Abstract
SOD2 is an antioxidant protein that protects cells against mitochondrial superoxide. Hematopoietic stem cells (HSCs) lacking SOD2 are capable of rescuing lethally irradiated hosts, but reconstituted animals display a persistent hemolytic anemia characterized by increased oxidative damage to red cells, with morphologic similarity to human “sideroblastic” anemia. We report further characterization of this novel SOD2-deficiency anemia. Electron micrographs of SOD2-deficient reticulocytes reveal striking mitochondrial proliferation and mitochondrial membrane thickening. Peripheral blood smears show abundant iron-stainable granules in mature red cells (siderocytes). Fluorescence-activated cell sorting (FACS) analysis of cells labeled with oxidation-sensitive dyes demonstrates enhanced production of superoxide and hydrogen peroxide by SOD2-deficient cells. Oxidative damage to proteins is increased in SOD2-deficient cells, with much of the damage affecting membrane/insoluble proteins. Red cell proteome analysis demonstrates that several proteins involved in folding/chaperone function, redox regulation, adenosine triphosphate (ATP) synthesis, and red cell metabolism show altered expression in SOD2-deficient cells. This data, combined with information on how protein expression levels change upon antioxidant therapy, will aid in identification of proteins that are sensitive to oxidative damage in this model, and by extension, may have a role in the regulation of red cell lifespan in other hemolytic disorders.
Introduction
Hematopoiesis in mice reconstituted with Sod2–/– hematopoietic stem cells (HSCs) is notable for a persistent hemolytic anemia with morphologic similarity to sideroblastic anemia (SA).1 Several genetic lesions have now been identified as causes of both hereditary and acquired SA.2 Of interest, most lesions affect proteins localized to mitochondria and/or the mitochondrial genome itself. There are 2 X-linked forms of SA—one caused by mutations of an erythroid-specific form of the heme biosynthetic enzyme aminolevulinoc acid (ALA)–synthase (ALAS2),3 and one caused by mutation of a putative mitochondrial iron transport protein, ABC7.4 Mutation of another mitochondrial membrane protein, flexed-tail, causes a transient neonatal form of SA in mice.5 Lesions in mitochondrial DNA have also been implicated in acquired and congenital SA. Pearson marrow-pancreas syndrome results from a large deletion in mitochondrial DNA,6 somatic point mutations in cytochrome c oxidase subunit 1 (COX1) have been found in patients with myelodysplastic syndromes with ringed sideroblasts (eg, refractory anemia with ringed sideroblasts),7 and there are reports of mutations affecting other portions of the mitochondrial DNA in isolated cases of SA.8,9 Some cases of acquired SA are associated with specific drugs such as chloramphenicol, an inhibitor of protein synthesis in mitochondria,10 or isoniazid,11,12 which interferes with pyridoxine metabolism, a cofactor for ALAS2. Together, these reports suggest that mitochondrial dysfunction is a common theme important for understanding the etiology of SA, and may be both necessary and sufficient to create a convergent phenotype of pathologic iron deposition in mitochondria of red cell progenitors from a diverse group of primary or secondary genetic or toxin-induced lesions. In murine models of SA, morphologic abnormalities appear more prominent in mature red cells (siderocytes) than in marrow erythroid progenitors (sideroblasts). This may reflect a species difference, as similar observations have been made when comparing the phenotype associated with pyridoxine deficiency in mice (siderocytes) and humans (sideroblasts).13 Despite differences in the maturational stage at which abnormalities occur, a common finding in SA is accumulation of excess iron in erythroid cells, primarily within mitochondria.
SOD2-deficiency anemia is another example of mitochondrial dysfunction resulting in an erythroid-specific SA-like phenotype. SOD2 deficiency is known to result in severe mitochondrial dysfunction in neurons and muscle in vivo leading to neonatal lethality in homozygous knockout animals.14,15 Mitochondrial function in Sod2–/– tissues is characterized by decreased activity of (oxidation sensitive) iron-sulfur cluster enzymes, including complexes I and II, as well as the tricarboxylic acid cycle enzyme aconitase.16 In SOD2-deficient hematopoietic cells in vivo, dysfunction is most prominent in erythroid cells. Tissue pathologies, mitochondrial enzyme damage, and anemia can be partially alleviated by treatment with synthetic catalytic antioxidants,1,17,18 a family of related compounds that possess both catalase and superoxide dismutase activity. The most effective such compound in mice, a lipophilic analog known as EUK-189,18 was employed in the present study. In this report we further characterize abnormal erythropoiesis in irradiated wild-type mice with repopulation of hematopoietic tissues by transplanted fetal liver cells from Sod2+/+ or Sod2–/– donors, and describe alterations in protein expression that provide insight into cellular pathways affected by oxidative stress.
Materials and methods
Measurement of ROS production by red cells
Red cells from Sod2–/– and control Sod2+/+ animals that received transplants of fetal liver (chimeric)1 were collected into phosphate-buffered saline (PBS) containing EDTA (ethylenediaminetetraacetic acid) and washed 3 times to remove residual serum proteins. Red cells were then resuspended at a concentration of 107 cells/mL in prewarmed RPMI (no phenol red). Samples were divided and stained as follows: 4 μM dihydroethidium (DHE; Molecular Probes, Eugene, OR) and 4 μM chloromethyl, dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Molecular Probes) were added and samples were placed in a humidified 5% CO2 incubator for 30 minutes. Samples were then diluted into cold PBS containing 2 mM EDTA with 0.5% bovine serum albumin (BSA) and spun and resuspended in same buffer for FACS analysis.
EM sample preparation and microscopy
Red cells were harvested into PBS containing 2 mm EDTA and washed twice to remove serum proteins before being diluted in Karnovsky fixative (Figure 1B). Alternatively (Figure 1C), fixed red cells were washed 3 times in PBS, stained with 10% potassium ferricyanide in 1.2 N HCl for 20 minutes, washed in PBS 3 times, and returned to Karnovsky electron microscope (EM) fixative before being routinely processed with osmium tetroxide and embedded in plastic. Sections (60-nm thick) were observed without further staining. Spleen and bone marrow specimens expressed intact from mouse tibia were placed into formalin, followed by paraffin embedding and sectioning for hematoxylin-eosin or Perl iron stain. Images were acquired on a Nikon Eclipse E600 microscope with a 100×/0.30 oil immersion or 20×/0.50 lens and an RT Slider SPOT 2.3.1 camera (Diagnostic Instruments) using SPOT Advanced software (v. 3.5.9).
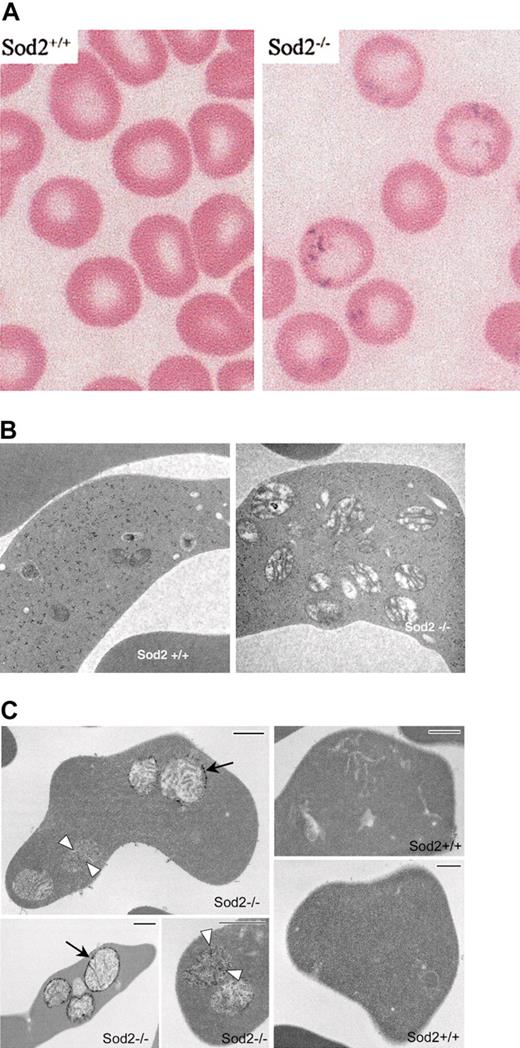
Iron deposition and morphology on EM of SOD2-deficient red cells. (A) Modified Perl stain of peripheral blood from recipients of Sod2+/+ or Sod2–/– fetal liver transplants (as a source of hematopoietic stem cells) demonstrating excess iron on SOD2-deficient cells. (B) Electron micrographs of Sod2+/+ and Sod2–/– reticulocytes reveal an increased number of mitochondria with prominent intramitochondrial membranes in SOD2-deficient cells. No obvious foci of iron deposition (electrondense material) were seen within SOD2-deficient cells with standard EM processing. (C) Electron micrographs of reticulocytes prestained with potassium ferricyanide prior to processing. Small arrows show diffuse electron-dense deposits in matrix of mitochondria, and large arrows show strong electron-dense deposits in the outer mitochondrial membrane of Sod2–/– cells. Bar length is 500 nm.
Iron deposition and morphology on EM of SOD2-deficient red cells. (A) Modified Perl stain of peripheral blood from recipients of Sod2+/+ or Sod2–/– fetal liver transplants (as a source of hematopoietic stem cells) demonstrating excess iron on SOD2-deficient cells. (B) Electron micrographs of Sod2+/+ and Sod2–/– reticulocytes reveal an increased number of mitochondria with prominent intramitochondrial membranes in SOD2-deficient cells. No obvious foci of iron deposition (electrondense material) were seen within SOD2-deficient cells with standard EM processing. (C) Electron micrographs of reticulocytes prestained with potassium ferricyanide prior to processing. Small arrows show diffuse electron-dense deposits in matrix of mitochondria, and large arrows show strong electron-dense deposits in the outer mitochondrial membrane of Sod2–/– cells. Bar length is 500 nm.
Antioxidant treatment and red cell half-life measurement
Euk-189 was prepared as a sterile suspension (3 mg/cc) in 5% dextrose. Animals received an intraperitoneal injection of 30 mg/kg weight 3 times per week. For the red cell half-life experiment, pretreated groups received Euk-189 for 4 weeks prior to red cell labeling. Red cell labeling was performed using the in vivo biotinylation method19 modified as previously described.1 Assessment of the percent of biotinylated red cells was performed by harvesting a small amount of blood (< 5 μL) from the retro-orbital sinus using an insulin syringe preloaded with a small volume of PBS containing EDTA (3 mg/cc). Red cells were washed in PBS and then incubated with streptavidin–phycoerythrin (PE) prior to FACS analysis.
Preparation of RBC proteins for oxyblot, 2-D gel separation, and mass spectrometry analysis of selected proteins
Red cells were collected by cardiac puncture of anesthetized animals and diluted into 2 volumes of PBS containing EDTA (3 mg EDTA/cc). An equal volume of 2% dextran T-500 in PBS was added, and red cells were allowed to sediment by gravity. Supernatant containing leukocytes was discarded, and red cells were diluted again in 2 volumes of PBS containing EDTA for a second round of sedimentation prior to further processing. Red cells were washed several times in cold PBS prior to hypotonic lysis of red cell pellet in 3 volumes of cold lysis buffer (20 mM Tris pH 7.5, 2 mM EDTA, 10 mM dithiothreitol [DTT] plus protease inhibitors). Red cell lysates were spun 10 minutes at 20 000g to separate cytosolic/soluble from membrane/insoluble fractions. Membrane/insoluble fractions were washed and spun 5 times with cold lysis buffer before solubilization in 3 volumes of lysis buffer containing 1.5% Triton X-100. For oxyblot analysis, 5 μg membrane protein or 20 μg cytosolic protein were derivatized according to the manufacturer's protocol (Oxyblot kit; Intergen, Purchase, NY).
K-Cl cotransport
Heparinized blood collected from the inferior vena cava was processed as described after removal of an aliquot for measurement of red cell indices using a hemoanalyzer ADVIA (Bayer, Tarrytown, NY). Plasma and buffy coat were removed after passing through cotton and centrifugation at 2500 rpm for 4 minutes. Cells were washed 4 times with ice-cold choline wash solution (CWS) containing 172 mM choline chloride, 1 mM MgC12, 10 mM Tris-MOPS, pH 7.40 at 4° C. An aliquot of cells was then suspended in an approximately equal volume of CWS, and determinations of hematocrit, cell Na (1:50 dilution), and K (1:500 dilution) were performed. Erythrocyte Na+ and K+ contents were quantified by atomic absorption spectrophotometry with standards made in double-distilled water. The maximal rate of K-Cl cotransport from fresh cells was measured as volume-dependent K+ efflux. Volume-induced K+ flux was calculated as the difference between K+ efflux in NaCl hypotonic (100 mM) and in NaCl isotonic (160 mM) media. Incubation times at 37° C for flux measurements were 5 and 25 minutes.
2-D gel electrophoresis, quantitation of protein expression, and mass spectrometry determination of protein identity
Red cell membrane (200 μg) or red cell cytosolic protein (500 μg) was prepared and run on 2-D gels as previously described.20,21 Gel images were acquired on a laser scanner and analyzed for differential protein expression using BioImage software (BioImage, Ann Arbor, MI) as described.22 The BioImage software calculates protein quantity and expresses it as integrated intensity or volume. Gel images were normalized to each other with respect to total intensity. Once the differentially expressed protein spots had been located, the proteins were excised manually from the gels, digested with trypsin, and identified using matrix-assisted laser desorption and ionization (MALDI) time-of-flight (TOF) mass spectrometric analysis as described.21
Statistics
Analysis of variance (ANOVA) was performed (Table 1 and Figure 3) using GraphPad Prism software (GraphPad, San Diego, CA).
K-Cl cotransport activity in Sod2+/+, Sod2-/-, and antioxidant-treated Sod2-/- red cells
. | Sod2+/+ . | Sod2-/- . | P . | Sod2-/- Euk . | P . |
|---|---|---|---|---|---|
| MCV, fL | 52.5 ± 3.34 | 45.1 ± 0.53 | < .05 | 46.2 ± 0.60 | NS |
| Reticulocytes, % | 2.9 ± 0.6 | 18.2 ± 1.5 | < .01 | 11.0 ± 0.4 | < .01 |
| Na+, mmol/kg Hb | 25.5 ± 2.6 | 51.6 ± 3.1 | < .01 | 40.0 ± 3.4 | < .05 |
| K+, mmol/kg Hb | 740 ± 28 | 1285 ± 141 | < .05 | 1265 ± 184 | < .05 |
| Mg2+, mmol/kg Hb | 14.1 ± 1.7 | 42.1 ± 8.4 | < .05 | 29.8 ± 6.1 | NS |
| K-Cl cotransport, mmol/1013 cell × h | 8.03 ± 0.6 | 19.7 ± 3 | < .05 | 14.4 ± 3.4 | NS |
. | Sod2+/+ . | Sod2-/- . | P . | Sod2-/- Euk . | P . |
|---|---|---|---|---|---|
| MCV, fL | 52.5 ± 3.34 | 45.1 ± 0.53 | < .05 | 46.2 ± 0.60 | NS |
| Reticulocytes, % | 2.9 ± 0.6 | 18.2 ± 1.5 | < .01 | 11.0 ± 0.4 | < .01 |
| Na+, mmol/kg Hb | 25.5 ± 2.6 | 51.6 ± 3.1 | < .01 | 40.0 ± 3.4 | < .05 |
| K+, mmol/kg Hb | 740 ± 28 | 1285 ± 141 | < .05 | 1265 ± 184 | < .05 |
| Mg2+, mmol/kg Hb | 14.1 ± 1.7 | 42.1 ± 8.4 | < .05 | 29.8 ± 6.1 | NS |
| K-Cl cotransport, mmol/1013 cell × h | 8.03 ± 0.6 | 19.7 ± 3 | < .05 | 14.4 ± 3.4 | NS |
Data presented are mean ± standard error. ANOVA with Dunnett multiple comparison test was used to compare Sod2+/+ with Sod2-/- and Sod2-/- treated with Euk-189 for 28 days. For ion measurements: Sod2+/+, n = 5; Sod2-/-, n = 7; Sod2-/- Euk-189, n = 5; except for Mg2+: Sod2+/+, n = 4; and Sod2-/-, n = 6. For MCV and reticulocyte count, n = 4 for all groups. Red cell ion content was measured in red cell hemolysate using atomic absorption spectrophotometry. The activity of the K-Cl cotransport was measured by Cl-dependent K efflux in intact red blood cells as described.36
NS indicates not significant.
Zinc protoporphyrin levels. ZPP levels are higher in Sod2–/– peripheral red cells compared with Sod2+/+. Treatment of SOD2-deficient (transplant recipient) mice with the antioxidant EUK-189 (duration 4 weeks) leads to a further increase in ZPP levels. The increase occurs despite a concomitant increase in hematocrit (from 0.237 ± 0.02 [23.7% ± 2%] without treatment to 0.305 ± 0.03 [30.5% ± 3%] after therapy) and a decrease in reticulocyte count (from 0.175 ± 0.06 [17.5% ± 6%] prior to therapy to 0.072 ± 0.007 [7.2% ± 0.7%] after therapy). Data presented are mean ± standard deviation; ANOVA was used for statistical analysis with Tukey-Kramer multiple comparison test between the 3 groups (n = 5 animals per group). In a second experiment (right), the effect of Euk-189 on Sod2+/+ red cells was assessed following therapy for 21 days (n = 4 animals per group). Although these data are suggestive of an effect of Euk-189 on ZPP in Sod2+/+ cells, results from this trial were not statistically significant (2-tailed t test, P = .07).
Zinc protoporphyrin levels. ZPP levels are higher in Sod2–/– peripheral red cells compared with Sod2+/+. Treatment of SOD2-deficient (transplant recipient) mice with the antioxidant EUK-189 (duration 4 weeks) leads to a further increase in ZPP levels. The increase occurs despite a concomitant increase in hematocrit (from 0.237 ± 0.02 [23.7% ± 2%] without treatment to 0.305 ± 0.03 [30.5% ± 3%] after therapy) and a decrease in reticulocyte count (from 0.175 ± 0.06 [17.5% ± 6%] prior to therapy to 0.072 ± 0.007 [7.2% ± 0.7%] after therapy). Data presented are mean ± standard deviation; ANOVA was used for statistical analysis with Tukey-Kramer multiple comparison test between the 3 groups (n = 5 animals per group). In a second experiment (right), the effect of Euk-189 on Sod2+/+ red cells was assessed following therapy for 21 days (n = 4 animals per group). Although these data are suggestive of an effect of Euk-189 on ZPP in Sod2+/+ cells, results from this trial were not statistically significant (2-tailed t test, P = .07).
Results
SOD2-deficient red cells contain free iron and abundant mitochondria
The hallmark of SA is abnormal iron accumulation within mitochondria of erythroid progenitors. Iron staining demonstrates coarse iron granules that correspond to intramitochondrial electron-dense deposits on electron microscopy. Figure 1A shows coarse Prussian blue–positive inclusions within mature SOD2-deficient peripheral blood erythrocytes. The appearance and location of these iron deposits is similar to the basophilic inclusions previously demonstrated in SOD2-deficient cells on Wright stain.1 Nucleated erythroid progenitors in bone marrow or spleen do not have similar deposits (data not shown), suggesting that accumulation of iron occurs following enucleation and release from the marrow. Significant accumulation of iron in reticuloendothelial cells in marrow, spleen, and liver is seen in Sod2–/– recipients, but not in Sod2+/+ recipients, and is likely related to chronic hemolytic anemia (data not shown). Figure 1B shows electron micrographs of Sod2+/+ and Sod2–/– reticulocytes. There is a dramatic increase in mitochondrial number and mitochondrial size in SOD2-deficient reticulocytes. Morphologically, Sod2–/– mitochondria have prominent internal membranes and appear swollen. Mitochondria in Sod2+/+ reticulocytes are smaller, and many appear to degenerate as they mature to definitive erythrocytes.
Mitochondrial localization of excess iron in Sod2–/– cells is demonstrated in Figure 1C through enhancement of iron staining by treatment of fixed cells with potassium ferricyanide prior to embedding. The 3 Sod2–/– panels show variations observed in the density and localization of iron within mitochondria, perhaps representing different stages in the process of deposition. Diffuse deposition of iron can be seen in the matrix (white arrowheads), as well as strong deposition (black arrows) along the periphery of the mitochondria. No comparable electron densities can be found in Sod2+/+ reticulocytes (note presence of intracellular organelles) prepared in parallel.
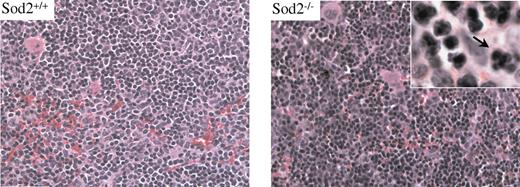
Erythropoiesis in spleen and marrow characterized by mild dysplasia
Hematoxylin and eosin (H&E)–stained sections of spleen and marrow from Sod2–/– animals reveal an expanded erythroid compartment with full maturation. Figure 2 compares sections of spleen from Sod2+/+ and Sod2–/– animals. Nuclear dysplasia (inset) is evident in a small percentage of Sod2–/– erythroid progenitors (arrow). FACS analysis of bone marrow cell suspensions from Sod2–/– and Sod2+/+ animals stained for the erythroid marker Ter119 and annexin V to assess the fraction of apoptotic nucleated erythroid cells show no excess of annexin V–positive cells in the Sod2–/– marrow (not shown). These results provide little evidence for significant intramedullary destruction (ie, ineffective erythropoiesis) of developing red cells deficient in SOD2.
Sod2+/+ and Sod2–/– spleen cells stained with H&E. Erythroid hyperplasia is evident in SOD2-deficient samples. Occasional nuclear clefting is evident in Sod2–/– erythroid precursors (arrow), suggesting mild dysplasia.
Sod2+/+ and Sod2–/– spleen cells stained with H&E. Erythroid hyperplasia is evident in SOD2-deficient samples. Occasional nuclear clefting is evident in Sod2–/– erythroid precursors (arrow), suggesting mild dysplasia.
Sod2–/– red cells have elevated ZPP levels
The ineffective mitochondrial iron utilization seen in SA may be due to either a lack of protoporphyrin IX, or failure to deliver iron in appropriate valency (Fe2+) for incorporation. One can distinguish between earlier biosynthetic lesions such as ALAS2 deficiency (no protoporphyrin IX) and later defects in iron incorporation by measuring zinc protoporphyrin (ZPP) levels. Many forms of SA not due to ALAS2 deficiency have high levels of ZPP. Figure 3 demonstrates that Sod2–/– red cells have high levels of ZPP, characteristic of a defect in iron incorporation rather than an earlier defect in heme biosynthesis. We have also investigated the effect of antioxidant therapy on ZPP levels using the SOD/catalase mimetic compound Euk-189, a member of a family of catalytic antioxidants that have previously been shown to partially correct SOD2-deficiency anemia,1 and to ameliorate the severity of SOD2 deficiency in whole animals.23 Unexpectedly, catalytic antioxidant therapy leads to a further increase in ZPP levels in treated red cells. This added increase in ZPP level with antioxidant therapy occurs coincident with an improvement in hematocrit and a decrease in reticulocyte count, as previously described,1 and is not due to direct interference of the antioxidant with the technique used for measurement of ZPP, as samples spiked with antioxidant do not show increased ZPP levels (data not shown). Of interest, catalytic antioxidant therapy of Sod2+/+ animals (3 weeks of treatment) also increases ZPP levels (Figure 3). However, prior experiments did not show a significant effect on hematocrit or reticulocyte count when Sod2+/+ animals were treated with catalytic antioxidant for 2 months.1 Whereas the net effect of catalytic antioxidant therapy on erythropoiesis is strongly positive in Sod2–/– mice, this appears to be the sum of positive and negative effects on erythropoiesis.
Distribution of oxidized protein in red cells and effect of catalytic antioxidants on protein oxidation
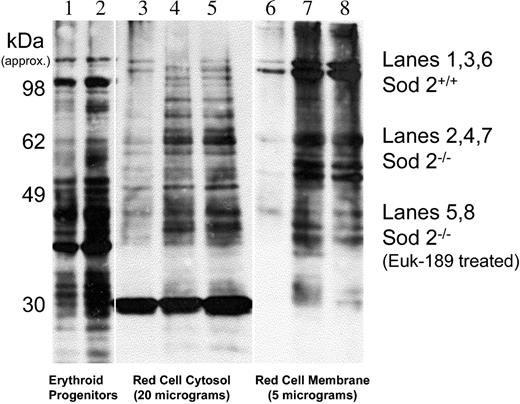
Protein oxidation can be measured through indirect detection on Western blot of carbonyls that result from reactive oxygen species (ROS) reacting with side chains of lysine, arginine, proline, and threonine.24,25 Prior characterization of SOD2-deficiency anemia revealed increased protein carbonyl content in Sod2–/– red cells.1 Figure 4 shows that most oxidized protein in SOD2-deficient red cells partitions to a membrane or insoluble fraction.
Protein oxidation in marrow progenitors and peripheral red cells. Protein carbonyl content in Sod2+/+ and Sod2–/– samples was compared by Oxyblot. Lane 1 (Sod2+/+) and lane 2 (Sod2–/–) compare whole-cell lysate protein from 5 × 106 Ter119+ marrow progenitor cells isolated by FACS. Lane 3 (Sod2+/+), lane 4 (Sod2–/–), and lane 5 (Sod2–/– Euk-189–treated) contain 20 μg red cell cytosolic protein, and lane 6 (Sod2+/+), lane 7 (Sod2–/–), and lane 8 (Sod2–/– Euk-189–treated) contain 5 μg membrane protein from the corresponding red cell samples.
Protein oxidation in marrow progenitors and peripheral red cells. Protein carbonyl content in Sod2+/+ and Sod2–/– samples was compared by Oxyblot. Lane 1 (Sod2+/+) and lane 2 (Sod2–/–) compare whole-cell lysate protein from 5 × 106 Ter119+ marrow progenitor cells isolated by FACS. Lane 3 (Sod2+/+), lane 4 (Sod2–/–), and lane 5 (Sod2–/– Euk-189–treated) contain 20 μg red cell cytosolic protein, and lane 6 (Sod2+/+), lane 7 (Sod2–/–), and lane 8 (Sod2–/– Euk-189–treated) contain 5 μg membrane protein from the corresponding red cell samples.
Some oxidized protein is evident in Sod2+/+ membranes, but the signal intensity is less than that observed for Sod2–/– samples. Increased protein oxidation is also evident in soluble proteins from Sod2–/– red cells when compared with Sod2+/+. In vivo antioxidant therapy with the SOD/catalase mimetic Euk-189 does not significantly reduce protein oxidation (Figure 4, lanes 5 and 8) in either soluble or membrane proteins from SOD2-deficient red cells, although such therapy does significantly increase hematocrit, reduce reticulocytosis (Figure 3 legend), and reduce spleen weight in treated animals (data not shown). Increased oxidation can also be detected in whole-cell lysate protein from Sod2–/– nucleated marrow erythroid progenitor cells (Ter 119+), although the difference compared with Sod2+/+ cells (Figure 4, lanes 1 and 2) is less pronounced than that observed in peripheral red cells.
Effect of catalytic antioxidant therapy on red cell survival
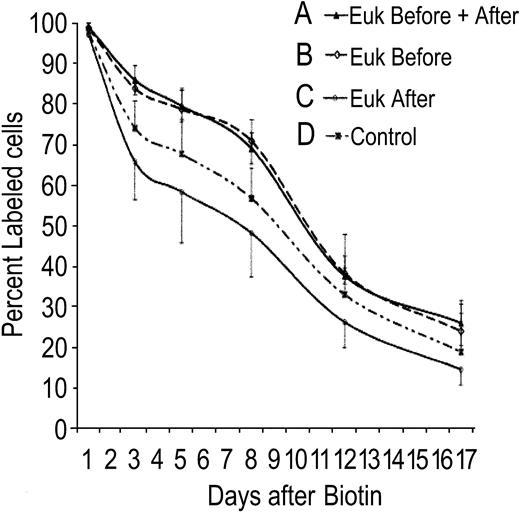
Catalytic antioxidant therapy partially ameliorates anemia secondary to SOD2 deficiency. Antioxidant treatment was combined with concurrent measurement of red cell lifespan to determine whether antioxidant therapy exerts its effect during red cell development, protects mature red cells in the periphery, or both. In Figure 5, 4 groups of animals (all received transplants of Sod2–/– stem cells; n = 5 animals per group) were treated as follows: group A, 4 weeks of antioxidant therapy prior to red cell labeling and continuation of therapy during measurement of half-life; group B, 4 weeks of antioxidant therapy prior to red cell labeling, at which time antioxidant was stopped; group C, antioxidant therapy commenced at the time of red cell labeling and continued during measurement of half-life; and group D, no antioxidant therapy. The 2 groups that received antioxidant prior to red cell labeling show an extended red cell lifespan (groups A and B). There is no increase in red cell half-life when antioxidant treatment is begun at the same time as red cell labeling (group C), nor is there an additional benefit of continuing antioxidant therapy once cells have been labeled (comparing groups A and B). Thus, SOD/catalase mimetic therapy results in increased red cell survival when given during red cell formation, but does not extend survival of red cells already in the peripheral circulation. Treatment starting coincident with red cell labeling (group C) reproducibly results in a measurable decrease in survival of peripheral cells. As with the ZPP data in Figure 3, whereas the net effect of catalytic antioxidant therapy is protective, this appears to be the sum of both positive and negative effects on developing and mature red cells.
Effect of antioxidant therapy on red cell lifespan. Mice that received transplants of Sod2–/– fetal liver stem cells were divided into 4 groups for measurement of red cell lifespan in the presence or absence of SOD/catalase antioxidant therapy. Group A was pretreated with antioxidant for 4 weeks and continued treatment during measurement of red cell lifespan. Group B was pretreated as Group A, but stopped antioxidant at the time of red cell labeling with biotin. Group C commenced Euk-189 therapy coincident with red cell labeling, whereas group D received sham injections. Error bars = SD.
Effect of antioxidant therapy on red cell lifespan. Mice that received transplants of Sod2–/– fetal liver stem cells were divided into 4 groups for measurement of red cell lifespan in the presence or absence of SOD/catalase antioxidant therapy. Group A was pretreated with antioxidant for 4 weeks and continued treatment during measurement of red cell lifespan. Group B was pretreated as Group A, but stopped antioxidant at the time of red cell labeling with biotin. Group C commenced Euk-189 therapy coincident with red cell labeling, whereas group D received sham injections. Error bars = SD.
Steady-state ROS production is elevated in SOD2-deficient red cells
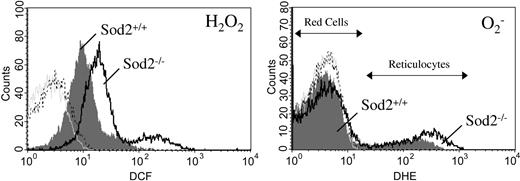
Oxidation-sensitive dyes can be used to measure ongoing production of ROS by actively respiring cells in vitro.26 Sod2–/– red cells have elevated ROS production as measured by the peroxide-sensitive dye dichlorodihydrofluorescein diacetate (H2DCF-DA), when compared with Sod2+/+ red cells, and Sod2–/– reticulocytes have both increased peroxide and increased superoxide as measured by the oxidation-sensitive dye dihydroethidium (DHE) when compared with Sod2+/+ reticulocytes (Figure 6).
ROS generation by reticulocytes and mature red cells. Peripheral blood cells from Sod2+/+ and Sod2–/– transplant recipients were incubated in the presence of the superoxide-sensitive dye DHE and the peroxide-sensitive dye H2DCFDA for 30 minutes at 37° C, followed by FACS analysis. (Left) Peroxide production by Sod2+/+ red cells and reticulocytes (shaded gray); and Sod2–/– red cells and reticulocytes (black line). (Right) Superoxide production by Sod2+/+ (shaded gray) and Sod2–/– (black line) red cells and reticulocytes. Red cells are the major peak for each histogram, with reticulocytes forming a tail or secondary peak with higher ROS production. Plots shown are representative of 5 mice for each genotype. For each histogram: light gray tracing indicates unstained Sod2+/+ cells; broken black tracing indicates unstained Sod2–/– cells.
ROS generation by reticulocytes and mature red cells. Peripheral blood cells from Sod2+/+ and Sod2–/– transplant recipients were incubated in the presence of the superoxide-sensitive dye DHE and the peroxide-sensitive dye H2DCFDA for 30 minutes at 37° C, followed by FACS analysis. (Left) Peroxide production by Sod2+/+ red cells and reticulocytes (shaded gray); and Sod2–/– red cells and reticulocytes (black line). (Right) Superoxide production by Sod2+/+ (shaded gray) and Sod2–/– (black line) red cells and reticulocytes. Red cells are the major peak for each histogram, with reticulocytes forming a tail or secondary peak with higher ROS production. Plots shown are representative of 5 mice for each genotype. For each histogram: light gray tracing indicates unstained Sod2+/+ cells; broken black tracing indicates unstained Sod2–/– cells.
In Figure 6, the predominant peak for each FACS histogram represents mature red cells, with a positive shoulder or smaller secondary peak representing reticulocytes, which produce higher steady-state levels of ROS (verified by cell sorting of DCF- and DHE-positive cells followed by H&E staining; data not shown). Reticulocytosis (induced or constitutive) does not lead to elevated ROS production by red cells from Sod2+/+ animals made anemic through serial phlebotomy (Figure 6) or mice with hereditary microcytic anemia27 (mk/mk; not shown). Increased ROS production of SOD2-deficient cells may be secondary to increased mitochondrial mass (as respiring mitochondria are likely the locus of superoxide generation) or decreased mitochondrial efficiency. The increase in DCF fluorescence (peroxide sensitive) may result from leakage of superoxide out of mitochondria where cytosolic SOD1 can then utilize it for peroxide production, or via spontaneous dismutation of superoxide. Mature red cells show limited oxidation of DCF during a 30-minute incubation, but do demonstrate a reproducible increase in DCF fluorescence produced by SOD2-deficient red cells (Figure 6 left), suggesting ongoing higher peroxide production. Oxidation of DCF has been demonstrated to be dependent on the presence of intracellular iron, suggesting that similar increases in DCF signal may occur in a variety of iron overload conditions.28 In the absence of nucleic acid (mitochondrial DNA and RNA in reticulocytes), DHE oxidation to ethidium may not be detectable; thus, we cannot assess ongoing superoxide production by mature red cells using this method.
Ion content and K-Cl cotransport in Sod2-deficient cells
The decreased membrane deformation and reduced volume previously described in Sod2–/– red cells1 may in part be due to alteration in volume regulatory ion transport systems in the red cell membrane. To investigate this possibility, we assessed K-Cl cotransport (KCC) activity in Sod2–/– red cells, and the effect of catalytic antioxidant treatment on its activity (Table 1). Sod2–/– red cells were significantly dehydrated as indicated by a decrease in the MCV when compared with control. Furthermore, we found that Sod2–/– red cells have an approximately 2-fold elevation of intracellular Na+, K+, and Mg2+ ion concentrations. The activity of the K-Cl cotransport system was also increased approximately 2-fold when comparing Sod–/– to Sod2+/+ red cells. These results are consistent with prior studies that have demonstrated increased K-Cl cotransport activity secondary to oxidative stress.29 Treatment of Sod2–/– cells with Euk-189 shifts ion and K-Cl cotransport activities toward values seen in Sod2+/+ cells, but only the change in Na+ content reaches statistical significance when compared with untreated Sod2–/– cells.
Proteomic analysis comparing Sod2+/+-, Sod2–/–-, and antioxidant-treated Sod2–/– red cell proteins
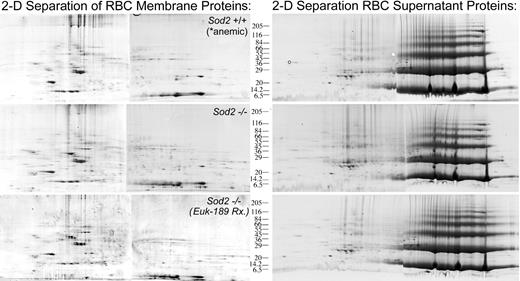
Comparison of protein expression in peripheral red cells from Sod2+/+-, Sod2–/– -, and antioxidant-treated Sod2–/– (transplantation subject) animals was performed in order to identify patterns of altered protein expression in response to oxidative stress. Control (Sod2+/+; n = 5) animals were phlebotomized to produce a reticulocytosis (10%-15% retics) and hematocrit (0.25-0.30 [25%-30%]) comparable to that of Sod2–/– animals prior to collection of peripheral red cells. Figure 7 shows 2-D gel analysis of “membrane” (left) and “supernatant” (right) proteins from each group. Image analysis software (BioImage) was used to determine the integrated intensity or volume of specific spots relative to total intensity of all spots on each gel. The relative intensity of each spot was then compared between samples. Three sets of gels were compared in this fashion to identify reproducibly differentially expressed proteins. Based on this analysis, individual spots were selected for semiautomated processing involving manual spot excision, transfer to microplates, trypsin digestion, and protein identification by MALDI-TOF mass spectrometry using peptide mass fingerprinting.21 Table 2 lists 41 proteins identified by MALDI mass spectrometry as differentially expressed when comparing red cell membrane preparations from Sod2+/+-, Sod2–/– -, and antioxidant-treated Sod2–/– blood cells. Many of the proteins identified as being differentially expressed are involved in mitochondrial functions. Based on electron micrographs and Western blots, mitochondrial mass is increased in SOD2-deficient cells, likely representing an important response by cells to compensate for poor mitochondrial function. To control for differences in mitochondrial content in our samples, protein expression data were normalized against the mitochondrial structural protein porin (voltage-dependent anion channel). Proteins in Table 2 are loosely grouped according to subcellular localization or function. For some of the protein spots in SOD2-deficient preparations we did not locate a corresponding spot in the control sample, and thus cannot determine a fold-differential expression. Failure to identify a corresponding spot may be secondary to differences in posttranslational modifications affecting protein mobility (within either preparation) or because the corresponding species is present below the level of our detection methods. For these proteins, relative expression data in SOD2-deficient cells in the presence or absence of antioxidant therapy is presented. Because we did not compare protein expression from antioxidant-treated Sod2+/+ samples, changes that are documented in the presence of antioxidant treatment may not be unique to Sod2–/– samples. Proteins that were identified more than once are marked with an asterisk. Presumably, differences in migration reflect either posttranslational modifications or splicing variants.
Representative 2-D gel analysis of membrane and soluble protein preparations from Sod2+/+, Sod2–/–, and Euk-189–treated Sod2–/– red cells. Membrane (200 μg) or soluble red cell protein (400 μg) was separated by 2-D gel electrophoresis. Sod2+/+ red cell samples were obtained from serially phlebotomized animals (in order to induce reticulocytosis similar to that seen in Sod2–/– samples). Each sample was prepared from pooled red cells obtained from 5 donor animals.
Representative 2-D gel analysis of membrane and soluble protein preparations from Sod2+/+, Sod2–/–, and Euk-189–treated Sod2–/– red cells. Membrane (200 μg) or soluble red cell protein (400 μg) was separated by 2-D gel electrophoresis. Sod2+/+ red cell samples were obtained from serially phlebotomized animals (in order to induce reticulocytosis similar to that seen in Sod2–/– samples). Each sample was prepared from pooled red cells obtained from 5 donor animals.
Proteomic analysis of differential protein expression in Sod2+/+, Sod2-/-, and antioxidant-treated Sod2-/- RBC membrane preparations
Protein ID . | SOD2+/+ versus SOD2-/- . | SOD2-/- (EUK-189) versus SOD2-/- . |
|---|---|---|
| Membrane/structural proteins | ||
| Anion exchanger 4b | -5.95 | -1.75 |
| Guanine nucleotide exchange factor | -19.17 | -1.03 |
| p97 membrane-bound transferrin-like protein | -54.20 | -1.01 |
| Actin-like protein | — | 1.96 |
| Rbcc 728 | -5.63 | 1.32 |
| Alpha2-macroglobulin receptor | — | 8.36 |
| HA receptor for hyaluronic acid | -25.62 | 1.41 |
| HA receptor for hyaluronic acid* | -18.59 | 1.99 |
| Surfeit locus protein 4 | — | 2.26 |
| Cytokeratin 10 | 3.79 | 17.16 |
| Kainate receptor GluR7 subunit | — | -1.50 |
| Stress response/chaperones | ||
| 60-kDa chaperonin mitochondrial | — | -2.40 |
| Heat-shock 60-kDa chaperonin | — | -1.93 |
| Sulfotransferase-related protein | — | 1.35 |
| Prohibitin | — | -1.82 |
| Prohibitin* | — | -1.56 |
| Peroxiredoxin 2 | 1.48 | 2.90 |
| Peroxiredoxin 2* | — | 2.75 |
| Peroxiredoxin 2* | — | 2.79 |
| DNAK-type molecular chaperone | -10.11 | -3.15 |
| Metabolism | ||
| Octanoyltransferase | — | 2.05 |
| Gamma enolase | -23.01 | 1.90 |
| Gamma enolase* | — | 1.67 |
| Microsomal tryglyceride transfer protein | — | 4.25 |
| Microsomal tryglyceride transfer protein* | -5.12 | 3.50 |
| UDP glucuronyltransferase-P | — | 2.06 |
| Carbonic anhydrase I | 1.55 | 5.54 |
| (Similar to) biliverdinreductase B flavin reductase | — | 2.67 |
| Leucyl-tRNA synthetase | -4.66 | -1.00 |
| Nucleoside diphosphate kinase | — | 2.68 |
| Mitochondrial/bioenergetics | ||
| GFAP | — | 1.38 |
| MECA32 | — | 1.39 |
| Delta subunit ATPase | — | -2.17 |
| Cytochrome C oxidase, subunit Va | — | -2.52 |
| ATP synthase beta subunit | -1.60 | -1.95 |
| ATP synthase Beta chain | — | 4.01 |
| ATP synthase D chain | -24.02 | -2.03 |
| ATP synthase D chain* | -1.96 | -1.32 |
| ATP synthase alpha, mitochondrial | — | -4.67 |
| Voltage-dependent anion channel† | 1.00 | 1.00 |
| Not classified | ||
| Calreticulin | — | 1.46 |
| Cleavage and polyadenylation-specific factor 2 | -2.96 | 2.43 |
| Thrombospondin | -8.87 | 1.36 |
| Thrombospondin* | -5.34 | 1.25 |
| Cytochrome B5 | — | -1.11 |
| Apolipoprotein A-I | — | 2.44 |
| Retinoic acid-binding protein II | — | 2.44 |
| Ubiquitin B | -1.02 | 1.17 |
| Trophoblast protein-1 | -1.34 | -1.98 |
Protein ID . | SOD2+/+ versus SOD2-/- . | SOD2-/- (EUK-189) versus SOD2-/- . |
|---|---|---|
| Membrane/structural proteins | ||
| Anion exchanger 4b | -5.95 | -1.75 |
| Guanine nucleotide exchange factor | -19.17 | -1.03 |
| p97 membrane-bound transferrin-like protein | -54.20 | -1.01 |
| Actin-like protein | — | 1.96 |
| Rbcc 728 | -5.63 | 1.32 |
| Alpha2-macroglobulin receptor | — | 8.36 |
| HA receptor for hyaluronic acid | -25.62 | 1.41 |
| HA receptor for hyaluronic acid* | -18.59 | 1.99 |
| Surfeit locus protein 4 | — | 2.26 |
| Cytokeratin 10 | 3.79 | 17.16 |
| Kainate receptor GluR7 subunit | — | -1.50 |
| Stress response/chaperones | ||
| 60-kDa chaperonin mitochondrial | — | -2.40 |
| Heat-shock 60-kDa chaperonin | — | -1.93 |
| Sulfotransferase-related protein | — | 1.35 |
| Prohibitin | — | -1.82 |
| Prohibitin* | — | -1.56 |
| Peroxiredoxin 2 | 1.48 | 2.90 |
| Peroxiredoxin 2* | — | 2.75 |
| Peroxiredoxin 2* | — | 2.79 |
| DNAK-type molecular chaperone | -10.11 | -3.15 |
| Metabolism | ||
| Octanoyltransferase | — | 2.05 |
| Gamma enolase | -23.01 | 1.90 |
| Gamma enolase* | — | 1.67 |
| Microsomal tryglyceride transfer protein | — | 4.25 |
| Microsomal tryglyceride transfer protein* | -5.12 | 3.50 |
| UDP glucuronyltransferase-P | — | 2.06 |
| Carbonic anhydrase I | 1.55 | 5.54 |
| (Similar to) biliverdinreductase B flavin reductase | — | 2.67 |
| Leucyl-tRNA synthetase | -4.66 | -1.00 |
| Nucleoside diphosphate kinase | — | 2.68 |
| Mitochondrial/bioenergetics | ||
| GFAP | — | 1.38 |
| MECA32 | — | 1.39 |
| Delta subunit ATPase | — | -2.17 |
| Cytochrome C oxidase, subunit Va | — | -2.52 |
| ATP synthase beta subunit | -1.60 | -1.95 |
| ATP synthase Beta chain | — | 4.01 |
| ATP synthase D chain | -24.02 | -2.03 |
| ATP synthase D chain* | -1.96 | -1.32 |
| ATP synthase alpha, mitochondrial | — | -4.67 |
| Voltage-dependent anion channel† | 1.00 | 1.00 |
| Not classified | ||
| Calreticulin | — | 1.46 |
| Cleavage and polyadenylation-specific factor 2 | -2.96 | 2.43 |
| Thrombospondin | -8.87 | 1.36 |
| Thrombospondin* | -5.34 | 1.25 |
| Cytochrome B5 | — | -1.11 |
| Apolipoprotein A-I | — | 2.44 |
| Retinoic acid-binding protein II | — | 2.44 |
| Ubiquitin B | -1.02 | 1.17 |
| Trophoblast protein-1 | -1.34 | -1.98 |
Table 2 lists 41 proteins identified by MALDI-MS as differentially expressed when comparing membrane preparations from Sod2+/+, Sod2-/-, and antioxidant-treated Sod2-/- RBCs. Sod2+/+ mice were made anemic through serial phlebotomy in order to ensure that reticulocyte counts were comparable to those in Sod2-/- samples. Data have been normalized to content of the mitochondrial protein voltage-dependent anion channel (VDAC), in bold. Most of the identified proteins are expressed at higher levels in Sod2-/- RBCs than in Sod2+/+ RBCs (negative values in the “SOD2+/+ versus SOD2-/-” column indicate fold decrease in protein expression). Changes in protein expression in response to Euk-189 therapy vary (“SOD2-/- (EUK-189) versus SOD2-/-” column). Some proteins were not detected in the Sod2+/+ sample, and data for these proteins is limited to comparison of plus and minus Euk-189 treatment. — indicates protein not detected in Sod2+/+ preparation.
Several proteins migrated as 2 or more unique spots, suggesting differences in posttranslational modification or variant splicing.
This row used for normalization.
Discussion
SOD2-deficient cells manifest a primary lesion in detoxification of mitochondrial superoxide. This defect is demonstrated by measurement of increased ROS production in nucleated marrow cells (data not shown) as well as reticulocytes using ROS-sensitive dyes and FACS (Figure 6). Increased protein oxidation can be detected in marrow progenitors but is more prominent in peripheral red cells (Figure 4). This suggests that damage in nucleated cells may be ameliorated by protein turnover, whereas damage accumulates in definitive red cells, chiefly in the membrane/insoluble fraction. Alternatively, the rate of damage to red cells may be accelerated due to exposure to higher oxygen concentrations in the arterial circulation.
Alterations in ion transport in accord with oxidative stress in red cell membrane were observed between control and Sod2–/– mice. Such an increase in K-Cl cotransport activity secondary to red cell oxidative damage has been previously reported in rats treated in vivo with dapsone,30 in human red cells exposed to oxidative agents in vitro,31 and in both sickle cell disease and β-thalassemia.32 Increased activity of this system promotes cell shrinkage, which might explain the relative dehydration of Sod2–/– red cells (Table 1). The measured stimulation of K-Cl cotransport in SOD2-deficient cells appears to result from both an increase in reticulocyte content and from membrane oxidative damage to the red cells. As with other indices of red cell damage, K-Cl cotransport activity decreased, but was not completely reduced to basal levels by treatment with the antioxidant EUK-189.
In addition to effects on ion transport, SOD/catalase mimetic therapy of SOD2-deficient stem cell transplant recipients significantly improves hematocrit and decreases reticulocyte count.1 Our evidence suggests that SOD/catalase mimetics act primarily on red cell progenitors. First, SOD/catalase mimetics do not extend the survival of mature red cells (acute administration appears to slightly decrease red cell survivall; Figure 5, group C). Cells that develop in the presence of drug exhibit extended peripheral survival, with no further enhancement of survival with continued presence of drug once cells are circulating (Figure 5, groups A and B). Second, neither protein carbonyl formation (a marker of accumulated red cell protein damage; Figure 4) nor measurement of steady-state ROS production by reticulocytes or mature red cells reveal dramatic changes with antioxidant treatment (not shown).
Phenotypic manifestations of SOD2 deficiency are found in reticulocytes, which contain large numbers of mitochondria (Figure 1B), and in mature red cells, which contain basophilic inclusions on Wright Stain1 and free iron (Figure 1A). The similarity between SOD2-deficiency anemia and SA is confirmed by the finding of iron deposition in mitochondria of SOD2-deficient cells (Figure 1C). Elevation of ZPP level (Figure 3) indicates a late defect in heme biosynthesis involving iron incorporation. It may also indicate functional iron deficiency analogous to prior explanation for increased ZPP levels in disorders such as thalassemia, although we have not explored this possibility.33 The further increase in ZPP level observed upon antioxidant therapy may result from an additional heme biosynthetic defect involving protoporphyrin IX synthesis or may be a direct effect of the drug as Sod2+/+ mice also show a slight increase in ZPP levels upon treatment with Euk-189. We did not find significant phenotypic abnormalities affecting nucleated red cell precursors in marrow or spleen by light microscopy (Figure 2) or EM (not shown), although we did detect increased iron content in reticuloendothelial cells of spleen and liver, likely secondary to increased red cell turnover (data not shown). The appearance of phenotypic abnormalities late in red cell development may be due to a breakdown in compensatory mechanisms in the terminal stages of differentiation. The identity of putative components of such a compensatory mechanism is provided by comparative protein analysis of red cells from normal and SOD2-deficient stem cell recipients.
SOD2-deficient red cells contain higher levels of DNAK-type chaperonin (a member of the HSP-70 family localized to mitochondria), a mitochondrial 60 kDa heat shock/chaperonin, and prohibitin,34 which function to aid refolding of newly imported or misfolded proteins in the mitochondrial matrix, suggesting that protein stability or folding is affected by SOD2 deficiency. These 3 proteins show a similar pattern with low or no expression detected in wild-type cells, and a decrease in expression in SOD2-deficient cells upon antioxidant therapy. A recent report35 suggests that the yeast hsp-60 (a mitochondrial chaperonin orthologue of hsp-70 family members) is directly involved in cellular responses to oxidative stress. Yeast hsp-60 function appears to be required to protect the integrity of iron containing mitochondrial proteins against oxidative damage. Both yeast hsp-6035 and E coli DNAK37 have been described as direct targets of protein oxidation under conditions of increased oxidative stress.
Proteins involved in pathways that reverse oxidant damage are increased in SOD2-deficient cells and show a further increase with antioxidant therapy—peroxiredoxin 2, sulfotransferase, and B flavin reductase, a protein involved in reduction of methemoglobin, along with cytochrome b5.38,39 This further increase with antioxidant therapy indicates that the turnover of some of these proteins may be directly tied to oxidative damage. Supporting this idea, we have identified peroxiredoxin 2 (PRDX-2) as one of the major oxidized proteins (as measured by protein carbonyl content) in SOD2-deficient cells. PRDX-2 is a member of a family of proteins also known as thioredoxin peroxidases.40 In tissue culture, expression of PRDX-2 is induced by oxidative stress, and under such conditions a portion of the protein becomes oxidized in neuronal cells and T cells.41,42 More recently, mice with a targeted deletion of PRDX-2 have been shown to have a mild hemolytic anemia.43 It will be of interest to determine whether alterations in expression of PRDX-2 affect severity of SOD2 deficiency or response to therapy with antioxidants.
Multiple subunits of mitochondrial ATP synthase are expressed at elevated levels in Sod2–/– red cells and show a decrease in expression with drug therapy. This may reflect increased metabolic demands in response to oxidative stress, or poor mitochondrial efficiency secondary to oxidative damage, either of which is improved by antioxidant therapy. A highly differentially expressed protein is p97 transferrin-like protein,44 also called melanotransferrin, found in Sod2–/– cells both in the presence or absence of drug. This protein has been extensively studied as a tumor-associated antigen in melanoma. p97 is a glycosylphosphatidylinositol (GPI)–linked surface molecule capable of delivering iron to cells, but the process appears to be inefficient, and not mediated by specific internalization via a surface receptor as in the case of transferrin.45,46 It will be important to determine whether increased expression of p97 plays a pathologic role in iron delivery to SOD2-deficient cells. Cytochrome c oxidase (COX) subunit 5a is expressed at higher levels in Sod2–/– red cells (not detected in Sod2+/+) and decreases with antioxidant therapy. Mutations in COX subunit I have been identified in some cases of SA, and COX activity has been postulated to be involved in reduction of iron prior to incorporation into heme.7 It will be of interest to investigate whether alteration in COX function affects iron utilization in SOD2-deficient cells.
Differential protein expression analysis has the potential to pick up “acquired” proteins that stick to the surface of red cells, but were not differentially expressed by red cell progenitors. Thrombospondin, found at higher levels on Sod2–/– cells, may be an instructive example of this phenomenon. Increased adherence of sickle red cells or phosphatidyl-serine exposing red cells to thrombospondin has been reported,47,48 suggesting that a similar phenomenon dependent on recognition of alterations of the red cell surface may occur in SOD2-deficient cells. HA receptor for hyaluronic acid is found at higher levels in Sod2–/– membrane preparations and may affect the adhesive properties of SOD2-deficient red cells. Higher “expression” of these proteins on the surface of SOD2-deficient red cells may either accelerate their removal from the circulation or be a marker of cells destined for early removal.
Some of the proteins in Table 2 have not previously been described as components of red cells. As with thrombospondin, some of these proteins may be associated with SOD2-deficient red cells, but not produced by the red cell progenitors. It will be important to corroborate proteomic data directly with protein expression data through Western blot (when antibodies are available) or indirectly through gene expression analysis. Analysis of mRNA expression from red cell progenitors in marrow or spleen will help to determine whether these genes are expressed during normal and/or SOD2-deficient erythropoiesis. Gene expression profiling of progenitor cells will help to determine in more detail the alterations in gene expression secondary to increased oxidative stress, which ultimately result in SOD2-deficiency anemia, and will reveal details of how antioxidant therapy affects this pathologic expression pattern.
Prepublished online as Blood First Edition Paper, June 17, 2004; DOI 10.1182/blood-2003-11-3858.
Supported by grants from the Ellison Medical Foundation (J.S.F.) and the National Institutes of Health (NIDDK RO1 DK62473 [J.S.F.], NCI PO1 CA39542 [S.J.B.], and NIDDK RO1 DK62474 [M.D.F.]).
S.R.D. is an employee of Eukarion Inc, whose potential product was studied in the present work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors wish to thank Terry Law of The Children's Hospital Clinical Lab and Howard Mulhern of the Children's Hospital Department of Pathology Electron Microscope Facility.

![Figure 3. Zinc protoporphyrin levels. ZPP levels are higher in Sod2–/– peripheral red cells compared with Sod2+/+. Treatment of SOD2-deficient (transplant recipient) mice with the antioxidant EUK-189 (duration 4 weeks) leads to a further increase in ZPP levels. The increase occurs despite a concomitant increase in hematocrit (from 0.237 ± 0.02 [23.7% ± 2%] without treatment to 0.305 ± 0.03 [30.5% ± 3%] after therapy) and a decrease in reticulocyte count (from 0.175 ± 0.06 [17.5% ± 6%] prior to therapy to 0.072 ± 0.007 [7.2% ± 0.7%] after therapy). Data presented are mean ± standard deviation; ANOVA was used for statistical analysis with Tukey-Kramer multiple comparison test between the 3 groups (n = 5 animals per group). In a second experiment (right), the effect of Euk-189 on Sod2+/+ red cells was assessed following therapy for 21 days (n = 4 animals per group). Although these data are suggestive of an effect of Euk-189 on ZPP in Sod2+/+ cells, results from this trial were not statistically significant (2-tailed t test, P = .07).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/104/8/10.1182_blood-2003-11-3858/6/m_zh80200467770003.jpeg?Expires=1767732046&Signature=m8LuEd24jpp7ODia-1a2jCeg7a~bz7whcjHngiDvfNfEaYJBvECuHawBx2UHIAlCF7WCbpjrcZe-r~xXkQsifEs20D396xCexSWNGFScFg4BgNHGab~9OMJxY6vMqzKmj85ZCqm8fQC~98jYHOMpvTv3~Tc2ju-AXkT22v2FBnKV4eQb4tmL~EiR4cymFoF0qNU0gWvMLPSEbZsnN7QTPs3l6KeHru~EXKWedhLF0FO3biDx2-UXpsFUJTuk3xvZ1O61hSydhpWANWeDKmC93eKD10lIskuqQpvf7Vk2XtkGPBhvBxTzDHbloGcv-RjWU~1ID9PW8A6CtjhbyyzY3Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal