Abstract
Iron (Fe) is critical for proliferation, but its precise role in cell cycle progression remains unclear. In this study, we examined the mechanisms involved by assessing the effects of Fe chelators on the expression of molecules that play key roles in this process. In initial studies, gene arrays were used to assess gene expression after incubating cells with 2 Fe chelators, namely, desferrioxamine (DFO) and 2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311), or the DNA-damaging agent, actinomycin D. From the genes assessed, only the N-myc downstream-regulated gene 1 (Ndrg1) was specifically up-regulated by Fe chelation. Although the function of Ndrg1 is unclear, previous studies showed it markedly slows tumor growth and acts as a potent metastasis suppressor. Incubation of cells with chelators markedly increased Ndrg1 mRNA and protein expression, but this was not found with their Fe complexes or when the Fe-binding site had been inactivated. Increased Ndrg1 expression following Fe chelation was related to the permeability and antiproliferative activity of chelators and could be reversed by Fe repletion. Moreover, Ndrg1 up-regulation after chelation occurred at the transcriptional level and was mediated by hypoxia inducible factor-1α (HIF-1α)-dependent and -independent mechanisms. Our investigation suggests Ndrg1 is a novel link between Fe metabolism and the control of proliferation.
Introduction
Iron (Fe) is fundamental for many important cellular processes, such as DNA synthesis (for reviews see Andrews1 and Le and Richardson2 ). Studies using Fe chelators such as desferrioxamine (DFO), have shown that Fe deprivation results in G1/S arrest and apoptosis.2-11 Tumor cells are far more sensitive than normal cells to Fe depletion, probably because of their increased Fe requirements that are reflected by marked expression of transferrin receptor 1 (TfR1).12-15 Furthermore, neoplastic cells express higher levels of the Fe-containing enzyme, ribonucleotide reductase (RR), which is a critical rate-limiting step for DNA synthesis and is an important molecular target of chelators.10,11,16-22
A wide variety of in vitro3,5,8,10,22-28 and in vivo7,29,30 investigations as well as clinical trials4,6,31,32 have shown that DFO and other Fe chelators are effective antitumor agents.11,16,33 However, the use of DFO as an anticancer agent is limited by its modest antiproliferative activity that is related to its poor membrane permeability and short half-life.23,33 In contrast, novel aroylhydrazone chelators, such as 2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311), demonstrated far greater antiproliferative activity and Fe chelation efficacy than DFO and show promise as an antitumor agent.8-10,23-25 This chelator belongs to the pyridoxal isonicotinoyl hydrazone (PIH) group, which shows high affinity and selectivity for Fe(III) that is comparable to DFO (for reviews see Lovejoy and Ridardson,16 Richardson33 , and Buss et al34 ).
The G1/S arrest after Fe chelation was thought to be due to the ability of Fe chelators to inhibit RR that prevents the production of deoxyribonucleotides for DNA synthesis.11,17 However, the effect of Fe chelators is complex, with these agents affecting numerous molecular targets in addition to RR.8,10,22,25,35-40 Despite considerable advances in understanding Fe metabolism,1 to date, the molecular mechanisms involved in the G1/S arrest41,42 observed after Fe chelation remain poorly understood. Hence, it is important to examine this, considering the current interest in the use of Fe chelators as antitumor agents.3-11,16,22-33,43-47 To further investigate the effects of chelators on the expression of molecules involved in cell-cycle progression, gene array studies were initiated, comparing Fe chelators (DFO and 311) to the well-characterized DNA-damaging agent, actinomycin D (Act D). These experiments showed that of all genes assessed, the N-myc downstream-regulated gene 1 (Ndrg1), was markedly up-regulated at the mRNA level by both Fe chelators but not Act D. Considering our interest in the role of Fe in gene expression and proliferation,8-10,23-25,35-38 it was important to examine this molecule further because its expression has been shown to markedly inhibit tumor cell growth and metastasis.48-57
Ndrg1 has been identified under a variety of conditions, and the protein has previously been termed RTP, Drg1, TDD5, Cap43, Rit42, and Ndr1.48-58 In this study, it will be referred to as Ndrg1, because this terminology has been agreed to by a recent consortium.58 Ndrg1 has been shown to be down-regulated in tumor cell lines as well as in breast, prostate, and invasive colorectal cancers.49,55 In contrast, the expression of Ndrg1 was increased when human colon cancer cells were induced to differentiate.49 Further studies suggested that Ndrg1 was induced by Ni(II) and hypoxia by way of hypoxia inducible factor-1α (HIF-1α).52-54
Significantly, Ndrg1 up-regulation decreased tumor growth rate, and its expression was increased by DNA-damaging agents such as Act D.55 Moreover, induction of Ndrg1 was shown to be dependent on p53 by using cell lines transfected with this tumor suppressor.55 Considering these findings, it was suggested Ndrg1 has a growth inhibitory role, and its down-regulation contributes to the malignant phenotype.55 Recent studies in prostate cancer demonstrated Ndrg1 protein levels were inversely correlated with patient survival.56 Further, Ndrg1 expression prevented metastatic prostate cancer cell colonization in the lungs of mice, indicating that this gene encoded a metastasis suppressor protein.56 The fact Ndrg1 is down-regulated in neuroblastomas with amplified N-myc could translate to aggressive growth and poor clinical outcome.58
In the current study, we investigated the mechanism(s) involved in up-regulating Ndrg1 expression after Fe depletion by using specific Fe chelators.
Materials and methods
Cell treatments
The chelators, 311, di-2-pyridylketone-4,4,-dimethyl-3-thiosemicarbazone (dp44mT), and di-2-pyridylketone 2-methyl-3-thiosemicarbazone (dp2mT), were prepared as described.24,59,60 DFO was from Novartis (Basel, Switzerland), L1 (deferiprone; was from Roger Dean, Heart Research Institute, Sydney, Australia). All other chemicals were purchased from Sigma (St Louis, MO).
Cell culture
All cell lines were obtained from the American Type Culture Collection (ATCC; Rockville, MD) and cultured as described previously.13,14 Fibroblasts from homozygous HIF-1α knock-out (HIF-1α-KO) mice were obtained from Dr R. Johnson (University of California, San Diego).
The effect of the chelators on cellular proliferation was examined by using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium) assay.24
RNA isolation and gene expression analysis using array membranes
Total RNA was isolated from cells by using Total RNA Isolation Reagent from Advanced Biotechnologies Ltd (Surrey, United Kingdom).35 In gene array studies, RNA was used to generate cDNA with the True Labeling-Reverse Transcriptase kit (Superarray, Frederick, MD). This procedure resulted in the generation of specific radioactive cDNA probes for hybridization onto GEArray Q series membranes (Superarray). The gene arrays used in this study represented 192 genes that are involved with cell cycle progression and the p53 tumor suppressor pathway.
Semiquantitative RT-PCR
Isolated RNA was used to perform reverse-transcriptase polymerase (RT-PCR). Briefly, 2.5 μg RNA was incubated with gene-specific oligonucleotides (Tables 1 and 2) in a reaction mixture containing 1 × reverse transcriptase (RT) reaction buffer, 312 μM dNTP (deoxynucleoside triphosphate), 25 units RNAsin, and 200 units M-MLV (Moloney murine leukemia virus) RT (Promega, Madison, WI) at 42°C for 1 hour. Following cDNA synthesis, M-MLV was heat-inactivated at 70°C for 10 minutes. Between 0.5 and 2 μL cDNA was used for PCR analysis with a reaction mixture consisting of 10 pmol of each oligonucleotide pair (Tables 1 and 2), 1 × PCR reaction buffer, 200 μM dNTP, and 2.5 units Taq Polymerase (Promega). This reaction mixture containing the cDNA was initially denatured at 94°C for 2 minutes and subjected to 20 to 25 PCR cycles that included a 94°C denaturation step for 30 seconds, 60°C annealing step for 30 seconds, a 72°C extension step for 30 to 60 seconds, with a final extension time of 7 minutes. As an internal control, β-actin was amplified from the same cDNA samples.
Sequences and Accession number of primers used to amplify human transcripts by RT-PCR
Gene . | Oligonucleotide sequences . | GenBank Accession no. . |
|---|---|---|
| Ndrg1 | 5′-CCCTCGCGTTAGGCAGGTGA-3′ FWD | NM_006096 |
| 5′-AGGGGTACATGTACCCTGCG-3′ REV | ||
| TfR1 | 5′-TCAGGTCAAAGACAGCGCTCAAAACTC-3′ FWD | X01060 |
| 5′-AGTCTCCTTCCATATTCCCAAACAGCTTTT-3′ REV | ||
| VEGF-1 | 5′-AGTGCTCCAGCGCGCGCGCTCCC-3′ FWD | AF022375 |
| 5′-CTCTATCTTTCTTTGGTCTGCATTCACATT-3′ REV | ||
| WAF1 | 5′-CTCAGAGGAGGCGCCATGTC-3′ FWD | U03106 |
| 5′-GAGTGGTAGAAATCTGTCATGCTGG-3′ REV | ||
| β-actin | 5′-CCCGCCGCCAGCTCACCATGG-3′ FWD | X00351 |
| 5′-AAGGTCTCAAACATGATCTGGGTC-3′ REV |
Gene . | Oligonucleotide sequences . | GenBank Accession no. . |
|---|---|---|
| Ndrg1 | 5′-CCCTCGCGTTAGGCAGGTGA-3′ FWD | NM_006096 |
| 5′-AGGGGTACATGTACCCTGCG-3′ REV | ||
| TfR1 | 5′-TCAGGTCAAAGACAGCGCTCAAAACTC-3′ FWD | X01060 |
| 5′-AGTCTCCTTCCATATTCCCAAACAGCTTTT-3′ REV | ||
| VEGF-1 | 5′-AGTGCTCCAGCGCGCGCGCTCCC-3′ FWD | AF022375 |
| 5′-CTCTATCTTTCTTTGGTCTGCATTCACATT-3′ REV | ||
| WAF1 | 5′-CTCAGAGGAGGCGCCATGTC-3′ FWD | U03106 |
| 5′-GAGTGGTAGAAATCTGTCATGCTGG-3′ REV | ||
| β-actin | 5′-CCCGCCGCCAGCTCACCATGG-3′ FWD | X00351 |
| 5′-AAGGTCTCAAACATGATCTGGGTC-3′ REV |
FWD indicates forward; REV, reverse.
Sequences and Accession number of primers used to amplify mouse mRNA by RT-PCR
Gene . | Oligonucleotide sequences . | GenBank Accession no. . |
|---|---|---|
| Ndrg1 | 5′-TGCTTGCTCATTAGGTGTGTGATAGC-3′ FWD | NM_010884 |
| 5′-CCATCCTGAGATCTTAGAGGCAGC-3′ REV | ||
| HIF-1α | 5′-CTGGATGCCGGTGGTCTAGACAGT-3′ FWD | NM_010431 |
| 5′-CGAGAAGAAAAAGATGAGTTCTGAACGTCG-3′ REV | ||
| VEGF-1 | 5′-CCATGCCAAGTGGTCCCAG-3′ FWD | M95200.1 |
| 5′-GTCTTTCTTTGGTCTGCATTCACAT-3′ REV | ||
| β-actin | 5′-CTTCCTTCTTGGGTATGGAATCCTGT-3′ FWD | NM_00739 |
| 5′-CTCAGGAGGAGCAATGATCTTGATCTTC-3′ REV |
Gene . | Oligonucleotide sequences . | GenBank Accession no. . |
|---|---|---|
| Ndrg1 | 5′-TGCTTGCTCATTAGGTGTGTGATAGC-3′ FWD | NM_010884 |
| 5′-CCATCCTGAGATCTTAGAGGCAGC-3′ REV | ||
| HIF-1α | 5′-CTGGATGCCGGTGGTCTAGACAGT-3′ FWD | NM_010431 |
| 5′-CGAGAAGAAAAAGATGAGTTCTGAACGTCG-3′ REV | ||
| VEGF-1 | 5′-CCATGCCAAGTGGTCCCAG-3′ FWD | M95200.1 |
| 5′-GTCTTTCTTTGGTCTGCATTCACAT-3′ REV | ||
| β-actin | 5′-CTTCCTTCTTGGGTATGGAATCCTGT-3′ FWD | NM_00739 |
| 5′-CTCAGGAGGAGCAATGATCTTGATCTTC-3′ REV |
FWD indicates forward; REV, reverse.
Promoter and secondary structure analysis
Western blot analysis
Western blot analysis was performed as described previously.36 The mouse monoclonal antihuman β-actin antibody was from Sigma (clone AC-15) and used at a 1:5000 dilution. The mouse antihuman TfR1 antibody was from Zymed Laboratories (San Francisco, CA) and used at 0.16 μg/mL. Antibodies against Ndrg1 and HIF-1α were purchased from Santa Cruz (Santa Cruz, CA) and used at 0.4 μg/mL. The secondary antibodies used were antirabbit antibody (1:8000 dilution; Sigma) or antimouse antibody (1:8000 dilution; Sigma) conjugated with horseradish peroxidase.
Statistical analysis
Experimental data were compared by using Student t test. Results were considered statistically significant at a P value less than .05.
Results
Ndrg1 mRNA is up-regulated by chelators but not the DNA-damaging agent, actinomycin D
To further understand the role of Fe in cell cycle progression, we initially used gene arrays to assess the effects on gene expression of 2 well-characterized Fe chelators, DFO (250 μM) and 311 (25 μM),8-11,16,22-25,35-38 compared with the DNA-damaging agent, Act D (5 nM).63,64 At the low concentration used, Act D (5 nM) induces DNA damage without significantly inhibiting transcription.63,64 These studies were performed to dissect out the potential ability of Fe chelators to induce DNA damage (eg, through inhibition of RR),10,18,19 compared with their effect on other Fe-requiring metabolic pathways. We used a 10-fold greater concentration of DFO than 311 because of the lower membrane permeability and Fe chelation efficacy of the former ligand.23-25,35
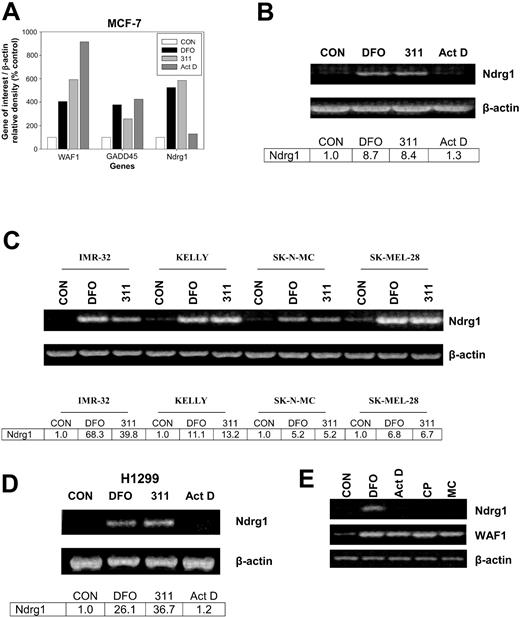
The densitometric analysis in Figure 1A illustrates the expression of 3 genes of interest from the array, namely WAF1, GADD45, and Ndrg1. Incubation of MCF-7 cells for 24 hours with the chelators, DFO (250 μM) or 311 (25 μM), increased the expression of WAF1, GADD45, and Ndrg1 to between 280% and 595% of the control (Figure 1A). The effect of Fe chelators at increasing WAF1 and GADD45 mRNA expression was described previously by our laboratory,9,35-38 and these genes were included as positive controls. In contrast to the Fe chelators, Act D increased GADD45 and WAF1 expression to 400% and 910% of the control, respectively, but had no marked effect on the expression of Ndrg1 (Figure 1A). The up-regulation of GADD45 and WAF1 mRNA by Act D is a well-described response to DNA damage and served as another positive control.38 The fact that Act D at 5 nM markedly increased WAF1 and GADD45 expression (Figure 1A) also indicates that this Act D concentration did not inhibit transcription, because these genes respond to DNA damage through the p53 transcription factor pathway.64 Indeed, it takes at least 0.4 μM Act D to inhibit WAF1 transcription.38 The incubation of cells with other DNA-damaging agents such as cisplatin (CP; 20 μM) or mitomycin C (MC; 30 μM) also did not increase Ndrg1 mRNA levels despite an increase in WAF1 expression (Figure 1E) which is known to respond to DNA damage.37,38,64 The interpretation of these data suggested that the induction of Ndrg1 expression by Fe chelation was by way of a mechanism that did not involve DNA damage.
The effect of Fe chelators and DNA-damaging agents on Ndrg1 expression. (A) Densitometric analysis of gene array data demonstrating marked up-regulation of Ndrg1 mRNA expression after incubation of MCF-7 cells with the chelators desferrioxamine (DFO) and 2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311) but not the DNA-damaging agent actinomycin D (Act D). (B) Semi-quantitative RT-PCR demonstrates that Ndrg1 mRNA is markedly up-regulated after incubation of MCF-7 cells with DFO or 311. (C) Chelators up-regulate the expression of Ndrg1 mRNA in multiple cell lines. (D) The p53-independent induction of Ndrg1 mRNA following Fe chelation with DFO or 311 in the H1299 p53 mutant cell line. (E) The chelator DFO, but not Act D, cisplatin (CP), or mitomycin C (MC), increases Ndrg1 mRNA expression. Cells were incubated for 24 hours at 37°C with either 311 (25 μM), DFO (250 μM), Act D (5 nM), CP (20 μM), or MC (30 μM), and the RNA was isolated and then used for (A) gene array analysis using GEArray Q series membranes (Superarray), or (B-E) semiquantitative RT-PCR analysis (see “Materials and methods” for details). (A-D) Densitometry was performed and gene expression then calculated relative to the β-actin control. Results in panel A show a typical experiment from 2 performed, and panels B to E show a typical experiment from 3 performed.
The effect of Fe chelators and DNA-damaging agents on Ndrg1 expression. (A) Densitometric analysis of gene array data demonstrating marked up-regulation of Ndrg1 mRNA expression after incubation of MCF-7 cells with the chelators desferrioxamine (DFO) and 2-hydroxy-1-naphthylaldehyde isonicotinoyl hydrazone (311) but not the DNA-damaging agent actinomycin D (Act D). (B) Semi-quantitative RT-PCR demonstrates that Ndrg1 mRNA is markedly up-regulated after incubation of MCF-7 cells with DFO or 311. (C) Chelators up-regulate the expression of Ndrg1 mRNA in multiple cell lines. (D) The p53-independent induction of Ndrg1 mRNA following Fe chelation with DFO or 311 in the H1299 p53 mutant cell line. (E) The chelator DFO, but not Act D, cisplatin (CP), or mitomycin C (MC), increases Ndrg1 mRNA expression. Cells were incubated for 24 hours at 37°C with either 311 (25 μM), DFO (250 μM), Act D (5 nM), CP (20 μM), or MC (30 μM), and the RNA was isolated and then used for (A) gene array analysis using GEArray Q series membranes (Superarray), or (B-E) semiquantitative RT-PCR analysis (see “Materials and methods” for details). (A-D) Densitometry was performed and gene expression then calculated relative to the β-actin control. Results in panel A show a typical experiment from 2 performed, and panels B to E show a typical experiment from 3 performed.
To validate the gene array results found with Ndrg1 (Figure 1A), semiquantitative RT-PCR was performed (Figure 1B). As found using the gene arrays, both DFO and 311 significantly (P < .00001) increased Ndrg1 mRNA expression to 8.7- and 8.4-fold of the control, respectively, whereas Act D had no effect (P > .05) (Figure 1B).
Effect of chelators at increasing Ndrg1 expression is found in many cell types and is independent of p53 status
To determine whether Ndrg1 up-regulation by DFO (250 μM) and 311 (25 μM) were not restricted to MCF-7 cells, we examined the effects of Fe chelators on Ndrg1 mRNA expression in a range of tumor cell lines (Figure 1C). Incubation of IMR-32 neuroblastoma, Kelly neuroblastoma, SK-N-MC neuroepithelioma, and SK-Mel-28 melanoma cell lines with DFO (250 μM) or 311 (25 μM) for 24 hours resulted in robust up-regulation of Ndrg1 mRNA levels compared with the control (Figure 1C).
A previous study using cells stably transfected with p53 indicated Ndrg1 up-regulation could be by way of this transcription factor.55 Considering DFO and 311 up-regulate the p53 tumor suppressor protein,37,38,40,64 we examined whether increased Ndrg1 mRNA levels were dependent on this molecule. To test this hypothesis, we incubated p53-deficient H1299 lung carcinoma cells65 with DFO (250 μM) or 311 (25 μM) for 24 hours. As found for all other cell types (Figure 1B-C), incubation with DFO or 311 resulted in marked Ndrg1 mRNA expression compared with the control (Figure 1D). This result demonstrated that the increase in Ndrg1 mRNA after Fe deprivation was mediated through a p53-independent mechanism. Our observations were supported by studies using SK-N-MC cells that possess mutant p53,66 whereby Ndrg1 mRNA levels were elevated after incubation with 311 or DFO (Figure 1C).
Up-regulation of Ndrg1 mRNA and protein levels after iron chelation is rapid
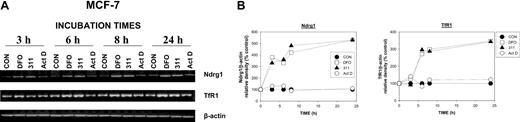
To examine the kinetics of Ndrg1 induction, we incubated MCF-7 cells for 3 to 24 hours with DFO (250 μM) or 311 (25 μM) and compared their effects with Act D (5 nM; Figure 2A-B). We also assessed TfR1 mRNA expression over the same times as a positive control for Fe depletion.33,35
Up-regulation of Ndrg1 mRNA levels after Fe chelation with DFO and 311 is rapid but is not induced by the DNA-damaging agent Act D. (A) MCF-7 cells were incubated with DFO (250 μM), 311 (25 μM), or Act D (5 nM) for 3 to 24 hours at 37°C. (B) The expression of Ndrg1 and TfR1 mRNA was then assessed by semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results show a typical experiment from 3 performed.
Up-regulation of Ndrg1 mRNA levels after Fe chelation with DFO and 311 is rapid but is not induced by the DNA-damaging agent Act D. (A) MCF-7 cells were incubated with DFO (250 μM), 311 (25 μM), or Act D (5 nM) for 3 to 24 hours at 37°C. (B) The expression of Ndrg1 and TfR1 mRNA was then assessed by semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results show a typical experiment from 3 performed.
The incubation of MCF-7 cells for up to 24 hours with Act D (5 nM) had no significant effect on Ndrg1 or TfR1 mRNA expression when compared with the control (Figure 2A-B). Our results are in contrast to previous studies by others using MCF-7 cells, whereby marked up-regulation of Ndrg1 mRNA was observed after 12 to 48 hours of incubation with 9 nM Act D.55
The induction and up-regulation of Ndrg1 mRNA after Fe chelation was rapid compared with the relevant control at each time point and occurred after a 3-hour incubation with both DFO or 311 (Figure 2A-B). This increase in Ndrg1 mRNA after incubation with Fe chelators continued up until 8 hours and plateaued at 24 hours (Figure 2A-B). Examining the kinetics of TfR1 expression, we demonstrate that the up-regulation of TfR1 mRNA by DFO or 311 was not as rapid as Ndrg1 (Figure 2A-B). In fact, increased TfR1 mRNA expression was only evident after 6 hours of incubation with the Fe chelators and became more marked as the incubation progressed to 24 hours when compared with the control (Figure 2A-B). Incubation of MCF-7 breast cancer cells with Act D did not result in any significant change in TfR1 mRNA expression when compared with the untreated controls. Collectively, our results demonstrate that the up-regulation of Ndrg1 after Fe chelation was rapid, occurring 3 hours earlier than the TfR1.
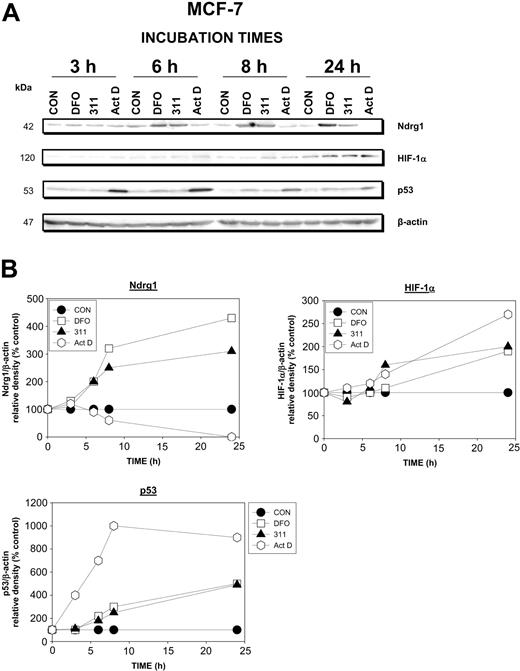
The rapid up-regulation of Ndrg1 mRNA (Figure 2A-B) resulted in an increase of Ndrg1 protein levels compared with the control after a 6-hour incubation with chelators and appeared to plateau after 8 to 24 hours (Figure 3A-B). As found at the mRNA level (Figure 2A), Act D did not up-regulate Ndrg1 protein levels, and after a 24-hour incubation ablated its expression (Figure 3A-B). Considering the possible role of HIF-1α in up-regulating Ndrg1,52-54 it is of interest that increased HIF-1α protein levels in the presence of the chelators were only evident after 24 hours (Figure 3A-B).
Ndrg1 protein levels were up-regulated after Fe chelation with DFO and 311 but was not induced by the DNA-damaging agent Act D. (A) MCF-7 cells were incubated with DFO (250 μM), 311 (25 μM), or Act D (5 nM) for 3 to 24 hours at 37°C. Protein was then isolated and Western blot analysis was performed (see “Materials and methods” for details). (B) Densitometry was performed, and protein expression was then calculated relative to the β-actin control. Results show a typical experiment from 3 performed.
Ndrg1 protein levels were up-regulated after Fe chelation with DFO and 311 but was not induced by the DNA-damaging agent Act D. (A) MCF-7 cells were incubated with DFO (250 μM), 311 (25 μM), or Act D (5 nM) for 3 to 24 hours at 37°C. Protein was then isolated and Western blot analysis was performed (see “Materials and methods” for details). (B) Densitometry was performed, and protein expression was then calculated relative to the β-actin control. Results show a typical experiment from 3 performed.
To further assess the role of the p53 protein in Ndrg1 upregulation, we examined the kinetics of its expression following Fe chelation and DNA damage (Figure 3A-B). After a 6-, 8-, and 24-hour incubation, DFO and 311 increased p53 protein levels 1.5- to 4-fold compared with the control, and this was somewhat similar to the effects of these agents on Ndrg1 (Figure 3A-B). In contrast, Act D markedly increased p53 protein levels at all incubation times (Figure 3A-B), and this did not correlate with the effect of this agent on Ndrg1 protein levels. Indeed, the Ndrg1 levels were markedly decreased in the presence of Act D (Figure 3A-B). Together with the results examining cells with mutant p53 (Figure 1D), these studies demonstrate that p53 and DNA damage were not involved in up-regulating Ndrg1 expression in MCF-7 cells.
Different structural classes of iron chelators induce Ndrg1 mRNA expression, whereas Fe complexes of DFO and 311 have no effect
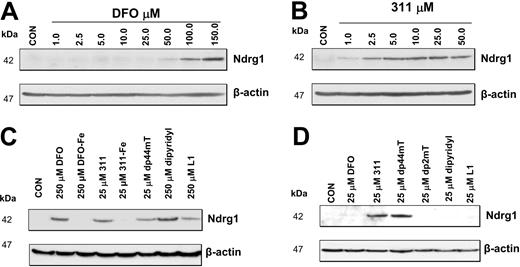
The increased expression of Ndrg1 mRNA (Figure 2A-B) and protein (Figure 3A-B) were observed with 2 structurally different high-affinity Fe chelators, namely DFO and 311.24,25,35-38 Hence, the ability for these ligands to induce Ndrg1 expression may be related to their ability to permeate plasma membranes to directly induce intracellular Fe depletion.8,10,24,35,67 Indeed, we have previously shown that 311 is markedly more permeable than DFO, with the former being far more effective at inducing 59 Fe release and preventing 59 Fe uptake from 59Fe-Tf.10,23-25,35,38 To assess the role of chelator permeability in Ndrg1 induction, we directly compared the ability of DFO or 311 to up-regulate Ndrg1 protein levels (Figure 4A-B). In these experiments, cells were incubated for 24 hours with control media or media containing DFO (1-150 μM) or 311 (1-50 μM). Our results showed that Ndrg1 protein expression was increased only at DFO concentrations more than 50 μM (Figure 4A). In contrast, 311 clearly increased Ndrg1 protein levels at only 2.5 μM with maximal induction occurring at 10 μM (Figure 4B). These results demonstrate that 311 was more efficient than DFO at inducing Ndrg1 protein expression. This observation correlates with the far lower membrane permeability of hydrophilic DFO compared with the highly lipid-soluble and membrane-permeable chelator, 311.10,23-25,35,38
The effect of Fe chelators or Fe complexes on Ndrg1 protein levels in MCF-7 cells. (A) High concentrations of DFO (100-150 μM) were required to markedly increase Ndrg1 protein levels. (B) Concentrations of 311 of only 2.5 μM or more were required to increase Ndrg1 protein levels. (C) Membrane-permeable chelators such as DFO, 311, Dp44mT, and L1 increase Ndrg1 protein levels. (D) Only Fe chelators that show high-antiproliferative activity and Fe chelation efficacy increase Ndrg1 protein levels when examined at 25 μM. MCF-7 cells were incubated for 24 hours at 37°C with the chelators or Fe complexes, and the expression of Ndrg1 and β-actin protein levels was assessed by Western blotting (see “Materials and methods” for details). Results show a typical experiment from 3 performed.
The effect of Fe chelators or Fe complexes on Ndrg1 protein levels in MCF-7 cells. (A) High concentrations of DFO (100-150 μM) were required to markedly increase Ndrg1 protein levels. (B) Concentrations of 311 of only 2.5 μM or more were required to increase Ndrg1 protein levels. (C) Membrane-permeable chelators such as DFO, 311, Dp44mT, and L1 increase Ndrg1 protein levels. (D) Only Fe chelators that show high-antiproliferative activity and Fe chelation efficacy increase Ndrg1 protein levels when examined at 25 μM. MCF-7 cells were incubated for 24 hours at 37°C with the chelators or Fe complexes, and the expression of Ndrg1 and β-actin protein levels was assessed by Western blotting (see “Materials and methods” for details). Results show a typical experiment from 3 performed.
Permeability of the chelator to induce intracellular Fe depletion appears essential to induce Ndrg1 expression. To evaluate if induction of Ndrg1 protein levels is related to the antiproliferative activity of the chelators we compared the permeable Fe chelators, DFO, L1, dipyridyl, 311, and dp44mT, which show low- or high-antiproliferative activity.8,9,24,25,60 The chelators with low-antiproliferative activity against MCF-7 cells included DFO (IC50 [concentration that inhibits proliferation by 50%] = 20 μM), L1 (deferiprone; IC50 = 180 μM), and dipyridyl (IC50 > 250 μM) and were examined at 25 and 250 μM (Figure 4C-D). The chelators, 311 and dp44mT, that showed high-antitumor efficacy against MCF-7 cells (IC50 = 0.6 μM and 0.1 μM, respectively) were assessed at 25 μM (Figure 4C-D). All permeable Fe chelators, including DFO, dipyridyl, and L1, that have low-antiproliferative activity were able to up-regulate Ndrg1 protein at 250 μM (Figure 4C) but not 25 μM (Figure 4D) when compared with the relevant control. To demonstrate that the increase in Ndrg1 expression was due to effective Fe depletion, we incubated cells with the Fe complexes of DFO or 311 (Figure 4C). Our results indicate that both Fe complexes were unable to increase Ndrg1 expression, suggesting Fe deprivation was required (Figure 4C). As an appropriate negative control for the highly active ligand, dp44mT, we incubated cells with its structural analog, dp2mT, which cannot effectively bind Fe.60 As shown in Figure 4D, dp2mT had no effect, indicating the importance of Fe chelation in up-regulating Ndrg1.
Increase in Ndrg1 expression following iron deprivation can be reversed by incubation with iron(III) salts
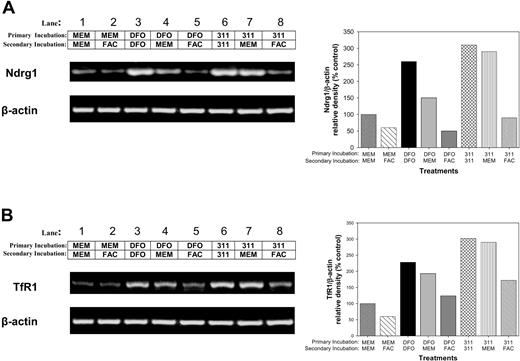
Further investigations were undertaken to determine whether Ndrg1 mRNA up-regulation after Fe chelation could be subsequently down-regulated by reincubating cells with Fe (Figure 5A). In these experiments, MCF-7 cells were incubated with control medium (MEM), DFO (250 μM), or 311 (25 μM) for 10 hours (primary incubation; Figure 5). This medium was then removed, and the cells were secondarily reincubated for 14 hours with either MEM, DFO (250 μM), 311 (25 μM), or the Fe donor, ferric ammonium citrate (FAC; 100 μg/mL). The mRNA levels of Ndrg1 (Figure 5A) and TfR1 (Figure 5B) were then assessed.
The increase in Ndrg1 and TfR1 mRNA following Fe deprivation with DFO and 311 can be reversed with the Fe donor, ferric ammonium citrate. MCF-7 cells were incubated with control medium (minimum essential medium; MEM), DFO (250 μM), or 311 (25 μM) for 14 hours at 37°C (primary incubation). This medium was then removed, and the cells were then reincubated for 14 hours at 37°C with either control media (MEM) or media containing either the Fe donor ferric ammonium citrate (FAC; 100 μg/mL), DFO (250 μM), or 311 (25 μM) (secondary incubation). The expression of (A) Ndrg1 and (B) TfR1 mRNA levels (positive control) were then assessed by using semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results show a typical experiment from 3 performed.
The increase in Ndrg1 and TfR1 mRNA following Fe deprivation with DFO and 311 can be reversed with the Fe donor, ferric ammonium citrate. MCF-7 cells were incubated with control medium (minimum essential medium; MEM), DFO (250 μM), or 311 (25 μM) for 14 hours at 37°C (primary incubation). This medium was then removed, and the cells were then reincubated for 14 hours at 37°C with either control media (MEM) or media containing either the Fe donor ferric ammonium citrate (FAC; 100 μg/mL), DFO (250 μM), or 311 (25 μM) (secondary incubation). The expression of (A) Ndrg1 and (B) TfR1 mRNA levels (positive control) were then assessed by using semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results show a typical experiment from 3 performed.
Reincubation of MCF-7 cells with FAC after primary incubation with MEM slightly decreased Ndrg1 mRNA levels (Figure 5A; lane 2) compared with cells in which the primary and secondary incubations were in MEM alone (Figure 5A; lane 1). Both primary and secondary incubations in the presence of DFO markedly and significantly (P < .001) up-regulated Ndrg1 expression (Figure 5A; lane 3) compared with the control (Figure 5A; lane 1). When cells primarily incubated with DFO were then secondarily incubated with MEM (Figure 5A; lane 4) or FAC (Figure 5A; lane 5), there was a decrease in Ndrg1 expression when compared with cells incubated with DFO alone (Figure 5A; lane 3). However, the down-regulation of Ndrg1 mRNA levels was more marked in cells reincubated with FAC (P < .001) than with MEM (P < .05) (Figure 5A; compare lane 5 with lane 4). Conducting similar experiments with 311 revealed that only FAC was able to significantly (P < .001) reverse Ndrg1 up-regulation (Figure 5A; lane 8) when compared with cells treated with 311 alone (Figure 5A; lane 6). The differing MEM reincubation results following Fe deprivation by DFO or 311 may be related to the efficiency of these ligands at inducing Fe depletion.24,25,35 These data were similar to those found for the TfR1 under the same conditions (Figure 5B), and their interpretation shows that the Ndrg1 expression, like the TfR1, was Fe responsive. In contrast to FAC (100 μg/mL) or FeCl3 (300 μM), reincubation with CuCl2 (300 μM) or ZnCl2 (300 μM) did not reverse induction of Ndrg1 mRNA expression by 311 (data not shown).
Ndrg1 is transcriptionally up-regulated after incubation with DFO or 311
Our above-mentioned repletion studies demonstrate that like the TfR1,1,2,67 the regulation of Ndrg1 was dependent on intracellular Fe levels. Considering that TfR1 up-regulation is dependent on the 3′ binding of iron regulatory proteins (IRPs) to iron responsive elements (IREs) to increase mRNA stability,1,2,67 we assessed the 3′ and 5′ untranslated regions (UTRs) of Ndrg1 mRNA for IREs. The secondary structure for these sequences was predicted by using RNA structure 3.71,62 but they did not contain the consensus IRE.
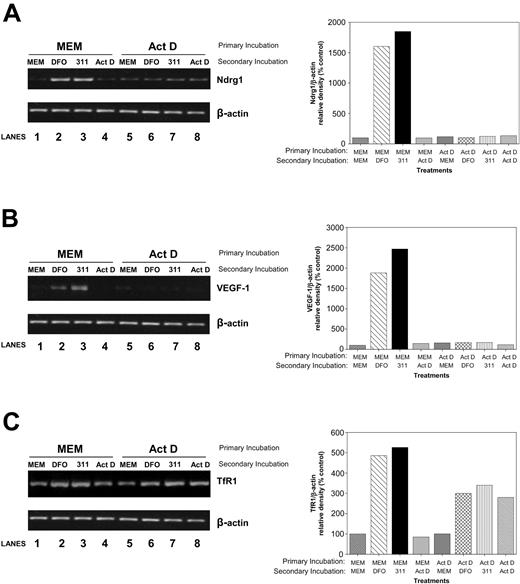
To determine whether Ndrg1 is transcriptionally regulated by Fe depletion, in the primary incubation, we incubated cells with control media (MEM) or medium containing the transcription inhibitor Act D (4 μM) for 2 hours. It should be noted that at low Act D concentrations (ie, 5 nM), such as those used in Figures 1A-B,D; 2; and 3, the agent acts as a well-described DNA-damaging agent but does not appreciably prevent transcription.64,68 In contrast, at much higher concentrations (ie, 4 μM), Act D markedly inhibits transcription.69 After incubation with Act D (4 μM), cells were reincubated with DFO, 311, or Act D for an additional 6 hours. As a positive control for transcriptional regulation, we also assessed the mRNA expression of vascular endothelial growth factor-1 (VEGF-1) gene.70-72 Primary incubation in MEM followed by a secondary incubation with DFO or 311, but not Act D, resulted in a marked accumulation of Ndrg1 mRNA (Figure 6A, lanes 2-3) when compared with the untreated control (Figure 6A, lane 1). In contrast, primary incubation of cells with Act D followed by a secondary incubation with the chelator totally inhibited Ndrg1 up-regulation (Figure 6A, lanes 6-7) when compared with the control (Figure 6A, lane 5). Similar results were found for VEGF-1 expression (Figure 6B). These results suggest Ndrg1 expression is transcriptionally regulated by cellular Fe levels. In contrast to Ndrg1 and VEGF-1, TfR1 up-regulation was not totally ablated by Act D, confirming at least some posttranscriptional regulation by Fe in this cell type (Figure 6C).
Studies using the transcriptional inhibitor, Act D, demonstrate that the induction of Ndrg1 is transcriptionally regulated following Fe chelation. MCF-7 breast cancer cells were preincubated for 2 hours at 37°C with either control media or media containing the transcriptional inhibitor, Act D (4 μM). This media were then subsequently removed and replaced with control media (MEM) or media containing either DFO (250 μM), 311 (25 μM), or Act D (4 μM) for an additional 6 hours at 37°C. The mRNA levels for Ndrg1 (A), VEGF-1 (B), and TfR1 (C) were then assessed by using semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results are a typical experiment from 3 performed.
Studies using the transcriptional inhibitor, Act D, demonstrate that the induction of Ndrg1 is transcriptionally regulated following Fe chelation. MCF-7 breast cancer cells were preincubated for 2 hours at 37°C with either control media or media containing the transcriptional inhibitor, Act D (4 μM). This media were then subsequently removed and replaced with control media (MEM) or media containing either DFO (250 μM), 311 (25 μM), or Act D (4 μM) for an additional 6 hours at 37°C. The mRNA levels for Ndrg1 (A), VEGF-1 (B), and TfR1 (C) were then assessed by using semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results are a typical experiment from 3 performed.
Up-regulation of Ndrg1 after iron chelation occurs by HIF-1α-dependent and -independent mechanisms
The above-mentioned experiments indicate that a transcriptional mechanism could be involved in the ability of chelators to up-regulate Ndrg1. Considering this indication, the HIF-1α transcription factor was suggested to be involved in Ndrg1 upregulation after incubation under hypoxic conditions.52 However, Masuda et al73 have suggested that no HIF-response element (HRE) exists in the Ndrg1 promoter up to -1122 bp. A similar analysis was performed during this study by using the promoter analysis program, Genematix Suite 3.0, which identified putative HREs upstream of the promoter at -1376 bp and -7503 bp. This finding suggests that HIF-1α may act as a transcriptional enhancer for efficient transactivation of Ndrg1. Interestingly, an enhancer role for HIF-1α has been reported for the erythropoietin gene following hypoxia.74
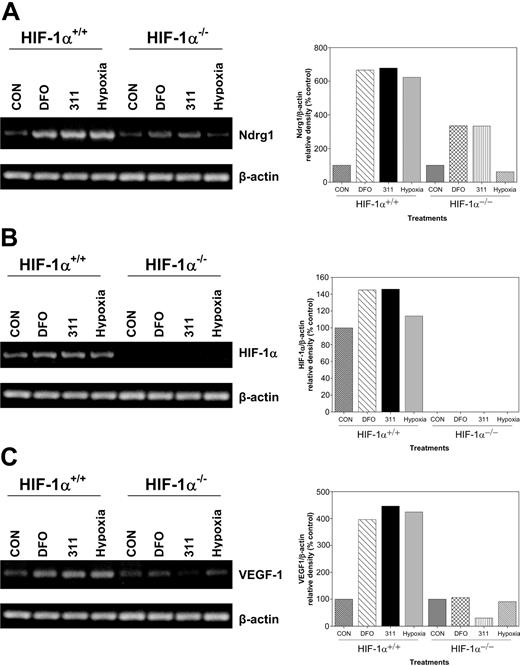
To directly examine the role of HIF-1α in the up-regulation of Ndrg1 after Fe chelation, we obtained HIF-1α-KO (HIF-1α-/-) murine embryo fibroblasts (MEFs).75,76 Previous studies showed that these cells do not express wild-type HIF-1α protein.75,76 In our experiments, wild-type or HIF-1α-KO MEFs were incubated with DFO (250 μM), 311 (25 μM), or hypoxia (0.5% O2) for 8 hours, and then Ndrg1, HIF-1α, and VEGF-1 mRNA expressions were assessed (Figure 7A-C). Incubation of wild-type (HIF-1α+/+) or HIF-1α-KO (HIF-1α-/-) cells with the Fe chelators increased Ndrg1 mRNA levels. However, the extent of Ndrg1 up-regulation was lower in KO cells than in the wild-type cells (Figure 7A). In contrast, under hypoxic conditions, only wild-type cells were able to increase Ndrg1 expression (Figure 7A).
The up-regulation of Ndrg1 mRNA occurs by a HIF-1α-independent mechanism in cells deficient in the expression of the latter molecule. Wild-type (HIF-1α+/+) and HIF-1α-knockout (KO; HIF-1α-/-) murine embryo fibroblasts (MEFs) were incubated with either DFO (250 μM), 311 (25 μM), or hypoxia (0.5% O2) for 8 hours at 37°C, and their RNA was extracted for the analysis of (A) Ndrg1, (B) HIF-1α, or (C) VEGF-1 by using semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results are a typical experiment from 3 performed.
The up-regulation of Ndrg1 mRNA occurs by a HIF-1α-independent mechanism in cells deficient in the expression of the latter molecule. Wild-type (HIF-1α+/+) and HIF-1α-knockout (KO; HIF-1α-/-) murine embryo fibroblasts (MEFs) were incubated with either DFO (250 μM), 311 (25 μM), or hypoxia (0.5% O2) for 8 hours at 37°C, and their RNA was extracted for the analysis of (A) Ndrg1, (B) HIF-1α, or (C) VEGF-1 by using semiquantitative RT-PCR (described in “Materials and methods”). Densitometry was performed, and gene expression was then calculated relative to the β-actin control. Results are a typical experiment from 3 performed.
To confirm the absence of HIF-1α in the KO cell types, we designed primers that amplified the full-length transcript. Incubation of wild-type MEFs with Fe chelators or hypoxia resulted in a slight increase in HIF-1α mRNA expression when compared with the relevant controls (Figure 7B). In contrast, no full-length HIF-1α transcripts were detected in HIF-1α-KO cells (Figure 7B). We also examined VEGF-1 expression which is a downstream target of HIF-1α after Fe chelation and O2 deprivation2 (Figure 7C). Our results using HIF-1α-KO cells show that VEGF-1 expression did not increase following Fe chelation or hypoxia (Figure 7C). In contrast, a significant (P < .001) increase in VEGF-1 mRNA was observed in wild-type MEFs following Fe chelation or hypoxia when compared with untreated controls (Figure 7C). Thus, unlike VEGF-1 expression, Ndrg1 transactivation occurred by HIF-1α-dependent and -independent mechanisms.
Discussion
Despite the well-known role of Fe in growth and proliferation, little is understood concerning the mechanisms involved in G1/S arrest observed after Fe deprivation.2,11,25,41,42 This lack of information about the role of Fe is surprising, particularly considering the well-characterized roles of p53, the cyclins, and cdks in neoplasia and cell cycle control.2 Understanding the role of Fe in cell cycle progression is vital in terms of designing chelators for the treatment of cancer.16,33
The present work is the first to conclusively demonstrate that Fe depletion results in the marked up-regulation of the metastasis suppressor gene, Ndrg1.55,56,58 This result was observed with DFO and 311 that are known to deplete intracellular Fe pools8-10,23-25 as well as other well-characterized membrane-permeable Fe chelators, eg, dipyridyl and L1.13,33 Interestingly, the ability of chelators to increase Ndrg1 expression was correlated with their antiproliferative activity and Fe chelation efficacy (Figure 4). Indeed, comparing DFO and 311, the latter chelator was far more effective at increasing Ndrg1 protein levels (Figure 4A-B). This finding correlated with the fact that 311 is far more active than DFO at inducing Fe depletion and inhibiting proliferation.24,25,35 Our current studies also showed that DFO and 311 increased TfR1 mRNA levels under the same conditions (Figure 2A-B), indicating these chelators resulted in Fe depletion. Although the chelators could induce Ndrg1 up-regulation (Figure 4A-C), their Fe complexes had no influence (Figure 4C), demonstrating that their effect was likely to be due to their ability to bind Fe. Furthermore, we showed that, although the Fe chelator, dp44mT, effectively up-regulated Ndrg1 expression, its analog dp2mT that does not efficiently bind Fe60 had no effect (Figure 4D). Although a previous study by others showed that DFO increased Ndrg1,54 this increase was not demonstrated to be due to Fe depletion or interpreted to be due to this effect.
A study by Kurdistani et al55 showed Ndrg1 expression was down-regulated in breast and prostate cancer cells compared with normal tissues. The Ndrg1 expression was also shown to be inhibited by the proto-oncogenes, N-myc and c-myc.58 In addition, Ndrg1 overexpression markedly decreased tumor growth and inhibited metastasis.55,56 These results led to Ndrg1 being described as a metastasis suppressor gene.56 The finding that Ndrg1 expression is regulated by Fe is important in terms of understanding its poorly characterized role in cell cycle progression. Indeed, it is becoming clear that the G1/S arrest observed after Fe chelation25,41 is probably due to the effects of these agents on a variety of targets9,10,35-40 in addition to RR.10,17-19 The proper place of Ndrg1 in the antiproliferative activity of Fe chelators will eventually be defined in future studies. Certainly, the multiple molecular targets of Fe chelators may be beneficial in inducing antitumor activity.18,19,33,35-38
A prior investigation by others55 showed that DNA damage induced by Act D played an important role in up-regulating Ndrg1. Interestingly, p53 expression was also implicated in Ndrg1 regulation.55 Clearly, this would be important in terms of understanding cell cycle control. In our study, Act D had no effect on increasing Ndrg1 expression, despite that p53 was markedly up-regulated (Figure 3A-B). Moreover, in the p53-null cell line, H1299, Fe chelators markedly increased Ndrg1 expression (Figure 1D), demonstrating that a p53-independent pathway exists for up-regulating this molecule. This is an important observation, because in more than 50% of cancers p53 is nonfunctional.77 Hence, the finding that Ndrg1 can be up-regulated in tumor cells by Fe chelators by way of a p53-independent mechanism could be vital in terms of their ability to inhibit cancer growth and metastasis. Along with the inhibition of RR,10,18,19,22 the Fe-dependent up-regulation of Ndrg1 represents another molecular target of chelators.
Previous studies suggested that Ndrg1 up-regulation by Ni(II) or hypoxia may be mediated by HIF-1α.52-54 This finding is important, because Fe chelators up-regulate HIF-1α protein.78,79 Hence, Ndrg1 up-regulation by chelators may be due to HIF-1α protein expression, and we have identified potential HREs upstream of the promoter. Clearly, further studies are required to determine whether these are functional. However, our results using a HIF-1α-KO cells demonstrated that Ndrg1 up-regulation occurred by HIF-1α--dependent and -independent mechanisms following Fe chelation (Figure 7). The finding that Ndrg1 expression can be regulated by an HIF-1α-independent mechanism after Fe chelation is intriguing and has not been previously reported. Considering the possible mechanism, it can be speculated that another transcriptional process is involved, as high Act D concentrations prevented Ndrg1 up-regulation after Fe chelation (Figure 6). However, we cannot rule out that HIF-1α may act as a transcriptional enhancer, because Ndrg1 up-regulation was less marked for HIF-1α-KO cells when compared with their wild-type counterparts (Figure 7A).
The exact function of Ndrg1 remains uncertain, although the studies performed indicate a role in growth and differentiation.49,55,56,58 Considering the physiologic role of Ndrg1 upregulation by Fe deprivation, it can be suggested that its ability to inhibit proliferation may be by way of a mechanism induced by Fe depletion that arrests growth. Hence, together with inhibition of RR activity after Fe depletion,10,18,19,22 multiple molecular events respond to Fe deprivation to inhibit cell cycle progression.
As shown in our previous investigations, 311 and DFO induced a G1/S arrest25 and resulted in marked Fe deprivation,10,23-25,60 and in this study these chelators resulted in up-regulation of Ndrg1. Critically, one could suggest that the effect of the chelators at inducing Ndrg1 expression was simply due to a nonspecific stress response unrelated to Fe deprivation per se. However, this is not the case, because of the following lines of evidence. (1) Up-regulation of Ndrg1 after Fe chelation was reversed by Fe supplementation (Figure 5A) but not by supplementation with copper or zinc. (2) Iron complexes of the chelators that cannot bind cellular Fe pools had no effect on increasing Ndrg1 (Figure 4C). (3) As a negative control for the highly active chelator, dp44mT, we incubated cells with its analog, dp2mT, which has an inactivated Fe binding site that does not bind Fe.60 As shown in Figure 4D, in contrast to dp44mT, dp2mT did not up-regulate Ndrg1, indicating that Fe chelation was critical. (4) Incubation of cells with potent DNA-damaging and stress-inducing reagents that inhibited proliferation such as Act D, mitomycin C, or cisplatin, had no effect on increasing Ndrg1 expression (Figure 1E). Collectively, these findings clearly demonstrate that the up-regulation of Ndrg1 is due to cellular Fe-depletion and is not a nonspecific stress response.
In conclusion, this is the first study to demonstrate that intracellular Fe levels regulate the expression of the metastasis suppressor gene, Ndrg1.56 Considering the role of Ndrg1 in inhibiting tumor growth and metastasis,55,56,58 the ability of a chelator to markedly up-regulate its expression could be important in the cell cycle arrest observed after Fe chelation. Furthermore, our studies suggest Ndrg1 is a novel link between intracellular Fe levels and cellular proliferation.
Prepublished online as Blood First Edition Paper, July 13, 2004; DOI 10.1182/blood-2004-05-1866.
Supported by a fellowship and project grant from the National Health and Medical Research Council of Australia.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
Children's Cancer Institute Australia for Medical Research is affiliated with the University of New South Wales and Sydney Children's Hospital. We thank Dr Richard Lock and members of the Iron Metabolism and Chelation Program for their comments on the manuscript prior to submission.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal