthesized by Chiron Mimotopes (Melbourne, Australia). Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG [GAR-HRP]) was from Jackson Laboratories (West Grove, PA). Prostaglandin I2 (PGI2), bovine serum albumin (BSA), fatty-acid-free human serum albumin (HSA), TRITC (tetramethylrhodamine isothiocyanate)-phalloidin, dithiothreitol (DTT), and protein G-Sepharose CL4B were from Sigma Chemicals (St Louis, MO). Apyrase was purified from potatoes as previously described.16 Cover glasses and coverslips were purchased from VWR (Strasbourg, France) and enhanced chemiluminescence (ECL) reagents and Hyper-film-ECL, from Amersham (Bucks, United Kingdom). ReoPro was from Lilly Corporate Center (Indianapolis, IN), complete protease inhibitor cocktail and calpain inhibitor 1 were from Roche Molecular Biochemicals (Mannheim, Germany), and 1-paraformaldehyde (PFA) was from Electron Microscopy Sciences (Washington, PA). Laemmli buffer, Tween 20, Ready gels (4%-15%), and immunoblotting polyvinylidene fluoride (PVDF) membranes were purchased from Bio-Rad (Ivry sur Seine, France) and NHS-Biotin, Sulfo-NHS-Biotin, and Restore Western blot stripping buffer, from Pierce (Rockford, IL).
Antibody production
The anti-phospho-GPIbα (pSer609) antibody was raised by immunizing New Zealand White rabbits with the phosphorylated CYSGHpSL peptide conjugated to keyhole limpet hemocyanin. The antipeptide antibody was affinity purified with the immunizing peptide conjugated to BSA and coupled on a 1:1 column of Affi-Gel 10/15. The antibody was then absorbed with the nonphosphorylated CYSGHSL peptide conjugated to BSA, also coupled on a 1:1 column of Affi-Gel 10/15. The specificity of the anti-pS609 antibody was verified by dot immunoblots against the CYSGH(pS)L and CYSGHSL peptides conjugated to albumin (1:10 wt/wt), versus antibody that bound to both columns, anti-Ibα+/-P, the reactivity of which was phosphorylation nonspecific.
Cell lines
CHO cell lines stably expressing the wild-type GPIb-IX complex or GPIbα deleted forms of GPIb-IX have been described previously.17-19 Stable cell lines with Ser to Ala mutations at positions 587, 590, and a double 587/590 mutation were obtained following transfection of the GPIbα cDNA into GPIbβIX expressing cells as previously described.18 Oligonucleotide-directed mutagenesis was performed by polymerase chain reaction amplification of PDXGPIbα vector with Pfu DNA polymerase (Stratagene, La Jolla, CA), digestion with DpnI (Roche Molecular Biochemicals), and transformation in TOP10 bacteria (Invitrogen, San Diego, CA).
Platelet preparation
Washed human platelets were prepared from acid citrate dextrose (ACD) anticoagulated blood obtained from aspirin-free healthy volunteers by sequential centrifugation as previously described.16
Biotinylation
Cell biotinylation has been described in detail elsewhere.19 Briefly, platelets were washed twice in phosphate-buffered saline (PBS), centrifuged at 1900g for 8 minutes, and incubated for 30 minutes at room temperature (RT) in PBS containing 100 μg/mL NHS-Biotin at a final concentration of 106 platelets/μL.
Lysate preparation
Adherent CHO cells were detached with PBS EDTA (ethylenediaminetetraacetic acid; 5 mM), and washed 3 times in PBS. The lysates were prepared by resuspending washed platelets (106 platelets/μL) or washed cells (20 × 106 cells/mL) in ice-cold 1% Triton X-100, 1 mM EDTA, 1 mM EGTA (ethylene glycol tetraacetic acid), 2.5 mM Na3VO4, 150 mM NaCl, 100 mM NaF, 10 mM Tris (tris(hydroxymethyl)aminomethane), pH 7.5, 17 μg/mL calpain inhibitor I, and 1X protease inhibitor cocktail. After 20 minutes, the lysates were centrifuged at 14 000g for 15 minutes to remove insoluble material. Lysates pretreated with potato alkaline phosphatase (PAP) were prepared under the same conditions but without added Na3VO4 or NaF.
Immunoprecipitation and immunodepletion
Cell lysates were cleared twice by incubation for 60 minutes at 4°C with 50 μL protein G-Sepharose CL4B (50% wt/vol suspension in lysis buffer). Proteins were immunoprecipitated from 50 μL cleared lysate by addition of 5 μg mAb and 80 μL protein G-Sepharose CL4B (50% wt/vol suspension) and rotation for 2 hours at 4°C. The beads were washed 3 times in PBS containing 1% Triton X-100 before addition of 45 μL of 1X Laemmli buffer (62.5 mM Tris, pH 6.8, 0.2% sodium dodecyl sulfate (SDS), 25% glycerol, 0.01% bromophenol blue) containing 10 mM DTT. Samples were boiled for 5 minutes and proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 4% to 15% gels and transferred to PVDF membranes. Biotinylated proteins were revealed with streptavidin-HRP as previously described,19 while nonbiotinylated proteins were identified by Western blotting as previously described.20
In immunodepletion experiments, a first immunoprecipitation was performed using a 2-μL aliquot of cleared platelet lysate to ensure an excess of anti-phospho-GPIb IgG (pSGPIb1 or pSer609) and 80 μL protein G-Sepharose CL4B (50% wt/vol suspension). The remaining supernatant was reprecipitated with a mAb against GPIbβ (RAM.1).
Enzyme immunoassays
Synthetic peptides corresponding to the GPIbα 580-590 domain were coated from 5 μg/mL solutions in 50 mM carbonate buffer (pH 9.5) onto the wells of 96-well microtiter plates (Maxi-Sorb; Nunc, Rochester, NY) for 2 hours at RT. After 3 washes in PBS-T, the wells were blocked by incubation for 30 minutes at 37°C with 200 μL of 1% BSA in PBS. All subsequent incubations were carried out for one hour at RT, and each was followed by washing 3 times with PBS-Tween 0.05% (PBS-T). The wells were treated first with 100 μL diluted pSGPIb1 antiserum and then with GAR-HRP (1:1000 dilution). The secondary antibody was detected by addition of a chromogenic substrate (OPD: o-phenylenediamine dihydrochloride; Pierce) and measurement of optical densities at 490 nm in a THERMO-max microplate reader (Molecular Devices, Sunnyvale, CA).
Cell adhesion to VWF
Glass coverslips were coated with HVWF (10 μg/mL) in a humid chamber for 90 minutes at RT and then blocked for 60 minutes with PBS containing 1% BSA. CHO cells in PBS were incubated with the coverslips in the wells of microtiter plates (105 cells/well) at 37°C for 20 minutes. The coverslips were then washed 3 times in PBS, fixed for 20 minutes with 4% PFA in PBS, washed 3 times in PBS, and labeled for 30 minutes in the dark with 2 μg/mL TRITC-phalloidin. After 3 more washes in PBS and a last wash in water, the adherent cells were covered with 8 μL Mowiol 4-88 solution (France Biochem, Meudon, France), and the coverslips were mounted on microscope slides. Fluorescence was visualized by ultraviolet (UV) illumination at 570 nm under a Leica DMDL microscope (Leica Microsystems, Wetzlar, Germany). The cell count and morphology of the adherent cells were scored in 8 random fields (2.3 × 103 μm2/field).
Analysis of platelet adhesion by confocal microscopy
Glass coverslips were coated with HVWF as before, or coated with 0.01% polylysine for 5 minutes at RT and dried for one hour at 60°C. Washed platelets in Tyrode buffer were incubated with the coverslips in the wells of microtiter plates (3 × 106 platelets/well) for different times at 37°C. When indicated, 5 mM EDTA and 10 μg/mL ReoPro were added to block integrin activation. The coverslips were then washed 3 times in PBS, fixed for 15 minutes with 4% PFA in PBS, washed 3 times in PBS, and blocked with PBS containing 0.2% BSA and 1% normal goat serum (NGS) for one hour at RT. All subsequent incubations were followed by 3 washes in PBS-T. The samples were labeled for 45 minutes in the dark with 1 μg/mL ALMA.12-cyanin 3 (Cy3); permeabilized for one hour in PBS containing 0.2% BSA, 0.05% saponin, and 1% NGS; labeled for 45 minutes in the dark with pSGPIb1 or pSer609 antiserum diluted 1:10 000 in the blocking buffer; and finally counterstained with GAR-Cy5 (1:2000 dilution). The coverslips were then washed in PBS, rinsed with water, and mounted in Mowiol 4-88. The labeled platelets were examined under a Zeiss laser scanning microscope (LSM 510 invert; Carl Zeiss, Le Pecq, France) equipped with a Planapo oil-immersion lens (× 63, numerical aperture 1.4), and the Zeiss CLSM instrument software 2.8. Cy3 emission was excited with the 543-nm He/Ne laser line and signals were filtered with a long-pass 595-nm filter.
Shape change of CHO cells adhering to VWF
CHO cells (1 × 106/mL) suspended in PBS containing 2 mM Ca2+, 2 mM Mg2+, and 5 μg/mL botrocetin were allowed to adhere to an HVWF matrix (deposited from a 10 μg/mL solution) for 30 minutes at 37°C. After fixation, the adherent cells were stained with 2 μg/mL TRITC-phalloidin for 30 minutes, and the coverslips were mounted in Mowiol 4-88 solution on microscope slides. The cell morphology was examined by fluorescence microscopy (UV illumination at 570 nm, Leica DMDL microscope, PL Fluotar 40×/1.00-0.5 oil immersion objective) and visualized using a Micromax 1300Y digital camera (Princeton Instruments, Tucson, AZ) and Metamorph 4.5 software (Princeton Instruments), and numbers of adherent cells and spreading cells were scored in 5 random fields.
Statistical analyses
The statistical significance of differences between means was evaluated using Student t test for paired samples or Fisher exact test, and P values of less than .05 were considered significant.
Results
The GPIbα 580-590 intracytoplasmic sequence is required for the interaction between the GPIb-IX complex and 14-3-3ζ
Interaction of 14-3-3ζ with the GPIb-IX complex was studied in human platelets and transfected CHO cells, following biotinylation and immunoprecipitation with an antibody against GPIbα or GPIbβ. As shown in Figure 1A, coimmunoprecipitation of the 2 proteins was confirmed in human platelet lysates. To identify potentially novel 14-3-3ζ binding sites on GPIbα, this subunit was subjected to a series of truncations covering the 535-610 intracellular region and stably transfected into GPIbβ-IX CHO cells. The GPIb-IX-14-3-3ζ binding capacity was then evaluated by coimmunoprecipitation with an anti-GPIbβ mAb. Deletion of the known 605-610 binding site (Δ605-610) and larger truncations eliminating this domain (Δ518-610, Δ569-610, Δ576-610, and Δ595-610) prevented GPIb-IX-14-3-3ζ coprecipitation (Figure 1B). Conversely, deletion of the more central 535-569 region led to wild-type behavior and allowed efficient association of GPIb-IX with 14-3-3ζ (Figure 1B). To examine additional sites upstream of the 605-610 region, cells with shorter deletions spanning the 535-590 domain were evaluated (Figure 1C). All the deletions allowed for efficient GPIb-IX-14-3-3ζ binding with the exception of Δ580-590, which prevented association with 14-3-3ζ. These results suggested the existence of a second 14-3-3ζ binding site in addition to the previously identified 605-610 sequence.8
GPIb-IX-14-3-3ζ association in platelets and in GPIb-IX CHO cells with intracellular GPIbα deletions. Platelets (A) or CHO cells (B-C) were incubated with membrane-permeable EZ-link-NHS-Biotin and lysed in 1% Triton X-100. GPIb-IX complexes were immunoprecipitated with an anti-GPIb monoclonal antibody, separated by reduced 4% to 15% SDS-PAGE, and transferred to PVDF membranes. Coimmunoprecipitated 14-3-3ζ was detected using the anti-14-3-3ζ-specific polyclonal antibody C-16 and revealed by ECL. Membranes were then stripped and reprobed with streptavidin-HRP to reveal biotinylated GPIbα. The upper panels correspond to the region of the gel containing the approximately 29 kDa 14-3-3ζ band, and the lower panels correspond to the region containing the approximately 135 kDa GPIbα band. (A) In human platelets, 14-3-3ζ was coimmunoprecipitated with the GPIb-IX complex by an antibody against GPIbα or GPIbβ (for simplicity only the GPIbα band is shown). (B) In transfected CHO cells, 14-3-3ζ was coprecipitated with the wild-type (WT) GPIb-IX complex and with a complex containing a central (Δ535-569) deletion. Complete deletion of the intracellular domain of GPIbα (Δ518-610) or progressive truncations from the C-terminal end (Δ605-610, Δ595-610, Δ576-610, and Δ569-610) prevented GPIb-IX-14-3-3ζ association. (C) In cells containing a series of shorter deletions of the GPIbα 535-590 region, GPIb-IX-14-3-3ζ interaction was absent only for the Δ580-590 mutant receptor. Results are representative of 4 separate experiments.
GPIb-IX-14-3-3ζ association in platelets and in GPIb-IX CHO cells with intracellular GPIbα deletions. Platelets (A) or CHO cells (B-C) were incubated with membrane-permeable EZ-link-NHS-Biotin and lysed in 1% Triton X-100. GPIb-IX complexes were immunoprecipitated with an anti-GPIb monoclonal antibody, separated by reduced 4% to 15% SDS-PAGE, and transferred to PVDF membranes. Coimmunoprecipitated 14-3-3ζ was detected using the anti-14-3-3ζ-specific polyclonal antibody C-16 and revealed by ECL. Membranes were then stripped and reprobed with streptavidin-HRP to reveal biotinylated GPIbα. The upper panels correspond to the region of the gel containing the approximately 29 kDa 14-3-3ζ band, and the lower panels correspond to the region containing the approximately 135 kDa GPIbα band. (A) In human platelets, 14-3-3ζ was coimmunoprecipitated with the GPIb-IX complex by an antibody against GPIbα or GPIbβ (for simplicity only the GPIbα band is shown). (B) In transfected CHO cells, 14-3-3ζ was coprecipitated with the wild-type (WT) GPIb-IX complex and with a complex containing a central (Δ535-569) deletion. Complete deletion of the intracellular domain of GPIbα (Δ518-610) or progressive truncations from the C-terminal end (Δ605-610, Δ595-610, Δ576-610, and Δ569-610) prevented GPIb-IX-14-3-3ζ association. (C) In cells containing a series of shorter deletions of the GPIbα 535-590 region, GPIb-IX-14-3-3ζ interaction was absent only for the Δ580-590 mutant receptor. Results are representative of 4 separate experiments.
Deletion of the GPIbα 580-590 sequence does not prevent phosphorylation of Ser609
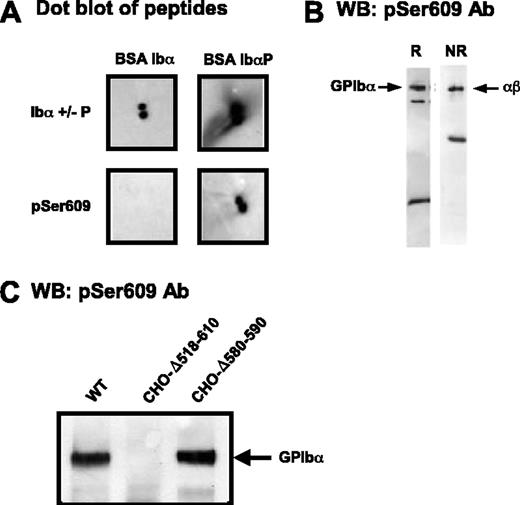
Phosphorylation of Ser609 in the 605-610 sequence has been reported to be critical for the interaction of GPIbα with 14-3-3ζ.8 Therefore, the possibility that the 580-590 deletion could indirectly block 14-3-3ζ interactions by interfering with Ser609 phosphorylation was explored. An antibody (pSer609) was raised and immunopurified against the YSGHpSL peptide. This antibody was specific for the phosphorylated form of the peptide in dot blot experiments (Figure 2A) and recognized GPIbα from lysates of resting platelets (Figure 2B) and CHO cells transfected with normal GPIb-IX (Figure 2C). pSer609 also recognized the 580-590 deleted form of GPIbα in CHO cell lysates, indicating normal phosphorylation at Ser609, whereas the 518-610 deleted form of GPIbα was not revealed by the antibody (Figure 2C).
Deletion of the GPIbα 580-590 sequence does not interfere with Ser609 phosphorylation. (A) Dot blots of BSA-CYSGHSL or BSA-CYSGH(pS)L (peptides corresponding to the nonphosphorylated and phosphorylated C-terminus of GPIbα) with IgG against either the phosphorylation-independent (anti-Ibα+/-P) or phosphorylated (pSer609) C-terminal peptide of GPIbα. Blots were visualized using standard ECL substrates. (B) Western blots (WB) of whole platelet lysates analyzed by SDS-PAGE under reducing (R) or nonreducing (NR) conditions and probed with anti-pS609 (pSer609). The major band at 135 kDa corresponds to GPIbα (disulfide-linked to GPIbβ, nonreduced); the band at approximately 20 kDa reduced or approximately 50 kDa nonreduced represents the membrane-associated calpain digestion fragment of GPIbα (minus the soluble extracellular glycocalicin fragment), which is linked to GPIbβ when nonreduced. (C) Triton X-100 lysates of GPIb-IX CHO cells were separated by 4% to 15% SDS-PAGE and immunoblotted with the pSer609 polyclonal antibody. Phosphorylated GPIbα was detected in cells expressing the wild-type complex or a complex containing the Δ580-590 deletion but not in cells lacking the entire GPIbα intracellular domain.
Deletion of the GPIbα 580-590 sequence does not interfere with Ser609 phosphorylation. (A) Dot blots of BSA-CYSGHSL or BSA-CYSGH(pS)L (peptides corresponding to the nonphosphorylated and phosphorylated C-terminus of GPIbα) with IgG against either the phosphorylation-independent (anti-Ibα+/-P) or phosphorylated (pSer609) C-terminal peptide of GPIbα. Blots were visualized using standard ECL substrates. (B) Western blots (WB) of whole platelet lysates analyzed by SDS-PAGE under reducing (R) or nonreducing (NR) conditions and probed with anti-pS609 (pSer609). The major band at 135 kDa corresponds to GPIbα (disulfide-linked to GPIbβ, nonreduced); the band at approximately 20 kDa reduced or approximately 50 kDa nonreduced represents the membrane-associated calpain digestion fragment of GPIbα (minus the soluble extracellular glycocalicin fragment), which is linked to GPIbβ when nonreduced. (C) Triton X-100 lysates of GPIb-IX CHO cells were separated by 4% to 15% SDS-PAGE and immunoblotted with the pSer609 polyclonal antibody. Phosphorylated GPIbα was detected in cells expressing the wild-type complex or a complex containing the Δ580-590 deletion but not in cells lacking the entire GPIbα intracellular domain.
A GPIbα 580-590 peptide competes with GPIb-IX for association with 14-3-3ζ
The GPIbα 580-590 sequence is well conserved between the human, mouse, and dog species, and its structure with the serines at positions 587 and 590 conformed to a consensus phosphorylation-dependent 14-3-3ζ binding domain, RX1-2(Sp)X2-3S (Figure 3C).21,22 To evaluate the ability of this sequence to bind 14-3-3ζ and the importance of Ser587 and Ser590, phosphorylated and nonphosphorylated peptides were synthesized: LVAGRRPpSALpS (P1P2) and LVAGRRPSALS (ØP). Addition of these peptides to platelet lysates demonstrated that the diphosphorylated peptide (P1P2) efficiently displaced 14-3-3ζ from GPIb-IX, relative to the nonphosphorylated peptide (ØP), irrelevant peptide sequence, and no peptide control (Figure 3B).
A phosphoserine GPIbα 580-590 peptide inhibits GPIb-IX-14-3-3ζ association. (A) Triton X-100 lysates of resting human platelets were immunoprecipitated with the mAb RAM.1 in the presence or absence of the nonphosphorylated ØP (LVAGRRPSALS) or diphosphorylated P1P2 (LVAGRRPpSALpS) GPIbα 580-590 synthetic peptide. Ctrl represents incubation with an unrelated peptide (TNRQPRDKNVKK) from the P2Y12 receptor sequence. RAM.1 coprecipitation of 14-3-3ζ was reduced in the presence of P1P2 and to a lesser extent in the presence of ØP, compared with incubation with no peptide or the Ctrl peptide. NI indicates nonimmune serum. (B) The coprecipitated 14-3-3ζ band was quantified with Quantity One Software (Bio-Rad, Hercules, CA) in 3 separate experiments, and results were expressed as the mean percentage (± SEM) of the band obtained in the absence of peptide (*P < .05). (C)Alignment of the amino acid sequences of the GPIbα 580-610 region in the human, mouse, and dog species. Ser residues are in boldface. The 580-590 domain (underlined) contains fully conserved serine residues with a spacing that corresponds to a consensus 14-3-3ζ binding motif.22 X indicates any amino acid; S, serine; pS, phosphorylated serine; R, arginine; and *, the pS609 residue. (GPIbα human, GPIbα mouse, and GPIbα canis have the GenBank accession numbers PO7359, AAC53320, and AAC14361, respectively.)
A phosphoserine GPIbα 580-590 peptide inhibits GPIb-IX-14-3-3ζ association. (A) Triton X-100 lysates of resting human platelets were immunoprecipitated with the mAb RAM.1 in the presence or absence of the nonphosphorylated ØP (LVAGRRPSALS) or diphosphorylated P1P2 (LVAGRRPpSALpS) GPIbα 580-590 synthetic peptide. Ctrl represents incubation with an unrelated peptide (TNRQPRDKNVKK) from the P2Y12 receptor sequence. RAM.1 coprecipitation of 14-3-3ζ was reduced in the presence of P1P2 and to a lesser extent in the presence of ØP, compared with incubation with no peptide or the Ctrl peptide. NI indicates nonimmune serum. (B) The coprecipitated 14-3-3ζ band was quantified with Quantity One Software (Bio-Rad, Hercules, CA) in 3 separate experiments, and results were expressed as the mean percentage (± SEM) of the band obtained in the absence of peptide (*P < .05). (C)Alignment of the amino acid sequences of the GPIbα 580-610 region in the human, mouse, and dog species. Ser residues are in boldface. The 580-590 domain (underlined) contains fully conserved serine residues with a spacing that corresponds to a consensus 14-3-3ζ binding motif.22 X indicates any amino acid; S, serine; pS, phosphorylated serine; R, arginine; and *, the pS609 residue. (GPIbα human, GPIbα mouse, and GPIbα canis have the GenBank accession numbers PO7359, AAC53320, and AAC14361, respectively.)
The 580-590 domain of GPIbα is phosphorylated in resting platelets
To further analyze the role of the phosphoserines of the GPIbα 580-590 sequence in 14-3-3ζ binding, antibodies were raised against the P1P2 peptide. The pSGPIb1 antiserum specifically bound to P1P2 as detected by enzyme-linked immunosorbent assay (ELISA) but did not recognize the nonphosphorylated peptide ØP (Figure 4A). pSGPIb1 also recognized the monophosphorylated P1 (LVAGRRPpSALS) and P2 (LVAGRRPSALpS) peptides. pSGPIb1 revealed a band corresponding to GPIbα in lysates from resting platelets, suggesting phosphorylation of this domain in the native protein (Figure 4B). The pSer609 antibody also recognized GPIbα, confirming previous findings that Ser609 is phosphorylated in resting platelets.8 The specificity of pSGPIb1 for the phosphorylated motif in the native sequence was demonstrated by a drop in reactivity in platelet lysates treated with alkaline phosphatase (Figure 4C). Similarly pSer609 labeling decreased following phosphatase treatment. Hence in the resting state, GPIbα appeared to be doubly phosphorylated at the 580-590 and 609 sites. The degree of phosphorylation at these 2 positions was then evaluated in resting platelets by immunodepletion of lysates with the pSGPIb1 antiserum or pSer609 antibody. After pSGPIb1 or pSer609 depletion, reprecipitation with a GPIbβ antibody yielded significantly less GPIbα than following nonimmune (Figure 4D), suggesting that GPIbα is similarly phosphorylated at high levels in the 580-590 and 609 domains. To confirm the importance of GPIbα phosphorylation for 14-3-3ζ binding, GST-14-3-3ζ pull-down experiments were performed using phosphatase-treated platelet lysates. The results demonstrate that dephosphorylation prevented GPIbα association with 14-3-3ζ (Figure 4E), an effect that correlated with the decrease in pSGPIb1 and pSer609 labeling.
Serine phosphorylation of the GPIbα 580-590 sequence in resting platelets and requirement for 14-3-3ζ binding. (A) The rabbit antiserum pSGPIb1 was obtained by immunization with the P1P2 phosphopeptide (LVAGRRPpSALpS) and its reactivity was analyzed by ELISA. Dilutions of the serum were incubated with immobilized GPIbα peptides and revealed with GAR-HRP. pSGPIb1 recognized P1P2 and the monophosphorylated P1 (LVAGRRPpSALS) and P2 (LVAGRRPSALpS) peptides but not the nonphosphorylated ØP (LVAGRRPSALS) peptide. Results are the mean (± SEM) of 3 separate experiments, and NI represents nonimmune serum from the same animal. (B) Triton X-100 lysates of resting platelets were separated by 4% to 15% SDS-PAGE and immunoblotted with pSGPIb1, pSer609, or an antibody against the 3 GPIb-IX subunits. GPIbα was heavily labeled by pSGPIb1, suggesting Ser phosphorylation of the 580-590 domain. In accordance with previous reports, labeling with pSer609 showed that GPIbα was also phosphorylated at Ser609. (C) Triton X-100 lysates of resting platelets were treated or not with 0.3 U/mL PAP for 30 minutes at 37°C. Proteins were separated by 4% to 15% SDS-PAGE, transferred to PVDF membranes, and probed with pSer609 and pSGPIb1. Reactivity to the antibodies was lost in samples treated with PAP, confirming phosphorylation at both sites. (D) The proportion of phosphorylated GPIbα was estimated by immunodepleting the cell lysates with an excess of the phosphospecific antibody pSGPIb1 (middle panel) or pSer609 (right panel) and performing a second immunoprecipitation with an anti-GPIbβ antibody. Products of the first and second immunoprecipitations were revealed with a polyclonal anti-GPIb-IX antibody. In both cases, a similar small proportion of GPIb-IX was revealed following the second immunoprecipitation, indicating that most GPIbα subunits were phosphorylated at both sites. The left panel corresponds to a control where the first depletion was performed in the presence of a nonimmune serum, and results are from 1 experiment representative of 4 independent assays. (E) In GST-14-3-3ζ pull-down experiments, proteins were precipitated from the same lysates as in panel C and probed with an antibody against the GPIb-IX complex. GST-14-3-3ζ precipitated GPIbα in the absence of PAP treatment but not after its dephosphorylation by PAP. A negative control using GST alone is shown in the left lane. Results in panels B-E are from 1 experiment representative of 3.
Serine phosphorylation of the GPIbα 580-590 sequence in resting platelets and requirement for 14-3-3ζ binding. (A) The rabbit antiserum pSGPIb1 was obtained by immunization with the P1P2 phosphopeptide (LVAGRRPpSALpS) and its reactivity was analyzed by ELISA. Dilutions of the serum were incubated with immobilized GPIbα peptides and revealed with GAR-HRP. pSGPIb1 recognized P1P2 and the monophosphorylated P1 (LVAGRRPpSALS) and P2 (LVAGRRPSALpS) peptides but not the nonphosphorylated ØP (LVAGRRPSALS) peptide. Results are the mean (± SEM) of 3 separate experiments, and NI represents nonimmune serum from the same animal. (B) Triton X-100 lysates of resting platelets were separated by 4% to 15% SDS-PAGE and immunoblotted with pSGPIb1, pSer609, or an antibody against the 3 GPIb-IX subunits. GPIbα was heavily labeled by pSGPIb1, suggesting Ser phosphorylation of the 580-590 domain. In accordance with previous reports, labeling with pSer609 showed that GPIbα was also phosphorylated at Ser609. (C) Triton X-100 lysates of resting platelets were treated or not with 0.3 U/mL PAP for 30 minutes at 37°C. Proteins were separated by 4% to 15% SDS-PAGE, transferred to PVDF membranes, and probed with pSer609 and pSGPIb1. Reactivity to the antibodies was lost in samples treated with PAP, confirming phosphorylation at both sites. (D) The proportion of phosphorylated GPIbα was estimated by immunodepleting the cell lysates with an excess of the phosphospecific antibody pSGPIb1 (middle panel) or pSer609 (right panel) and performing a second immunoprecipitation with an anti-GPIbβ antibody. Products of the first and second immunoprecipitations were revealed with a polyclonal anti-GPIb-IX antibody. In both cases, a similar small proportion of GPIb-IX was revealed following the second immunoprecipitation, indicating that most GPIbα subunits were phosphorylated at both sites. The left panel corresponds to a control where the first depletion was performed in the presence of a nonimmune serum, and results are from 1 experiment representative of 4 independent assays. (E) In GST-14-3-3ζ pull-down experiments, proteins were precipitated from the same lysates as in panel C and probed with an antibody against the GPIb-IX complex. GST-14-3-3ζ precipitated GPIbα in the absence of PAP treatment but not after its dephosphorylation by PAP. A negative control using GST alone is shown in the left lane. Results in panels B-E are from 1 experiment representative of 3.
Phosphorylation status of the GPIbα 580-590 and 609 domains in resting and adherent platelets
To evaluate phosphorylation of GPIbα in intact platelets, dual-label confocal analysis was performed using either pSGPIb1 or pSer609 (detected with fluorescein isothiocyanate [FITC]) and the pan-anti-GPIbα mAb ALMA.12 (coupled to Cy3). Platelets fixed in a resting discoid state and allowed to adhere to a polylysine surface were predominantly double labeled (yellow), indicating the colocalization of pSGPIb1 or pSer609 with ALMA.12 (Figure 5A,D). This confirmed that most of the GPIbα molecules were phosphorylated at the 580-590 and Ser609 sites in the resting state, in agreement with the biochemical data.
The GPIbα intracellular domain is dephosphorylated after platelet adhesion and shape change on a VWF matrix. Resting platelets were either fixed and allowed to adhere to a polylysine matrix (A,D), or allowed to adhere to a VWF matrix in the presence (B,E) or absence (C,F) of EDTAby incubation with the matrix for 20 minutes in the presence of botrocetin followed by washing and fixation. GPIbα was labeled with ALMA.12-Cy3 (red), and phosphorylated GPIbα was labeled with pSGPIb1 (A-C) or pSer609 (D-F), which was revealed with a secondary FITC-coupled antibody (green). Samples were analyzed by dual-label confocal microscopy. (A,D) Discoid platelets appeared predominantly yellow after staining with either pSGPIb1 or pSer609, indicating that GPIbα was mainly phosphorylated in resting platelets. (B,E) When platelets adhered to a VWF matrix and integrin activation was blocked with EDTA and ReoPro, they extended numerous filopodia. The staining of the cell body remained yellow, whereas the filopodia appeared in red, suggesting a lack of recognition by pSer609 or pSGPIb1 and dephosphorylation of pSer609 and pSer587/590 within the membrane extensions. (C,F) In the absence of ReoPro and EDTA, platelets spread on the VWF matrix and the labeling appeared as a mixture of yellow and red over the entire cell surface, pointing to the existence of separate pools of phosphorylated and dephosphorylated GPIbα. Shown is 1 representative experiment of 3 performed. Scale bars indicate 10 μm.
The GPIbα intracellular domain is dephosphorylated after platelet adhesion and shape change on a VWF matrix. Resting platelets were either fixed and allowed to adhere to a polylysine matrix (A,D), or allowed to adhere to a VWF matrix in the presence (B,E) or absence (C,F) of EDTAby incubation with the matrix for 20 minutes in the presence of botrocetin followed by washing and fixation. GPIbα was labeled with ALMA.12-Cy3 (red), and phosphorylated GPIbα was labeled with pSGPIb1 (A-C) or pSer609 (D-F), which was revealed with a secondary FITC-coupled antibody (green). Samples were analyzed by dual-label confocal microscopy. (A,D) Discoid platelets appeared predominantly yellow after staining with either pSGPIb1 or pSer609, indicating that GPIbα was mainly phosphorylated in resting platelets. (B,E) When platelets adhered to a VWF matrix and integrin activation was blocked with EDTA and ReoPro, they extended numerous filopodia. The staining of the cell body remained yellow, whereas the filopodia appeared in red, suggesting a lack of recognition by pSer609 or pSGPIb1 and dephosphorylation of pSer609 and pSer587/590 within the membrane extensions. (C,F) In the absence of ReoPro and EDTA, platelets spread on the VWF matrix and the labeling appeared as a mixture of yellow and red over the entire cell surface, pointing to the existence of separate pools of phosphorylated and dephosphorylated GPIbα. Shown is 1 representative experiment of 3 performed. Scale bars indicate 10 μm.
In previous studies it has been demonstrated that when platelets adhere to VWF in the presence of αIIbβ3 inhibitors, they become activated, change shape, and extend filopodia. The cell body of platelets adhering to a VWF matrix under these conditions remained double labeled (yellow). However, all the filopodia appeared in red (Figure 5B,E), indicating a lack of pSGPIb1 or pSer609 labeling and hence a significant dephosphorylation of Ser587/590 and Ser609. In the absence of integrin blockade, platelets can spread on a VWF surface and such conditions resulted in a mixture of red and yellow labeling over the entire cell surface (Figure 5C,F). These results show that a significant proportion of GPIbα molecules become dephosphorylated during the processes leading to integrin activation.
Critical role of Ser590 in the 589-590 domain for 14-3-3ζ binding and integrin activation
To more precisely define the role of the serine residues in the 580-590 region in promoting GPIbα association with 14-3-3ζ, Ser587 and Ser590 were substituted to Ala either individually or together and GPIbα was introduced into CHO cells containing GPIbβ-IX. Coimmunoprecipitation studies demonstrated a decreased 14-3-3ζ association for all the mutants that was almost completely abolished for the 2 cell lines containing the Ser590Ala substitution (Figure 6A). This defect in 14-3-3ζ binding was similar to that observed with the GPIbαΔ580-590 deletion, suggesting a major role for Ser590 in 14-3-3 binding with a secondary role contributed by Ser587. In order to then assess the functional importance of the 14-3-3ζ pool bound to the two 14-3-3 binding sites of GPIbα for integrin activation, GPIb-IX CHO cell lines containing either the deletion mutants (GPIbαΔ605-610 and GPIbαΔ580-590) or the specific alanine mutants (αA587, αA590, and αA587A590) were studied in VWF adhesion assays. Cells expressing wild-type GPIb/IX underwent extensive cell spreading, indicating that intact GPIb/IX is able to mediate efficient integrin activation, observations consistent with previous studies.12,13 A comparison with the deletion mutant cells, which had completely lost the ability to support GPIbα-14-3-3ζ association (Figure 1C), indicated that although these cells were still able to adhere as efficiently as wild-type control cells (Figure 6B) they were significantly less susceptible to spread on the VWF matrix (Figure 6B-C). This suggested inefficient integrin activation possibly due to an absence of mobilizable 14-3-3ζ. Similar assays were performed with the alanine mutants that demonstrated a profound and specific decrease in cell spreading for the 2 clones mutated at Ser590, reproducing the defect observed in GPIbαΔ 580-590 cells (Figure 6D). In contrast, the single Ser587Ala substitution did not prevent cell spreading. These observations suggested a central role for Ser590 to induce efficient integrin activation potentially as a result of a mobilizable pool of 14-3-3.
Involvement of serines in the 580-590 domain for 14-3-3ζ binding and integrin activation. (A) GPIb-IX-14-3-3ζ association was studied as described in Figure 1 in CHO GPIb-IX cells where Ser587 and Ser590 were individually substituted to Ala (αA587, αA590) or in cells containing a double mutation (αA587/590). A profound decrease in GPIb-IX-14-3-3ζ association was observed in both cells with a Ser590 to Ala substitution and a milder defect was observed for the single Ala587 mutant. The results are representative of 3 separate experiments. (B-C) GPIb-IX CHO cells expressing wild-type GPIbα (αβIX) or GPIbα with deletions of the 580-590 or 605-610 sequence were allowed to adhere to a VWF matrix in the presence of botrocetin (5 μg/mL) and left to spread for 30 minutes at 37°C. The cells were then fixed in 4% PFA and the actin cytoskeleton was labeled with TRITC-phalloidin. (B) Compared with αβIX cells, αΔ605-610βIX and αΔ580-590βIX cells adhered equally as efficiently but displayed a marked spreading defect. (C) Spreading cells were scored in 5 random fields and expressed as a percentage of total adherent cells. (***P < .001; Fisher exact test.) Results are the mean (± SEM) of 4 separate experiments. (D) Cells containing Ser to Ala substitutions as described in panel A were allowed to adhere and spread as described in panel B. Spreading cells were scored in 5 random fields and expressed as a percentage of total adherent cells. The level of spreading at 30 minutes in control αβIX cells was lower than in experiments presented in panel B due to a new batch of botrocetin with a lower activity. (***P < .001; *P < .05 Fisher exact test.) Results are the mean (± SEM) of 2 to 3 separate experiments.
Involvement of serines in the 580-590 domain for 14-3-3ζ binding and integrin activation. (A) GPIb-IX-14-3-3ζ association was studied as described in Figure 1 in CHO GPIb-IX cells where Ser587 and Ser590 were individually substituted to Ala (αA587, αA590) or in cells containing a double mutation (αA587/590). A profound decrease in GPIb-IX-14-3-3ζ association was observed in both cells with a Ser590 to Ala substitution and a milder defect was observed for the single Ala587 mutant. The results are representative of 3 separate experiments. (B-C) GPIb-IX CHO cells expressing wild-type GPIbα (αβIX) or GPIbα with deletions of the 580-590 or 605-610 sequence were allowed to adhere to a VWF matrix in the presence of botrocetin (5 μg/mL) and left to spread for 30 minutes at 37°C. The cells were then fixed in 4% PFA and the actin cytoskeleton was labeled with TRITC-phalloidin. (B) Compared with αβIX cells, αΔ605-610βIX and αΔ580-590βIX cells adhered equally as efficiently but displayed a marked spreading defect. (C) Spreading cells were scored in 5 random fields and expressed as a percentage of total adherent cells. (***P < .001; Fisher exact test.) Results are the mean (± SEM) of 4 separate experiments. (D) Cells containing Ser to Ala substitutions as described in panel A were allowed to adhere and spread as described in panel B. Spreading cells were scored in 5 random fields and expressed as a percentage of total adherent cells. The level of spreading at 30 minutes in control αβIX cells was lower than in experiments presented in panel B due to a new batch of botrocetin with a lower activity. (***P < .001; *P < .05 Fisher exact test.) Results are the mean (± SEM) of 2 to 3 separate experiments.
Discussion
The 96-amino acid GPIbα intracellular domain (residues 515-610) was originally described as a bridging sequence between the GPIb-V-IX complex and the platelet actin cytoskeleton via filamin.23 Deficient association with the cytoskeleton likely contributes to the giant platelet phenotype observed in Bernard-Soulier syndrome.24 It is now becoming apparent that this domain is multifunctional and can assemble additional partners potentially involved in signal transduction.25 GPIb/VWF signaling leads to the mobilization of intracellular Ca2+, platelet shape change, and activation of integrin αIIbβ3.12,26 The precise mechanism whereby binding of VWF to GPIb-IX triggers αIIbβ3-dependent functions such as platelet spreading and aggregation nevertheless remains unclear. Recruitment of the 14-3-3ζ adaptor protein to the GPIbα 605-610 C-terminal region has been proposed to play a role in integrin activation.7,12 The present study provides evidence for the existence of a second 14-3-3ζ binding site in the upstream 580-590 region and suggests the implication of both the 605-610 and 580-590 domains for functional GPIbα-14-3-3ζ association and in the control of integrin dependent spreading.
An interaction of 14-3-3ζ with the GPIb-V-IX complex was initially revealed by their coisolation during affinity chromatography purification of GPIb from platelet lysates.6 Subsequent synthetic peptide binding experiments and deletion mutagenesis in GPIb-IX-transfected CHO cells located a binding site for 14-3-3ζ in the last 6 amino acids of the GPIbα subunit (residues 605-610).7,10 A second binding site for 14-3-3ζ has been assigned to the 557-575 region of the GPIbα subunit, mainly from studies of synthetic peptide binding.10 More recently, the existence of yet another binding site upstream of the 605-610 domain was suggested from the lack of GPIbα-14-3-3ζ coprecipitation in cells deleted of the 570-590 region.9 In this work, a closer deletion scanning of the 535-610 region revealed defective GPIb-14-3-3ζ association only for the 580-590 deletion mutant, in addition to the C-terminal truncation. Together, this study and previous work19 indicate that in fact the 570-590 sequence contains separate functional sites involved in filamin (570-580) and 14-3-3ζ binding (580-590), respectively.
It was of interest that the 580-590 site contained serines at positions that agreed with a consensus 14-3-3ζ binding motif.22 The capacity of a phosphorylated 580-590 synthetic peptide to inhibit GPIbα-14-3-3ζ coimmunoprecipitation provided supporting evidence that the GPIbα 580-590 domain is involved in 14-3-3ζ binding, and also agreed with the mode of interaction of most 14-3-3ζ ligands, which require phosphorylation at key Ser or Thr residues for efficient binding.4 In a previous study, 2 large peptides (571-589 and 585-603) that partially overlapped the 580-590 sequence failed to bind purified 14-3-3ζ.10 This apparent discrepancy could be related to the use of nonphosphorylated peptides, as the nonphosphorylated 580-590 peptide competed less efficiently for 14-3-3ζ association (Figure 3). It can also be inferred from the present results that the 14-3-3ζ molecule can accommodate 2 peptide sequences from the GPIbα intracellular domain. This mode of interaction might be necessary to allow for efficient binding of the adaptor protein and for a fine control of its release. There are previous evidences that 14-3-3ζ can bind more than one peptide in other systems. For example, there was a report on the crystal structure of dimeric 14-3-3ζ in complex with 2 peptides from the kinase Raf-1,27 and, more recently, 3 separate binding sites were identified for the phosphatase cdc25B peptides.5 The C-terminal α-helix of 14-3-3ζ (helix I)12,28 appears to contain the GPIbα binding motifs, but the precise mode of interaction and in particular its capacity to interact with the 580-590 and 605-610 sequences remain to be determined.
Functional relevance of the 580-590 domain and its phosphorylation was indicated in a series of experiments in platelets with the use of an antibody against the diphosphorylated peptide. These studies revealed that the GPIbα 580-590 domain is phosphorylated in resting platelets. Phosphorylation was observed both in platelet lysates by immunoblotting and in intact cells by confocal microscopy analysis. Use of an antibody against the pSer609 sequence confirmed that this domain is likewise phosphorylated in resting platelets. Phosphorylation at both sites appears to be required to support efficient 14-3-3ζ-GPIb-V-IX association in resting platelets. Firstly, phosphatase treatment of platelet lysates prevented 14-3-3ζ-GPIb-V-IX association and was accompanied by dephosphorylation of pSer587/590 and pSer609. Studies in GPIb-IX-transfected CHO cells were also consistent with this model and showed that deletion of either binding site prevented association of GPIbα with 14-3-3ζ. Ala mutagenesis revealed that residues 587 and 590 were both required for correct association.
The occurrence of dephosphorylation of these 14-3-3ζ binding sites during the process leading to platelet spreading supports their role in a pathway leading to integrin activation. Platelets adhering to a VWF matrix initially extend thin filopodia in a process that has been found to be integrin independent.26 This is followed by platelet spreading, which requires integrin αIIbβ3 activation. Strikingly, the GPIbα molecules recruited to filopodial extensions were no longer recognized by pSer587/590 or pSer609 phosphospecific antibodies. This points to an early and localized dephosphorylation mechanism that precedes integrin activation and occurs concomitant with cytoskeletal reorganization. Deletion mutant cells lacking 14-3-3ζ binding are still able to extend filopodia, suggesting that the release of 14-3-3ζ from GPIbα is not required for this early cytoskeletal reorganization (data not shown). On the other hand, their inefficient spreading suggests a role in the late integrin-dependent shape change. In fully spread platelets, confocal microscopy indicated that a majority of the GPIbα subunits were dephosphorylated at this later integrin-dependent stage. Since phosphatase treatment leads to a decreased GPIbα-14-3-3ζ interaction, one likely consequence of the pSer587/590 and pSer609 dephosphorylation in platelets adhering to VWF will be the release of GPIbα-associated 14-3-3ζ. Consistent with this proposal, it has been reported that 14-3-3ζ was released from the GPIb-V-IX complex in shear activated platelets.9 Although it cannot be excluded that the loss of antibody labeling during shape change could be indirect, due for example to a change in GPIbα conformation or to the binding of other molecules that mask the epitopes, the fact that it was observed concomitantly for distinct GPIbα domains makes this appear less likely.
Deletion of the C-terminal end of GPIbα that contains the 605-610 14-3-3ζ binding site has led to discordant conclusions concerning the role of this adaptor protein in integrin activation. CHO cells transfected with integrin αIIbβ3 and GPIb-IX containing the same GPIbα 591-610 deletion were found to undergo defective12 or on the contrary normal or increased spreading13,14 following adhesion to a VWF matrix. These discrepancies are not easily explained but could be due to the use of larger deletions (591-610), which could affect other mechanisms, to the use of cells expressing different levels of integrin or 14-3-3, or to differences in the adhesive assays (adhesive proteins, botrocetin, time-course...). In the present study, deletion of the minimal GPIbα 605-610 binding site or the second binding motif in the 580-590 region prevented normal spreading of GPIb-IX-transfected cells on VWF. Furthermore, a single amino acid change of Ser590 to Ala, which has a low probability to alter the protein structure, yielded the same result. Collectively these findings favor a model in which a population of 14-3-3ζ molecules bound at the resting state to the intracellular face of GPIb-IX through 2 phosphorylated motifs is required for integrin activation following its release from the GPIb-IX complex upon GPIb/VWF interaction.
The concomitant phosphorylation/dephosphorylation of the 580-590 and 605-610 regions of GPIbα in resting versus VWF-activated platelets suggests the possibility of a coordinated control by specific kinases and phosphatases. PKA and protein kinase G (PKG) were recently implicated in the control of platelet activation following GPIb/VWF interaction.29,30 However, it has been reported that inhibitors of PKA and PKG have no effect on Ser609 phosphorylation.8 The 580-590 sequence and the Ser587 site display good homology with a consensus PKA phosphorylation site (RRXSX).30 However, incorporation of 32P was not observed in the presence of agents increasing cyclic adenosine monophosphate (cAMP) levels such as forskolin (data not shown), and phosphorylation of GPIbα could be documented only with phosphospecific antibodies. This was surprising in view of the large proportion of phosphorylated GPIbα detected with the phosphospecific antibodies. This could be related to a slow rate of phosphate exchange during the washing procedure, which is designed to avoid platelet activation.16
In conclusion, the present study describes a new 14-3-3ζ binding site located in the 580-590 region of GPIbα, which requires Ser phosphorylation in resting platelets. Adhesion of platelets to a VWF matrix led to dephosphorylation of GPIbα pSer587/590 and pSer609. This would seem to be a common regulatory mechanism mediating release of 14-3-3ζ from the GPIb-IX complex and thereby integrin activation and subsequent cell spreading.
Prepublished online as Blood First Edition Paper, March 30, 2004; DOI 10.1182/blood-2003-08-2881.
Supported in part by a grant from the Australia National Health & Medical Research Council and National Heart Foundation to S.L.C., and S.P.J., P.M., and T.D. were supported by the “Association de Recherche et de Développement en Médecine et Santé Publique” (ARMESA). M.C.B. was supported by National Institutes of Health (NIH) grant RO1 50744-02.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Martine Santer, Marie-Jeanne Baas, Sylvie Moog, and Julie Thomas for technical assistance; Sylvette Chasserot for help in confocal microscopy studies; and Juliette N. Mulvihill for reviewing the English of the manuscript.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal