Abstract
The CD11b/CD18 integrin plays a crucial role in cell-cell adhesion processes. Recently, we described a case of severe neonatal alloimmune neutropenia (NAIN) caused by an alloantibody against a variant of the CD11b subunit (Mart alloantigen). Allele-specific transfected cells allowed us to demonstrate that an H61R point mutation is directly responsible for the formation of Mart epitopes. No difference in the adhesion capability between H61 and R61 homozygous neutrophils was observed. Functional analysis showed that anti-Mart inhibited Mac-1–dependent adhesion of neutrophils and monocytic U937 cells to fibrinogen, intercellular adhesion molecule-1 (ICAM-1), receptor for advanced glycation end product (RAGE), and glycoprotein Ibα but not to junctional adhesion molecule-C or urokinase plasminogen activator receptor (uPAR). Accordingly, anti-Mart blocked neutrophil and U937 cell adhesion to endothelial cells and platelet-leukocyte aggregate formation in whole blood under high shear. Other sera of anti-Mart from mothers of infants without NAIN did not show inhibitory properties. We conclude that anti-Mart antibodies with different functional properties exist. This is supported by our findings that anti-Mart antibodies have different abilities to inhibit cell-cell adhesion, to enhance the respiratory burst of neutrophils, and to recognize different epitopes at the N-terminal region of CD11b. In conclusion, some anti-Mart alloantibodies interfere with Mac-1–dependent cellular functions of neutrophils, cause NAIN, and may be used as tools for studying Mac-1–dependent functions.
Introduction
Multicellular interactions between leukocytes and the blood vessel wall play an important role in inflammation, thrombosis, and immune responsiveness. This interaction is a multistep paradigm that is mediated by selectins, integrins, and junctional adhesion molecules.1,2 Signal-dependent alteration of β2 integrin family members is critical for the adhesion process, the arrest, and the transendothelial migration of leukocytes. The leukocyte β2 integrin family consists of 4 members sharing a common β-subunit (CD18), which is noncovalently associated with 4 different α-subunits (CD11a-d).3 CD11b/CD18, also known as Mac-1, CR3, or αMβ2-integrin, plays an important role in cell adhesion and phagocytotic processes. In this regard, CD11b/CD18 binds a wide range of ligands, including extracellular matrix proteins and coagulation proteins.4-6 Several data indicate that the CD11b/CD18 integrin is a major adhesion molecule in cellular interactions between leukocytes and endothelial cells or platelets. Leukocytes can be recruited to sites of platelet deposition,7,8 and platelet-induced neutrophil recruitment was shown to be of particular importance under high shear conditions in which leukocytes may attach to platelets rather than to endothelium.9 Recent studies indicate that glycoprotein Ibα (GPIbα) and junctional adhesion molecule-3 (now termed JAM-C) on platelets are potential counterreceptors for Mac-1,10,11 with JAM-C being important under low shear and GPIbα under high shear conditions. In this article, a consistent nomenclature proposed by Muller12 and agreed to by a committee of scientists in the JAM field will be used. Furthermore, it has been proposed that intercellular adhesion molecule-2 (ICAM-2) and αIIbβ3–associated fibrinogen mediate Mac-1–dependent leukocyte-platelet interactions13,14 ; however, the exact contribution of each system remains to be elucidated.
The CD11b subunit contains 7 repeating motifs of approximately 60 amino acids in the N-terminal region. A sequence of 200 amino acids, known as I domain (or A domain), is inserted between repeats 2 and 3. It has been shown that this I domain is important for ligand binding (eg, of iC3b, ICAM-1, fibrinogen).15 The binding is critically coordinated by Mg2+-cations that bind to the metal ion-dependent adhesion site (MIDAS). A region downstream of the I domain contains 3 repeats (V, VI, and VII) that resemble the EF-hand loop structure and that function as calcium-binding sites. The C-terminal region contains a sugar-binding site known as the lectin domain.5
The CD11b subunit is also known to be polymorphic and immunogenic in humans. The point mutation 230G>A in the CD11b gene, predicting an R61H dimorphism, is associated with the Mart alloantigen.16 Corresponding to the current nomenclature, this antigen is designated as the human neutrophil alloantigen (HNA) 4a.17 Carriers of the high-frequency G230 allele of CD11b (R61) appear to be Mart-positive, whereas homozygous persons with the A230 allele are Mart-negative (H61). Alloantibodies against the high-frequency R61 isoform, termed anti-Mart, were discovered almost 2 decades ago. By screening neutrophil reactive alloantibodies in 4800 multiparous women, 3 mothers with Mart alloantibodies were detected, but none of their infants had any apparent clinical signs of neonatal alloimmune neutropenia (NAIN).18 Recently, we described the first case of severe NAIN linked to Mart in a first-born neonate, demonstrating the potential clinical significance of Mart alloantigen.19 In this study we assessed the molecular and functional properties of Mart alloantigen and alloantibodies and investigated their roles in leukocyte adhesion.
Materials and methods
Monoclonal antibodies and purified proteins
Monoclonal antibodies (mAbs) bear-1 and ICRF-44 (anti-CD11b; Immunotech, Marseilles, France), blocking mAbs 2LPM19c (anti-CD11b; DAKO, Hamburg, Germany) and IB4 (anti-CD18; Alexis, Grünberg, Germany), mAb MY4 (anti-CD14; Beckman Coulter, Krefeld, Germany), mAb GA6 (anti-CD62P; Becton Dickinson, Heidelberg, Germany), and normal mouse immunoglobulin G (IgG; Becton Dickinson) were purchased. mAb CBRM1/32 was kindly provided by Dr T. A. Springer (Boston, MA). Hybridoma 7D8 producing mAb against CD177 was kindly provided by Dr D. Stroncek (Bethesda, MD). The following purified proteins were used in this study: I domain (provided by Dr D. Tuckwell, Manchester, United Kingdom), ICAM-1 (provided by Dr S. Bodary, Genentech, South San Francisco, CA), glycocalicin (provided by Dr K. Clemetson, Theodor Kocher Institute, Bern, Switzerland), recombinant receptor for advanced glycation end product20 (RAGE; provided by Dr M. Nagashima, Berlex Biosciences, Richmond, CA), recombinant urokinase plasminogen activator receptor21 (uPAR; provided by Dr D. B. Cines, Philadelphia, PA), and fibrinogen (purchased from Calbiochem, Schwalbach, Germany). The stable thromboxane A2 mimetic U46619 was provided by Dr Stegmeier (Roche, Mannheim, Germany).
Sera
Three sera containing neutrophil-specific Mart alloantibodies (sera 2-4; Table 2), described by Kline et al,18 were kindly provided by Dr D. Stroncek (University of Minnesota, Minneapolis). Another anti-Mart antibody (serum 1) was obtained from a mother who delivered a baby with severe NAIN and respiratory distress.19 IgG was isolated from these sera using protein G Sepharose columns (Pharmacia, Freiburg, Germany), as recommended by the manufacturer. The specificity of all sera and IgG preparations against the CD11b subunit was confirmed by mAb immobilization of the granulocyte antigen (MAIGA) assay using a panel of CD11b-specific mAbs as capture antibodies and granulocytes from donors who underwent Mart phenotyping.22
Human anti-Mart alloantisera
Serum . | Initial name . | Specificity . | NAIN . | Origin . | Reference . |
|---|---|---|---|---|---|
| 1 | Anti-Mart | CD11b (R61) | Yes | Australia | Fung et al19 |
| 2 | Anti-Piek | CD11b (R61) | No | United States | Kline et al18 |
| 3 | Anti-Hopk | CD11b (R61) | No | United States | Kline et al18 |
| 4 | Anti-Mart | CD11b (R61) | No | United States | Kline et al18 |
| Simsek et al16 |
Genotyping of Mart alloantigen by sequence-specific PCR (PCR-SSP)
Primers used to genotype Mart polymorphism on the CD11b gene were constructed according to the National Center for Biotechnology Information (NCBI database; accession number NT_024812). DNA was isolated from healthy donors, as described previously.23 In the population study, 360 unrelated healthy blood donors were assessed. All donors gave their informed consent. Aliquots of 60 ng DNA were amplified using 0.5 pmol allele-specific sense primers (5′-CTC ATG CGA GCC CAT CCG-3′ or 5′-CTC ATG CGA GCC CAT CCA-3′) and an intronic antisense primer (5′-ACAAGG AGG TCT GAC GGT G-3′), which is located 223 base pairs (bp) downstream from exon 3. Polymerase chain reaction (PCR) was performed with 0.2 mmol desoxynucleotide triphosphate (dNTP) and 2.0 units (U) TaqGold polymerase on a PCR Express Thermal Cycler (Thermo Life Sciences, Ulm, Germany) in a total volume of 20 μL. After heating at 95° C for 10 minutes, 2-step PCR was performed under the following conditions: denaturation (30 seconds, 95° C), annealing (40 seconds, 64° C), extension (30 seconds, 72° C) for 10 cycles, denaturation (30 seconds, 95° C), annealing (30 seconds, 61° C), extension (30 seconds, 72° C) for 20 cycles, and final extension (5 minutes, 72° C). As internal positive control, 0.4 pmol human growth hormone (hGH) primers amplifying a 439-bp fragment of the hGH gene were used. PCR products were analyzed on 1.6% agarose gels using Tris borate EDTA buffer (TBE-buffer; Gibco BRL, Karlsruhe, Germany).
Production of CD18 expression vector
Total leukocyte RNA was isolated from 10 mL EDTA (ethylenediaminetetraacetic acid)–anticoagulated blood using Roti Quick Kit (Roth, Karlsruhe, Germany), as recommended by the manufacturer. Leukocyte RNA (31 μL) was transcribed into cDNA with 10 μM random hexamer primer (Roche, Mannheim, Germany) using the Ready-to-Go Kit (Amersham Biosciences, Freiburg, Germany). To amplify the entire coding region of the CD18 subunit, 5 μL cDNA was amplified by PCR using 0.25 μmol sense primer (5′-CTC CAG CAC ACC GAG GGA CAT G -3′), 0.25 μmol antisense primer (5′-GTC TTC ACC AAG TGC TCC TAA C-3′), 175 μmol of each dNTP, and 2.5 U TaqGold polymerase in a total volume of 50 μL. Amplification was performed on a DNA thermal cycler for 30 cycles. Each cycle consisted of denaturation (30 seconds, 95° C), annealing (30 seconds, 59° C), extension (90 seconds, 72° C), and final extension (10 minutes, 72° C). After purification using the QIAquick kit (Qiagen, Hilden, Germany), PCR products were subcloned into the pCDNA4Zeo vector (Stratagene, Heidelberg, Germany). Plasmid DNA from positive clones was validated by nucleotide sequence analysis.
Construction of allele-specific CD11b expression vectors
Full-length cDNA encoding the wild-type CD11b isoform in the pcDNA3.1Neo expression vector was kindly provided by Dr A. Law (Nanyang Technological University, Singapore). Specific mutation G/A at position 230 was induced in the wild-type CD11b construct by site-directed mutagenesis using the QuickChange Mutagenesis Kit (Stratagene). For PCR amplification, a single nucleotide–mismatched sense primer (5′-CAT GCG AGC CCA TCC ACC TGC AGG TCC C-3′) and antisense primer (5′-GGG ACC TGC AGG TGG ATG GGC TCG CAT G-3′) were constructed. After 12 cycles of amplification (denaturation for 30 seconds at 95° C, annealing for 60 seconds at 55° C, and extension for 9 minutes at 68° C), PCR products were digested with DpnI endonuclease and transformed into XL1-Blue supercompetent Escherichia coli bacteria. Mutation was validated by nucleotide-sequence analysis.
Transient expression of allele-specific constructs in COS cells
COS-7 cells were transfected with allele-specific CD11b and CD18 constructs using Lipofectamine 2000 (Gibco BRL), as previously described.24 In brief, 2 μg each plasmid was mixed with 60 μL Lipofectamine in 1.5 mL Opti-MEM medium (Gibco BRL) and added to a subconfluent 10-cm2 plate of COS-7 cells (2 × 106 cells) for 5 hours. Nine mL Dulbecco modified Eagle medium (DMEM), supplemented with 10% fetal calf serum (FCS) and 0.5% penicillin/streptomycin (Pen/Strep), was then added, and the incubation continued for 48 hours. After 2 washes with PBS, cells were surface-labeled with 5 mM NHS-LC biotin (Pierce, Bonn, Germany) and were lysed in 1 mL solubilization buffer (50 mM Tris, 150 mM NaCl, I% Triton X-100, and 2 mM phenylmethylsulfonyl fluoride [PMSF]) for immunoprecipitation.24
Platelet adhesion to neutrophils in whole blood
Whole-blood adhesion experiments were carried out as recently described.11 Briefly, citrate-anticoagulated blood was subjected to shearing in a cone-plate rheometer (Haake, Karlsruhe, Germany). Purified IgG from sera 1, 2, and 3 or from AB serum derived from a healthy blood donor was added immediately before it was subjected to shearing (rates 20 and 2000 s–1) for 2.5 minutes at 37° C. After incubation with a fixative containing methacroleine (2:1 vol/vol), platelet-neutrophil aggregates were stained with fluorescein isothiocyanate (FITC)–labeled mAbs against CD41a and phycoerythrin (PE)–labeled mAb against CD45 (both from Beckman Coulter) for 10 minutes at room temperature and were analyzed by 2-color flow cytometry in a FACScan (Becton Dickinson).
Cell adhesion assay
The adhesion of myelomonocytic cells (U937; American Type Culture Collection, Wesel, Germany) and neutrophils to purified proteins was tested, as described previously.25 Briefly, microtiter plates were coated with 50 μL protein (10 μg/mL) or 0.2% gelatin in bicarbonate buffer, pH 9.6, and blocked with 3% bovine serum albumin (BSA) for 1 hour at room temperature. After 2 washes with 200 μL Hanks buffered salt solution (HBSS; PAA Laboratories, Coelbe, Germany), 100 μL cells (1 × 106/mL) in 150 mM NaCl, 20 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), in the absence or presence of 100 ng/mL phorbol 12-myristate-13 acetate (PMA), were plated onto precoated wells for 60 minutes at 37° C without or with purified antibody (final concentration, 100 ng/mL). Microtiter wells were then washed twice with HBSS, and adherent cells were fixed with 150 μL methanol-acetone (1:1) for 15 minutes at 4° C and stained with crystal violet (Sigma, Munich, Germany). Cell adhesion was quantified by measuring optical density (OD) at 595 nm using an enzyme-linked immunosorbent assay (ELISA) reader (Bio-Tek, NeuFahrn, Germany).
Adhesion of U937 cells and neutrophils to human umbilical vein endothelial cells (HUVECs) was tested as described previously.20 Briefly, HUVECs were grown to confluence on 96-well plates. Fluorescence-labeled neutrophils (105/well) or U937 cells were washed twice, followed by no pretreatment or stimulation with PMA (50 ng/mL). Cells were washed and added to HUVECs at 37° C for 30 minutes in the absence or presence of inhibitors. After washing, cell adhesion was quantified as the percentage of total cells added using a fluorescence microplate reader (Bio-Tek).
Flow cytometry
For competitive studies, U937 cells were washed twice and fixed with 2% paraformaldehyde (PFA) for 10 minutes at room temperature. Aliquots of 8 × 105 fixed cells were incubated with 20 μL anti-Mart IgG in the absence or presence of either purified I domain or mAb CBRM1/32. After 2 washes, the cells were labeled with 40 μL of an FITC-conjugated secondary antibody (1:80 dilution; DAKO), washed, and analyzed by flow cytometry (FACScalibur; Becton Dickinson).
Oxidative burst
Purified neutrophils (5 × 105) were incubated with 0.1 μg/mL IgG fractions from human anti-Mart (sera 1-3) or from control AB serum for 5 minutes in a total volume of 50 μL (neutrophil priming). N-formyl-MetLeuPhe (fMLP; final concentration 1 μM) was added; after 10 minutes of incubation, oxidative burst activity was measured by flow cytometry using the BurstTest kit from Becton Dickinson according to the manufacturer's instructions. mAb 7D8 specific for CD177 served as positive control.26 Oxidative burst activity of isolated neutrophils was controlled using either PMA, opsonized E coli, or no stimulus. All experiments were performed in triplicate.
Results
Characterization of cells and serum
We developed a PCR-SSP approach to determine the Mart genotype. Figure 1 shows the genotyping analysis of 3 donors. A visible PCR product of 249 bp indicates the presence of either the A230 allele (Mart-negative) or the G230 allele (Mart-positive). The PCR product of the hGH gene (439 bp) represents the internal control. In a population study of 360 healthy German blood donors, 5 were found to be A230 homozygous (Mart-negative), whereas 295 donors were G230 homozygous (Mart-positive). Sixty donors were heterozygous. The deviation of the observed numbers of genotypes from those expected by the Hardy-Weinberg equilibrium is statistically insignificant (P = .629). Similar gene frequencies were observed in the American and Australian populations (Table 1).
Genotyping of Mart alloantigens by allele-specific PCR (PCR-SSP).PCR was performed with a common intronic antisense primer and an allele-specific sense primer (G230 or A230) (top). Representative results of Mart genotypes determined by PCR-SSP of 3 donors: homozygous Mart-positive (G/G) (lane 2), heterozygous Mart-positive (G/A) (lane 3), and homozygous Mart-negative (A/A) (lane 4). Genomic DNA was amplified using primer for A230 or G230 and was analyzed by 1.8% agarose gel electrophoresis. The 249-bp bands represent the allele-specific product, and the hGH bands represent the internal control. DNA fragments VI (Roche) were used as standards (lane 1).
Genotyping of Mart alloantigens by allele-specific PCR (PCR-SSP).PCR was performed with a common intronic antisense primer and an allele-specific sense primer (G230 or A230) (top). Representative results of Mart genotypes determined by PCR-SSP of 3 donors: homozygous Mart-positive (G/G) (lane 2), heterozygous Mart-positive (G/A) (lane 3), and homozygous Mart-negative (A/A) (lane 4). Genomic DNA was amplified using primer for A230 or G230 and was analyzed by 1.8% agarose gel electrophoresis. The 249-bp bands represent the allele-specific product, and the hGH bands represent the internal control. DNA fragments VI (Roche) were used as standards (lane 1).
HNA-4a genotype and phenotype frequencies
. | Genotype frequency . | . | . | Phenotype frequency . | . | Antigen frequency . | |||
|---|---|---|---|---|---|---|---|---|---|
| Population . | GG . | GA . | AA . | Mart-positive . | Mart-negative . | . | |||
| Australian; n = 579 | 0.817 | 0.178 | 0.005 | 0.906 | 0.094 | 0.995* | |||
| American; n = 343 | NT | NT | NT | 0.906 | 0.094 | 0.991† | |||
| German; n = 360 | 0.819 | 0.166 | 0.013 | 0.903 | 0.097 | 0.986 | |||
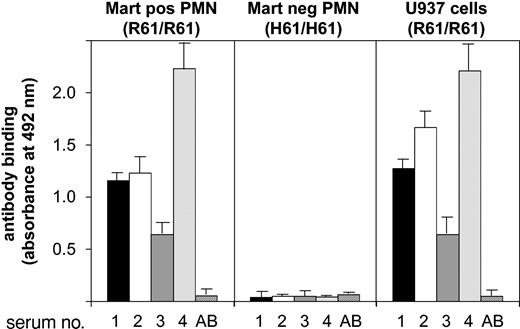
Only 4 sera containing anti-Mart antibodies are reported in the literature (Table 2). All sera were tested in the MAIGA assay to prove their specificity against the CD11b isoform. Positive reactions were obtained with neutrophils from Mart-positive (R61/R61) donors using mAb bear-1 against CD11b (Figure 2). In contrast, neutrophils from Mart-negative (H61/H61) donors did not react with any of the 4 sera. Similar results could be obtained when CD11b-specific mAb ICRF4 or 2LPM19c (against I domain) was used as capture antibodies. No reaction was observed with a mAb specific for CD11a (data not shown). These results demonstrate that all 4 sera do react with the Mart alloantigen residing on the CD11b subunit. We also evaluated the reactivity of anti-Mart sera with monocytic U937 cells in the MAIGA assay (Figure 2). Similarly positive reactions could be observed with different anti-Mart sera. Subsequently, we found using PCR-SSP that U937 cells are homozygous Mart-positive (not shown). These results demonstrate that U937 cells are comparable to neutrophils with respect to Mart epitopes.
Determining anti-Mart glycoprotein specificity in MAIGA assay.Granulocytes from donors who underwent Mart genotyping or from U937 cells were incubated with anti-Mart sera derived from 4 donors (donors 1-4) and mAb bear-1 specific for the CD11b subunit. Bound Mart alloantibodies on CD11b were measured by an antigen capture assay (MAIGA) using peroxidase-labeled antihuman IgG and enzyme substrate. Serum from a healthy blood donor with group AB blood served as a control (AB). All data are means ± SDs (n = 3) of 3 independent experiments.
Determining anti-Mart glycoprotein specificity in MAIGA assay.Granulocytes from donors who underwent Mart genotyping or from U937 cells were incubated with anti-Mart sera derived from 4 donors (donors 1-4) and mAb bear-1 specific for the CD11b subunit. Bound Mart alloantibodies on CD11b were measured by an antigen capture assay (MAIGA) using peroxidase-labeled antihuman IgG and enzyme substrate. Serum from a healthy blood donor with group AB blood served as a control (AB). All data are means ± SDs (n = 3) of 3 independent experiments.
R61H dimorphism is responsible for the Mart phenotype
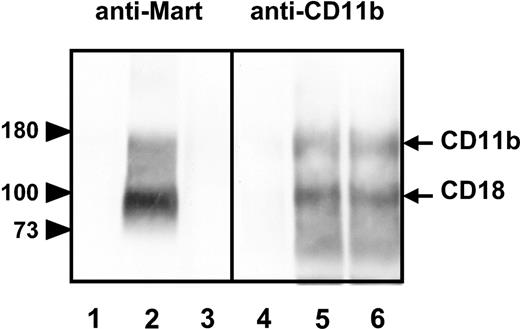
Although we and others have demonstrated that the single nucleotide substitution 230G>A is associated with the Mart phenotype,16 there is no direct evidence that this mutation controls the formation of Mart epitopes by itself. Analysis of the current gene database revealed at least 7 other amino-acid exchanges on CD11b (Table 3). We produced COS cells transiently expressing either the R61 or the H61 CD11b/CD18 complex isoform to test whether each of these mutations could possibly contribute to the formation of Mart epitopes. Immunoprecipitation analysis (Figure 3) showed that anti-Mart exclusively precipitated the R61 variant of CD11b/CD18 complex (lane 2), whereas mAb bear-1 precipitated both the R61 and the H61 isoform (lanes 5, 6). In the control experiment, neither Mart serum nor mAb bear-1 precipitated any protein from untransfected cells (lanes 1, 4). Thus, the formation of the Mart epitope is directly controlled by the amino acid exchange H>R at position 61.
Nonsynonymous mutations in CD11b
mRNA position . | Mutation . | Amino acid exchange, mature peptide . | Allele frequency . |
|---|---|---|---|
| 230 | CGC/CAC | Arg61His | G 0.90* |
| A 0.10* | |||
| 1029 | AGC/AGA | Ser327Arg | C 0.99 |
| A 0.01 | |||
| 1322 | ATG/ACG | Met425Thr | T 0.90 |
| C 0.10 | |||
| 1498-1499 | CAG (ins) | Gly483 Gln(ins) Arg484 | Unknown |
| 2573 | GCC/GTC | Ala842Val | C 0.68 |
| T 0.32 | |||
| 2894 | CCC/CTC | Pro949Leu | Unknown |
| 3436 | CCC/TCC | Pro1130Ser | C 0.68 |
| T 0.32 |
mRNA position . | Mutation . | Amino acid exchange, mature peptide . | Allele frequency . |
|---|---|---|---|
| 230 | CGC/CAC | Arg61His | G 0.90* |
| A 0.10* | |||
| 1029 | AGC/AGA | Ser327Arg | C 0.99 |
| A 0.01 | |||
| 1322 | ATG/ACG | Met425Thr | T 0.90 |
| C 0.10 | |||
| 1498-1499 | CAG (ins) | Gly483 Gln(ins) Arg484 | Unknown |
| 2573 | GCC/GTC | Ala842Val | C 0.68 |
| T 0.32 | |||
| 2894 | CCC/CTC | Pro949Leu | Unknown |
| 3436 | CCC/TCC | Pro1130Ser | C 0.68 |
| T 0.32 |
The public database (NCBI) was screened for nonsynonymous mutations in the coding region of CD11b. Allele frequencies are according to the single-nucleotide polymorphism database at NCBI (http://www.ncbi.nlm.nih.gov/SNP). mRNA position (underlined) applies to A in the ATG start codon (1).
This study.
Immunoprecipitation analysis of allele-specific recombinant CD11b isoforms. Recombinant forms of CD11b/CD18 were produced in COS cells transfected with either the CD11b R61 isoform (Mart-positive; lanes 2 and 5) or the CD11b H61 isoform (Mart-negative; lanes 3 and 6). After cotransfection with CD18 construct, COS cells were surface labeled with biotin and then lysed. Nontransfected COS cells were used as negative control (lanes 1 and 4). Cell lysates were precipitated with anti-Mart or mAb LPM-19c against CD11b. Immunoprecipitates were analyzed on 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reduced conditions, blotted, and visualized using streptavidin-horseradish peroxidase and chemiluminescence substrate.
Immunoprecipitation analysis of allele-specific recombinant CD11b isoforms. Recombinant forms of CD11b/CD18 were produced in COS cells transfected with either the CD11b R61 isoform (Mart-positive; lanes 2 and 5) or the CD11b H61 isoform (Mart-negative; lanes 3 and 6). After cotransfection with CD18 construct, COS cells were surface labeled with biotin and then lysed. Nontransfected COS cells were used as negative control (lanes 1 and 4). Cell lysates were precipitated with anti-Mart or mAb LPM-19c against CD11b. Immunoprecipitates were analyzed on 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reduced conditions, blotted, and visualized using streptavidin-horseradish peroxidase and chemiluminescence substrate.
Some anti-Mart antibodies interfere with platelet-neutrophil interaction
We and others have demonstrated that platelet JAM-C and GPIbα serve as the major counterreceptors for Mac-1.10,11 To determine whether human anti-Mart binding has any influence on the formation of platelet-leukocyte aggregates, we tried to detect ex vivo platelet-neutrophil aggregates from whole blood in a microcouette under conditions of low and high shear (Figure 4). Purified anti-Mart antibodies derived from sera 1 and 2 did not inhibit the formation of platelet-neutrophil aggregates under low shear, whereas a control mAb against P-selectin almost completely abolished aggregate formation. Interestingly, anti-Mart sera displayed different behavior under conditions of high shear. Aggregate formation was reduced by approximately two thirds in the presence of IgG derived from serum 1, whereas IgG from serum 2 had no effect. The inhibition observed with serum 1 was found to be dose dependent (data not shown). None of the sera influenced the formation of platelet-neutrophil aggregates when whole blood from homozygous Mart-negative donors was tested under conditions of high shear (Figure 4, right panel). Serum 3, also derived from the mother of a child without NAIN, displayed results similar to those obtained with serum 2 (not shown).
Influence of anti-Mart in platelet-neutrophil aggregate formation in whole blood from donors who underwent Mart genotyping. The effect of anti-Mart IgG (50 μL/mL) derived from 2 different sera (sera 1-2) and Fab fragments of a blocking mAb GA6* (10 μg/mL) against P-selectin on platelet-neutrophil adhesion in whole blood stimulated with the stable thromboxane A2 mimetic U46619 (5 μM) was tested. Adhesion experiments were carried out in a rheometer at shear rates of 20 s–1 (low shear) and 2000 s–1 (high shear). Data represent the mean ± SD of 3 independent experiments.
Influence of anti-Mart in platelet-neutrophil aggregate formation in whole blood from donors who underwent Mart genotyping. The effect of anti-Mart IgG (50 μL/mL) derived from 2 different sera (sera 1-2) and Fab fragments of a blocking mAb GA6* (10 μg/mL) against P-selectin on platelet-neutrophil adhesion in whole blood stimulated with the stable thromboxane A2 mimetic U46619 (5 μM) was tested. Adhesion experiments were carried out in a rheometer at shear rates of 20 s–1 (low shear) and 2000 s–1 (high shear). Data represent the mean ± SD of 3 independent experiments.
This demonstrates that all Mart alloantibodies are not alike— some interfere with platelet-neutrophil aggregate formation while others do not. In addition, anti-Mart interference in aggregate formation depends on the presence of the Mart antigen.
Influence of R61H dimorphism on adhesive function of neutrophils
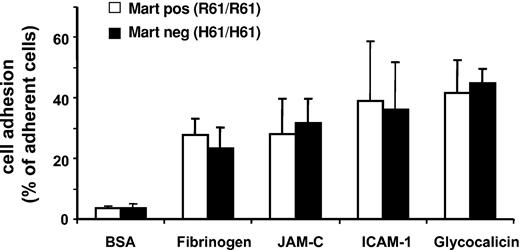
Static cell adhesion assays were performed with neutrophils from different donors to study the possible influence of the Mart phenotype on the adhesive functions of neutrophils. Cells were allowed to adhere to purified ligands and counterreceptors of Mac-1 (fibrinogen, ICAM-1, GPIbα, JAM-C) (Figure 5). There was no significant difference in the adhesion capacity to fibrinogen, ICAM-1, JAM-C, or glycocalicin between Mart-positive (R61/R61) and Mart-negative (H61/H61) donors, indicating that the R61H point mutation does not impair the adhesive function of neutrophils. These results support the clinical observation that all persons lacking the Mart antigen are apparently not ill.18
Influence of Mart phenotype on adhesive function of neutrophils. The adhesion of homozygous Mart-positive (R61) and Mart-negative (H61) granulocytes to immobilized BSA, fibrinogen, JAM-C, ICAM-1, and glycocalicin was assessed in a microtiter plate–based assay. All data are mean ± SD (n = 3) of 3 independent experiments.
Influence of Mart phenotype on adhesive function of neutrophils. The adhesion of homozygous Mart-positive (R61) and Mart-negative (H61) granulocytes to immobilized BSA, fibrinogen, JAM-C, ICAM-1, and glycocalicin was assessed in a microtiter plate–based assay. All data are mean ± SD (n = 3) of 3 independent experiments.
Influence of anti-Mart antibodies on the interaction between CD11b/CD18 and its ligands on platelets and endothelial cells
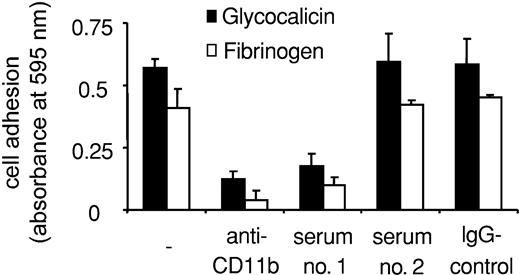
We then examined the influence of Mart alloantibodies on the interaction between neutrophils and CD11b/CD18 ligands and counterreceptors. Purified IgG derived from serum 1 significantly inhibited neutrophil adhesion to fibrinogen and glycocalicin (Figure 6). Again, serum 2 did not inhibit neutrophil adhesion, confirming the functional heterogeneity of Mart alloantibodies. Similar results could be observed by analyzing the adhesion of U937 cells to immobilized purified Mac-1 ligands. Anti-Mart (serum 1) blocked U937 cell adhesion to glycocalicin, fibrinogen, ICAM-1, and RAGE (Figure 7). In contrast, U937 cell adhesion to JAM-C was not affected by this serum (Figure 7A).
Effect of anti-Mart on granulocyte adhesion to purified glycocalicin and fibrinogen. Neutrophil adhesion to immobilized purified proteins is shown in the absence of antibody (–) or in the presence of Mac-1 blocking mAbs against CD11b, purified anti-Mart IgG (sera 1-2) and control IgG. Cell adhesion is expressed as absorbance at 590 nm. All data are mean ± SD (n = 3) of 3 independent experiments.
Effect of anti-Mart on granulocyte adhesion to purified glycocalicin and fibrinogen. Neutrophil adhesion to immobilized purified proteins is shown in the absence of antibody (–) or in the presence of Mac-1 blocking mAbs against CD11b, purified anti-Mart IgG (sera 1-2) and control IgG. Cell adhesion is expressed as absorbance at 590 nm. All data are mean ± SD (n = 3) of 3 independent experiments.
Effect of anti-Mart on adhesion of U937 cells to immobilized CD11b counterreceptors and ligands and to endothelial cells. (A-C) Immobilized CD11b counterreceptors and ligands. (D) Endothelial cells. Cell adhesion to immobilized purified proteins is shown in the absence of antibody (–) or in the presence of Mac-1–blocking mAbs against CD11b and CD18, purified anti-Mart IgG, and control IgG. Cell adhesion is expressed as absorbance at 590 nm. Cell adhesion to HUVEC monolayer (D) was quantified in the presence of sera 1 and 2 and of IgG as control. Anti-CD11b, anti–ICAM-1, and soluble RAGE were run in parallel. All data are mean ± SD (n = 3) of 3 independent experiments.
Effect of anti-Mart on adhesion of U937 cells to immobilized CD11b counterreceptors and ligands and to endothelial cells. (A-C) Immobilized CD11b counterreceptors and ligands. (D) Endothelial cells. Cell adhesion to immobilized purified proteins is shown in the absence of antibody (–) or in the presence of Mac-1–blocking mAbs against CD11b and CD18, purified anti-Mart IgG, and control IgG. Cell adhesion is expressed as absorbance at 590 nm. Cell adhesion to HUVEC monolayer (D) was quantified in the presence of sera 1 and 2 and of IgG as control. Anti-CD11b, anti–ICAM-1, and soluble RAGE were run in parallel. All data are mean ± SD (n = 3) of 3 independent experiments.
These findings agree with those of our previous study indicating that different regions of CD11b (I domain) are responsible for the recognition of GPIbα and JAM-C.11 Moreover, these data support the observation that anti-Mart inhibits aggregation between platelets and neutrophils under high but not under low shear (Figure 4). Furthermore, anti-Mart (serum 1) did not influence the adhesion of U937 cells to uPAR, which is an atypical (non–I-domain) ligand of Mac-1 (Figure 7).
We also investigated whether anti-Mart affects the adhesion of neutrophils or U937 cells to endothelium because ICAM-1 and RAGE are important counterreceptors for leukocyte Mac-1 on endothelial cells.20 Although serum 1 significantly blocked Mac-1–dependent leukocyte adhesion to HUVECs, serum 2 did not show any influence (Figure 7D).
Analysis of antibody recognition sites
To further analyze the location of Mart epitopes, we performed inhibition experiments using the purified I domain of CD11b. No inhibition of anti-Mart binding to U937 cells was observed in the presence of the I domain (Figure 8A). Comparable results were obtained with all 3 anti-Mart sera tested. In the control experiment, the binding of an I domain–specific mAb (LPM19c) was significantly inhibited. Accordingly, the I domain of CD11b is not involved in the formation of Mart epitopes.
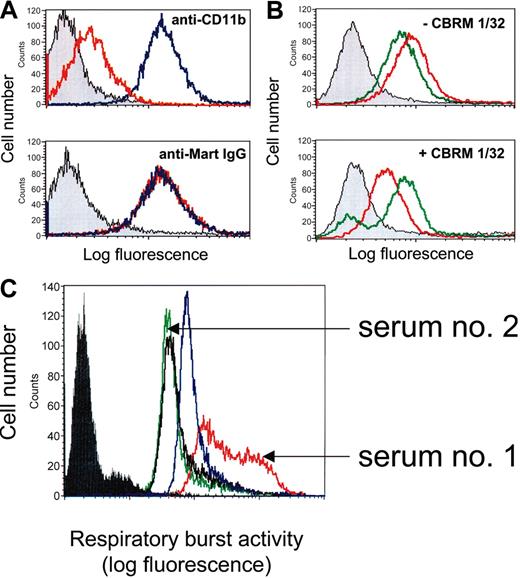
Competitive-binding studies by flow cytometry and effect of anti-Mart on respiratory burst activity of neutrophils. (A) U937 cells were incubated with mAb LPM-19c directed against the I domain of CD11b or with IgG from anti-Mart (serum 1) in the absence (blue) or in the presence of I domain (red). Gray histograms represent the isotype control. Bound antibodies were detected with fluorescein-labeled secondary antibodies. (B) U937 cells were incubated with anti-Mart IgG from serum 1 (red) or serum 2 (green) in the absence (top) or presence (bottom) of mAb CBRM1/32. Bound human IgG was detected with fluorescein-labeled secondary antibodies. (C) Neutrophils were incubated with control IgG (black line), mAb 7D8 against CD177 (blue), or IgG fractions derived from anti-Mart sera 1 and 2. ROS production after stimulation with fMLP was measured using flow cytometry. Filled histogram represents unstimulated control without fMLP. All sets of histograms represent the results of 1 of 3 separate experiments.
Competitive-binding studies by flow cytometry and effect of anti-Mart on respiratory burst activity of neutrophils. (A) U937 cells were incubated with mAb LPM-19c directed against the I domain of CD11b or with IgG from anti-Mart (serum 1) in the absence (blue) or in the presence of I domain (red). Gray histograms represent the isotype control. Bound antibodies were detected with fluorescein-labeled secondary antibodies. (B) U937 cells were incubated with anti-Mart IgG from serum 1 (red) or serum 2 (green) in the absence (top) or presence (bottom) of mAb CBRM1/32. Bound human IgG was detected with fluorescein-labeled secondary antibodies. (C) Neutrophils were incubated with control IgG (black line), mAb 7D8 against CD177 (blue), or IgG fractions derived from anti-Mart sera 1 and 2. ROS production after stimulation with fMLP was measured using flow cytometry. Filled histogram represents unstimulated control without fMLP. All sets of histograms represent the results of 1 of 3 separate experiments.
Because the R61H dimorphism is located at the N-terminus of CD11b, we sought to map Mart epitopes by blocking this region with mAb CBRM1/32. This mAb recognizes a discontinous epitope residing within the amino-terminal and cation-binding domains of CD11b and is known to interfere with cell adhesion processes.28-30 Anti-Mart derived from sera 1 and 2 showed comparable binding to U937 cells in the absence of mAb CBRM1/32 (Figure 8B; upper panel). These 2 sera behaved differently in the presence of mAb CBRM1/32 (Figure 8B; lower panel). Whereas a significant reduction in binding was detected with purified anti-Mart from serum 1 (ΔMFI [mean fluorescence intensity], 44.6% ± 4.2%), no reduction was observed with anti-Mart from serum 2 (ΔMFI, 1.2% ± 3.7%). The binding of anti-Mart derived from serum 3 was not hampered by CBRM1/32 either (data not shown). These results indicate that different anti-Mart sera recognize different epitopes on CD11b. One serum reacts with an epitope close to the CBRM1/32 recognition site, but the others do not. Interestingly, CBRM1/32 and anti-Mart from serum 1 are inhibitors of cell adhesion processes.
Influence of anti-Mart on neutrophil activation
Ligand binding to CD11b/CD18 is known to play a significant role in the generation of reactive oxygen species (ROS), which are important triggers of apoptosis in neutrophils.31-33 Therefore, we assessed the ability of different anti-Mart sera to stimulate ROS activity in isolated neutrophils. Serum 1, derived from a mother who gave birth to a child with severe NAIN, significantly increased ROS activity (Figure 8C). In contrast, serum 2, derived from a mother who gave birth to a child without clinical signs of NAIN, did not alter neutrophil response to fMLP when compared with control IgG. Increased ROS activity was also found in the control experiment with mAb against CD177.
Discussion
Although the Mart neutrophil alloantigen system (HNA-4) was discovered almost 2 decades ago, its clinical significance has remained uncertain. One important reason is that giving women alloimmunization against Mart is not regularly associated with NAIN of the newborn. In this study we were able to characterize 2 different types of Mart antisera: type 1 was derived from a mother who gave birth to a child with severe NAIN, and type 2 sera came from mothers who gave birth to children without clinical symptoms. Several observations underscore the evidence that anti-Mart sera are not always alike: (1) Type 1 Mart alloantibodies inhibited the formation of platelet-neutrophil aggregates in a whole-blood microcouette system under conditions of high shear, whereas type 2 Mart antibodies did not. This interference clearly depended on the presence of the Mart antigen, because type 1 Mart alloantibodies only inhibited platelet-neutrophil aggregation of Mart-positive, but not of Mart-negative, donors. (2) The same type 1 alloantibodies that inhibited the formation of platelet-neutrophil aggregates interfered with the adhesion of neutrophils to endothelial cells. In contrasting, type 2 alloantibodies did not impair these adhesive properties. (3) Functional studies reveal that type 1 alloantibodies, which interfere with cell adhesion processes, can also efficiently prime neutrophils for the generation of ROS. Type 2 alloantibodies did not display neutrophil priming activity. (4) Competitive-binding studies demonstrated that type 1 and type 2 Mart alloantibodies recognized different epitopes on CD11b. mAb CBRM1/32, an antibody that interferes with cell adhesion processes by recognizing a discontinous epitope within the amino-terminal and the cation-binding domains of CD11b, inhibited the binding of type 1 but not of type 2 Mart alloantibodies.28-30 These data suggest that the humoral response to the Mart alloantigen is heterogeneous, that is, it differs from one immunized person to another.
Valentin et al34 showed that antibodies against human platelet alloantigen (HPA)–1a on β3 integrin are heterogeneous and can differ between persons, though a single amino-acid exchange L33P controls the expression of HPA-1a epitopes. Some anti–HPA-1a epitopes recognize an epitope comprised solely of the aminoterminal part of β3 integrin, whereas others seem to react with a complex epitope that requires the “long-range” disulfide bond formed by cysteine residues 5 and 435. The heterogeneity of the epitopes recognized by alloantibodies may be reflected by different consequences on cellular function. For example, some anti–HPA-1a alloantibodies can induce a Glanzmann-like platelet dysfunction by inhibiting fibrinogen binding.35 Similarly, some anti-Mart can interfere with the adhesive properties of neutrophils, though it remains questionable whether this interference is important for neutrophil function in vivo.
However, some Mart alloantibodies (type 1) can also prime neutrophils for the production of ROS, whereas others (type 2) do not. Sustained ROS generation by NADPH oxidase has been implicated as the final common mediator of apoptosis in a variety of systems.33,36-38 Subsequent exposure of surface factors, such as phosphatidylserine, labels the cells for engulfment by macrophages.39 Whether neutrophil apoptosis represents a relevant mechanism for neutropenia has not yet been investigated in alloimmune disorders. However, recent evidence indicates that binding antibodies on their cellular receptors and inducing the apoptosis and the subsequent clearance of the affected cells is not an uncommon mechanism of cell removal. In systemic lupus erythematodes, binding anti-SSB/La antibodies to neutrophils was found to accelerate neutrophil apoptosis associated with neutropenia in affected patients.40 Neutrophil apoptosis could also be accelerated by antineutrophil cytoplasmic antibodies (ANCAs) by a mechanism dependent on the activation of ROS generation.41 In addition, Nardi et al42 have recently reported a novel mechanism of complement-independent immunologic platelet clearance in HIV-1 patients with immune thrombocytopenia. Binding of patients' IgG antibodies directed against platelet GPIIIa residues 49-66 caused platelet fragmentation because of the induction of ROS in the absence of complement. Interestingly, antibodies directed against 4 other regions of GPIIIa, and an antibody against GPIbα, failed to induce platelet fragmentation. These observations indicate that antibodies against specific regions of GPIIIa are capable of activating a peroxidase-generating pathway, with subsequent platelet clearance by other than classic Fc or complement receptors. Thus, a comparable mechanism of neutrophil clearance may be responsible for the ability of some Mart alloantibodies to induce severe neutropenia. This, in turn, suggests that Mart antibodies that do not prime neutrophils are unable to cause neutropenia, at least not to the same extent. Obviously, further investigation is required to determine the precise mechanism of neutropenia induced by neutrophil alloantibodies.
Additionally, we obtained some detailed information regarding the recognition site of anti-Mart. We could demonstrate that the single H61R point mutation is sufficient for the formation of Mart epitopes. This mutation is located between sheets 1 and 2 of the 7-bladed β-propeller of CD11b outside the I domain, which is localized between sheets 2 and 3 of the propeller.43 Our inhibition studies demonstrated that the I domain is not directly involved in the formation of Mart epitopes because soluble I domain was unable to block anti-Mart binding. However, multiple interactions between CD11b and its ligands (fibrinogen, ICAM-1) or counterreceptors (GPIbα, RAGE) mediated by the I domain can be blocked by type 1 anti-Mart. Diamond et al28 demonstrated that mAb CBRM1/32 recognizes epitopes outside the I domain of CD11b and abrogates the binding to several ligands, including fibrinogen and ICAM-1. It was suspected that the CBRM1/32 epitope was directly involved in ligand binding or in maintaining the functional conformation of the holoreceptor.29 We could demonstrate that mAb CBRM1/32 inhibited the binding of anti-Mart alloantibodies (type 1) to neutrophils. This indicated that these 2 antibodies recognize similar binding regions on CD11b and can, therefore, interfere with the binding of I domain ligands in a comparable manner. The binding of type 2 alloantibodies was not impaired by CBRM1/32. Thus, type 1 and type 2 anti-Mart alloantibodies recognize different epitopes on CD11b, which may lead to different functional properties.
Although type 1 anti-Mart blocked several CD11b-ligand interactions, the binding of 2 ligands, uPAR and JAM-C, remained unaffected. uPAR is an atypical CD11b ligand that does not interact with the I domain of CD11b but does interact with the region 424-440 located carboxy terminal to the I domain.44 The epitope recognizing uPAR is located in sheet 4 of the 7-bladed β-propeller, whereas the I domain is inserted between sheets 2 and 3.44 The interaction between uPAR and Mac-1, leading to the activation of Mac-1 and enhanced Mac-1–dependent cell adhesion, can be regulated by zinc cations.45 Accordingly, zinc-stimulated Mac-1–dependent neutrophil adhesion to fibrinogen in the presence of uPAR was not affected by anti-Mart (data not shown). Therefore, uPAR and anti-Mart regulate ligand binding to the I-domain by distinct mechanisms.
In addition to GPIbα, JAM-C has recently been shown to serve as one of the major platelet counterreceptors for neutrophil Mac-1 by interaction with the I domain.10,11 Here we provide evidence that GPIbα and JAM-C recognize different regions of the I domain: the adhesion of neutrophils to purified JAM-C was not inhibited by anti-Mart though neutrophil adhesion to GPIbα was impaired, and anti-Mart inhibited platelet-neutrophil aggregates only under conditions of high shear but not of low shear. Adhesion processes under high and low shear conditions are mainly ascribed to the interaction between Mac-1 and GPIbα or Mac-1 and JAM-C, respectively.11 Ehlers et al46 have recently identified amino acid residues 201-217 of CD11b as the binding region of GPIbα. The precise location of the JAM-C binding region is under investigation.
Evidence from several in vivo studies indicates that blocking Mac-1–dependent leukocyte-platelet and leukocyte-endothelial interactions by murine monoclonal antibodies against CD18 or CD11b might be beneficial in inflammatory disorders such as in atherosclerosis.47-49 Anti-Mart prevents platelet-leukocyte interaction and interferes with the adhesion of leukocytes to endothelium. Given that this human-derived alloantibody recognizes the high-frequency isoform of CD11b, it may represent a useful tool in targeting Mac-1–dependent leukocyte adhesion that promotes inflammatory processes in atherosclerotic disease.
Prepublished online as Blood First Edition Paper, April 8, 2004; DOI 10.1182/blood-2003-11-3809.
Supported by grants from the Fachbereich Humanmedizin der Justus-Liebig-Universität Giessen (U.J.H.S.); Deutsche Forschungsgemeinschaft (Ch279/1-1, SFB 405) (T.C.), (Bu770/3-6) (J.B.), and (SFB 547) (S.S.); and Novartis Foundation for Therapeutic Research (T.C.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr A. Law (Singapore) for kindly providing a CD11b expression vector and Dr A. Gardemann (Magdeburg, Germany) for statistical analysis. We also thank A. Athanasopoulos, O. Eva, C. Hofmann, Y. Mueller, D. Oehmichen, and S. Werth for their excellent technical assistance. This work contains parts of the doctoral thesis of A.L.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal