Abstract
The biosynthesis of aberrant immunoglobulin polypeptides by monoclonal plasma cells has been implicated in the pathogenesis of nonsecretory myeloma. Our studies of a patient with this disorder indeed have demonstrated the presence of abnormal κ light chains that resulted from a frameshift mutation in nucleotides encoding the constant region of the molecule. As a consequence of a 2-base deletion in codon 187 and loss of the normal stop codon, this portion of the κ chain was composed of 128 amino acids (rather than the expected 106), with a completely anomalous sequence after position 187 that included absence of the cysteines required for intrachain and interchain disulfide bonds. The unusual primary structure of this component was confirmed by mass spectrometric and amino acid sequence analyses of cytoplasmic protein extracts. Our studies provide the first evidence that human nonsecretory myeloma may result from an alteration in the light-chain constant region.
Introduction
Nonsecretory myeloma (NSM) is defined by the absence of serum or urinary (or both) monoclonal immunoglobulins in patients who otherwise manifest features typically found in multiple myeloma (MM).1,2 This condition, which occurs in about 1% to 5% of patients with MM, results either from the inability of malignant plasma cells to synthesize immunoglobulin (ie, nonproducer) or, more commonly, the failure of such components to be exported from the cells (ie, nonsecretor). In the latter case, it has been assumed that the protein undergoes intracellular proteolysis due to a structural defect in the immunoglobulin and, as a consequence, is not excreted. However, there is only limited information regarding the molecular factors that might account for this phenomenon.3-7 We now report our finding that the plasma cells of an individual with NSM indeed synthesized aberrant monoclonal light chains that resulted from a somatic mutation in the gene encoding the constant (CL) region of the molecule. Our studies provide the first conclusive evidence that an abnormality in this portion of the light chain may be implicated in the pathogenesis of this disorder.
Study design
Protein analyses
Serum IgG, IgA, and IgM proteins were quantitated with the Synchron LX20 Clinical System (Beckman Coulter, Fullerton, CA) and the concentration of free κ and λ light chains determined by enzyme-linked immunosorbent assay (ELISA), using our highly specific monoclonal antibodies (mAbs),8 as well as by nephelometry (Free Lite, The Binding Site, Birmingham, United Kingdom) with polyclonal reagents.9 For detection of monotypic immunoglobulins, serum was diluted 1:10 and subjected to immunofixation electrophoresis using the Paragon system (Beckman, Norcross, GA), according to the procedure specified by the manufacturer. Undiluted samples also were examined in similar fashion, as were unconcentrated urine specimens and 24-hour collections that had been dialyzed extensively against distilled water, lyophilized, and reconstituted to a protein concentration of 100 mg/mL.10
Immunocytochemistry
Bone marrow plasma cells were isolated and immunostained with our murine mAbs specific for human light-chain variable region (VL) subgroups11 and for total (heavy chain-bound) and free (unbound) κ and λ polypeptides,8 as well as with mouse antihuman plasma cell (Dako, Carpinteria, CA) and rabbit antihuman γ, α, μ, and δ heavy-chain.antisera (Biosource, Camarillo, CA).
RNA preparation and RT-PCR amplification
Total RNA was extracted from plasma cells with the PURESCRIPT RNA isolation kit (Gentra, Minneapolis, MN) and transcribed with both oligo (dT)15 and random primers, using reverse transcriptase (RT; Promega, Madison, WI) in a single reaction. First-strand DNA was amplified by forward and reverse primers specifying the first and last 7 amino acid residues of a prototypic κ1 light chain.12 The polymerase chain reaction (PCR) products were cloned using the perfectly blunt cloning kit with the pSTBlue-1 vector (Novagen, Madison, WI) and the colonies screened by PCR. Plasmids were isolated from candidate clones using the Quantum Miniprep kit (Bio-Rad, Richmond, CA), further screened by EcoRI digestion to confirm insert size, and sequenced in both directions. The nucleic acid and deduced protein sequences were compared to the GenBank database.
DNA analysis
gDNA was obtained from peripheral blood leukocytes using the PURE-GENE DNA isolation kit (Gentra) and the Cκ-encoding gene was amplified by PCR12 with upstream and downstream primers specifying the first 6 and last 7 amino acid residues, respectively.
Isolation and characterization of intracellular (nonsecreted) light chains
Total cell lysates were prepared13 from bone marrow–derived plasma cells and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS/PAGE). The light-chain–containing band identified by immunoblotting was reduced, alkylated, digested with trypsin, and the resultant peptides isolated and subjected to automated sequence analysis and tandem mass spectrometry (MS/MS).14
Approval for these studies was obtained from the institutional review boards of the University of Tennessee Graduate School of Medicine and Mayo Clinic. Informed consent was provided according to the Declaration of Helsinki.
Results and discussion
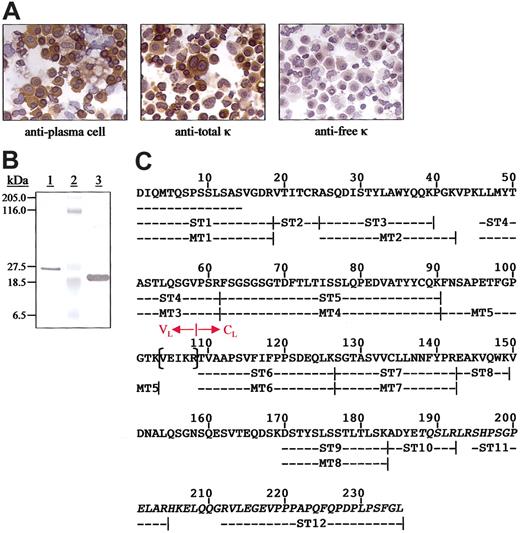
The patient was a 74-year-old man who fulfilled the principal diagnostic criteria for NSM; namely, no monoclonal immunoglobulins were detected even in undiluted sera or highly concentrated urines and he exhibited the salient bone marrow and skeletal abnormalities associated with MM (80% plasma cells and extensive osteolytic lesions), as well as reduced levels of normal IgG, IgA, and IgM (514, 46, and 33 mg/dL, respectively). The hemoglobin concentration was 9.2 mg/dL; serum calcium level, 15mg/dL; and creatinine level, 3.0 mg/dL. Values for serum-free κ and λ chains and the κ/λ ratio, as measured using both specific monoclonal8 and polyclonal9 antibodies, were within normal limits. The bone marrow was extensively infiltrated by CD56+, CD38+, and CD43+/CD19- plasma cells that had the t(11;14) (q13;q32) translocation. Their monotypic nature was apparent in immunohistochemical analyses using highly specific anti-Cκ and anti-Vκ mAbs that revealed the presence of cytoplasmic κ light chains that were classified as members of the Vκ1 gene family.11 Notably, the κ immunoreactivity was evidenced only by the mAb with specificity for an epitope present on κ chains linked covalently to immunoglobulin heavy chains by a disulfide bond. In contrast, these molecules were not recognized by the antifree κ-chain mAb (Figure 1A) and, with rare exception, were not immunostained by antiheavy-chain antisera.
Immunocytochemical analyses of plasma cells from the patient with NSM and chemical characterization of the nonsecreted κ light chains. (A) Reactivity of bone marrow–derived plasma cells. The primary reagents included a murine antiplasma cell mAb and those specific for free or bound human κ light chains. Microscopic analyses were performed using a Leitz DMRB microscope (Vashaw Scientific, Norcross, GA) and objectives that included Leica HC PL Fluotar 40× and 10× lenses. The digital photographs were taken with a SPOT RT Model 2.2.0 camera (Diagnostic Instruments, Sterling Heights, MI) using SPOT RT software, version 3.0. Original magnification, × 400. (B) Coomassie blue-stained SDS/PAGE gel of protein contained in the plasma cell lysates (lane 1), molecular mass standards (lane 2), and a κ1 Bence Jones protein isolated from the urine of a patient with MM (lane 3). (C) Primary structure of the κ1 light chain isolated from plasma cell lysates. The N-terminal amino acid sequence was established directly and the remainder from tryptic peptides subjected to Edman degradation (ST) and mass spectrometry (MT). The residues in parentheses were deduced from cDNA cloned from the patient's plasma cells. The junction between the VL and CL domains is as designated and the anomalous residues after position 187 are italicized.
Immunocytochemical analyses of plasma cells from the patient with NSM and chemical characterization of the nonsecreted κ light chains. (A) Reactivity of bone marrow–derived plasma cells. The primary reagents included a murine antiplasma cell mAb and those specific for free or bound human κ light chains. Microscopic analyses were performed using a Leitz DMRB microscope (Vashaw Scientific, Norcross, GA) and objectives that included Leica HC PL Fluotar 40× and 10× lenses. The digital photographs were taken with a SPOT RT Model 2.2.0 camera (Diagnostic Instruments, Sterling Heights, MI) using SPOT RT software, version 3.0. Original magnification, × 400. (B) Coomassie blue-stained SDS/PAGE gel of protein contained in the plasma cell lysates (lane 1), molecular mass standards (lane 2), and a κ1 Bence Jones protein isolated from the urine of a patient with MM (lane 3). (C) Primary structure of the κ1 light chain isolated from plasma cell lysates. The N-terminal amino acid sequence was established directly and the remainder from tryptic peptides subjected to Edman degradation (ST) and mass spectrometry (MT). The residues in parentheses were deduced from cDNA cloned from the patient's plasma cells. The junction between the VL and CL domains is as designated and the anomalous residues after position 187 are italicized.
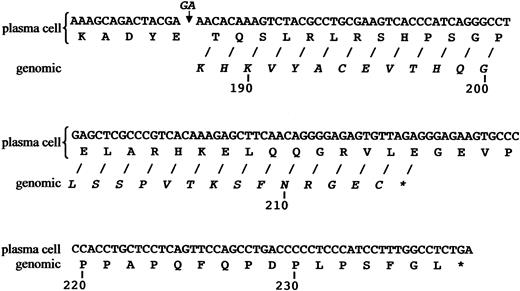
Analyses of cDNA cloned from the abnormal plasma cell population revealed that the deduced primary structure of the 108-residue VL was most closely homologous to that encoded by the A20 Vκ1 and the Jκ3 germline genes, differing by 5 and 1 amino acids, respectively (G28D, N31T, I48M, A50T, Y91F, D105E).12 The sequence of nucleotides specifying the Cκ was as expected until codon 187, where there was a 2-bp deletion (GA) that resulted in an extended, aberrant product due to loss of the normal TAG stop codon 215. Notably, this frameshift mutation was not present in the patient's gDNA (Figure 2) where the deduced protein product consisted of the anticipated 106 amino acids, including cysteines at positions 194 and 214.
Comparison of Cκ protein sequences after position 187, as predicted from plasma cell cDNA and gDNA. The 2-bp deletion (GA) present in the plasma cell cDNA and the positions of the stop codons are as indicated by the asterisks.
Comparison of Cκ protein sequences after position 187, as predicted from plasma cell cDNA and gDNA. The 2-bp deletion (GA) present in the plasma cell cDNA and the positions of the stop codons are as indicated by the asterisks.
The composition of the nonsecreted cytoplasmic κ light chains was established through chemical analyses of protein isolated from plasma cell lysates. As evidenced by SDS/PAGE, this component had an unusually high Mr of about 26 kDa, in contrast to a value of about 23 kDa found for a monomeric κ1 Bence Jones protein from a patient with MM (Figure 1B). In immunoblotting experiments, both light chains reacted with the antitotal κ-chain mAb, but only the MM-derived protein was recognized by the antifree κ monoclonal reagent (no heavy chains were detected in the plasma cell isolate). The complete primary structure of the nonsecreted molecule, as predicted from the cDNA data, was verified by amino acid sequencing and MS/MS (Figure 1C). Importantly, peptides ST10, ST11, and ST12 had anomalous sequences after position 187 that resulted from the frameshift mutation and confirmed that the Cκ extended 20 residues beyond position 214. Thus, a new Cκ reverse primer was prepared (5′-AGCTGGAGGACCGCAATAG-3′) and used to reclone plasma cell–derived cDNA where the 378 nucleotides encoding the entire Cκ, plus the stop codon (TGA), were identified. The deduced amino acid sequence of this gene product was identical to that of the isolated cytoplasmic protein (Figure 1C).
Heretofore, mutations in genes encoding human and murine VLs have been implicated as causal factors in NSM.3-6,15 Notably, our studies provide the first evidence that an aberrant product of a mutated CL gene also can be associated with this disorder. The primary sequence alterations that resulted from the frameshift mutation included loss of the cysteine residues normally present at positions 194 and 214 that are involved in formation of intrachain and interchain disulfide bonds, respectively. Based on x-ray crystallography, VL and CL domains typically are folded into independent compact structures.16 Although such data are not available on the patient's protein, it is likely that the absence of Cys194 profoundly disrupted the 3-dimensional features of this molecule, that is, lack of the internal Cys146-Cys194 disulfide bond prevented formation of a normal, stable CL (additionally, loss of Cys214 would render the light chain incapable of binding covalently with the cysteine in the first heavy-chain C domain). Because the intracellular fate and eventual secretion of light chains from plasma cells depend, in part, on the interaction of these molecules with heat shock protein 70 (BiP), an endoplasmic reticulum molecular chaperone that functions to facilitate the delivery of malfolded immunoglobulin polypeptides to the intracellular degradative machinery,17-20 we posit that the misfolded κ chains were retained within the plasma cell cytosol and underwent proteosome-mediated proteolysis.21,22 Whether immunoglobulin light chains from other cases of NSM exhibit similar structural features (abnormalities) remains to be established.
Prepublished online as Blood First Edition Paper, April 15, 2004; DOI 10.1182/blood-2004-02-0477.
Supported in part by United States Public Health Service research grant CA10056 from the National Cancer Institute and the Aslan Foundation. D.C. and M.E. were Harry and Elsa Jiler American Cancer Society International Visiting Scientists. A.S. is an American Cancer Society Clinical Research Professor.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Sallie D. Macy, Teresa K. Williams, Dennis A. Wofenbarger, and Craig Wooliver for technical assistance; Ronda L. Reed for manuscript preparation; the University of Tennessee Molecular Biology Resource Facility for nucleotide sequence analyses; and Dr Jerry A. Katzmann for the nephelometric free light chain assays.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal