The promise of modern molecular treatments for cancer is that the power of biotechnology will uncover new targets for therapy, killing more cancer while minimizing damage to normal tissues. Two articles in this issue of Blood suggest that these therapies live up to their promise.
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease consisting of a wide variety of genetic lesions, including translocations, hyperdiploidy, and even normal-appearing genotypes.1,2 Successful treatment is the norm for pediatric patients, but results in adults are far less satisfying. A major research pursuit is to understand how the different genetic lesions encountered in ALL map to differences in clinical outcome and provide insight into new therapeutic options. In this issue of Blood, 2 studies use novel molecular techniques to uncover new potentially therapeutic options for subsets of this disease.
Philadelphia chromosome–positive (Ph+) ALL occurs in 5% to 10% of pediatric cases and in approximately 25% of adult cases. In all populations, the outcomes following chemotherapy are especially dismal. Recently, the tyrosine kinase inhibitor imatinib has been shown to be effective in ALL, but responses are short-lived, usually lasting only about a month.3 Resistance in these cases often occurs from a mutation in the adenosine triphosphate (ATP)–binding domain of bcr-abl (the chimeric protein coded by the Ph chromosome), which effectively blocks the action of imatinib.4 There are some suggestions that increasing the imatinib dose may overcome resistance, at least temporarily,5 but increasing the dose too much invariably leads to toxicity, since imatinib also blocks the tyrosine kinase function of other nontarget enzymes necessary for normal organ function. How, then, can one increase the imatinib effectively delivered to the ALL cell without harmful effects on normal tissue?
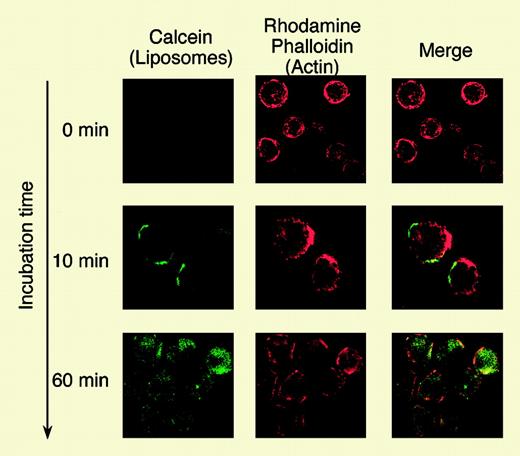
In this issue of Blood, Harata and colleagues report a clever and novel approach to target imatinib delivery using liposomes coupled to antibodies to the CD19 antigen. CD19 is a lymphoid-restricted surface protein, which is found on virtually all Ph+ ALL cells as well as normal B cells. It is not found on myeloid or CD34+ stem cells. The authors conjugated mouse monoclonal anti-CD19 antibodies to CD19 with small (100 nM) liposomes that were manufactured to encapsulate imatinib. Using ALL cell lines, they demonstrated that these CD19-targeted liposomes bound and entered CD19+ cells but not CD19– cells. The liposomal imatinib induced greater cell death than equal quantities of free imatinib, as demonstrated by in vitro culture and annexin V apoptotic assays. In addition, primary ALL cells from 4 patients were treated in vitro with the anti-CD19 liposomal imatinib as well as free imatinib, and these experiments again demonstrated the superiority of the antibody-liposome imatinib. It is instructive that 2 of these ALL samples had Abl point mutations yet still were susceptible to the liposomal-delivered imatinib, suggesting that high concentrations of intracellular imatinib can indeed overcome the resistance conferred by the point mutation. Lastly, the liposomal imatinib seemed to have little effect on normal CD34+ as measured in colony-forming assays.
Much may be learned from the “good risk” leukemia as well as the bad. The TEL-AML1 fusion gene occurs in approximately 25% of pediatric ALL cases and seems to identify a cohort with a favorable outlook.6 Why? It is known that de novo purine synthesis (DNPS) is higher in ALL compared with that of normal bone marrow cells and peripheral blood lymphocytes; among ALL cases, T-cell lineage ALL has greater DNPS than B-lineage ALL.7 Many drugs used in ALL (such as methotrexate and mercaptopurine) target DNPS and purine metabolism. Zaza and colleagues report on purine synthesis and metabolism in 113 pediatric ALL cases, focusing on the differences across different subtypes of ALL. Their findings are quite instructive. Among theFIG1 B-lineage ALL cases, those with the TEL-AML1 had a dramatically lower DNPS than other subtypes. The authors then used gene expression experiments to compare the expression of 82 genes involved in purine metabolism across the B-cell lineage cases. Sixteen genes were found to be significantly associated with the TEL-AML1 subgroup of ALL, and the expression of several genes (notably PAICS and IMPDH2) closely correlated with DNPS level. It is not clear mechanistically how the TEL-AML1 genetic lesion is associated with purine biochemistry, but the association may explain why this subgroup of ALL is especially sensitive to chemotherapy.
Internalization analysis of Cal-CD19-liposomes by confocal laser-scanning microscopy. See the complete figure in the article beginning on page 1442.
Internalization analysis of Cal-CD19-liposomes by confocal laser-scanning microscopy. See the complete figure in the article beginning on page 1442.
ALL is not a common disease, and given that these studies are investigating specific subsets of ALL, the skeptic may claim that the results are unlikely to change the face of cancer therapy. However, as models of clinical investigation, the results are quite interesting and energizing, showing how modern molecular biology can be applied in clever and powerful ways.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal