Abstract
Transferrin receptor 2 (TfR2) is a membrane glycoprotein that mediates cellular iron uptake from holotransferrin. Homozygous mutations of this gene cause one form of hereditary hemochromatosis in humans. We recently reported that homozygous TfR2(Y245X) mutant mice, which correspond to the TfR2(Y250X) mutation in humans, showed a phenotype similar to hereditary hemochromatosis. In this study, we further analyzed the phenotype as well as iron-related gene expression in these mice by comparing the TfR2-mutant and wild-type siblings. Northern blot analyses showed that the levels of expression of hepcidin mRNA in the liver were generally lower, whereas those of duodenal DMT1, the main transporter for uptake of dietary iron, were higher in the TfR2-mutant mice as compared to the wild-type siblings. Expression of hepcidin mRNA in the TfR2 mutant mice remained low even after intraperitoneal iron loading. In isolated hepatocytes from both wild-type and TfR2 mutant mice, interleukin-6 and lipopolysaccharide each induced expression of hepcidin mRNA. These results suggest that up-regulation of hepcidin expression by inflammatory stimuli is independent of TfR2 and that TfR2 is upstream of hepcidin in the regulatory pathway of body iron homeostasis. (Blood. 2005;105:376-381)
Introduction
Hereditary hemochromatosis (HH) is a group of genetic disorders that manifest iron deposition in a variety of organs such as the liver, pancreas, heart, and skin. If untreated, liver cirrhosis, heart failure, and diabetes can develop. Most HH is caused by mutations in the HFE gene.1 The frequency of homozygous C282Y mutation of this gene is estimated to be 1 in 150 in people of northern European descent,2 though its clinical penetrance is low.3 Mutations in several other genes also produce an HH phenotype, including hemojuvelin (HFE2/HJV),4 hepcidin (hepatic antimicrobial peptide: HAMP),5 and transferrin receptor 2 (TfR2).6 The phenotypes caused by mutations of these genes are similar, manifesting increased transferrin (Tf) saturations, periportal hepatic iron loading, and reticuloendothelial iron sparing. This observation suggests that the products of these genes are on a common pathway for regulation of iron homeostasis.
TfR2 protein is a membrane glycoprotein that can interact with Tf.7 Human TfR2 has at least 2 alternatively spliced transcripts, α and β. TfR2-α is the membrane-bound form predominantly expressed in the liver, whereas TfR2-β is a form that consists of only the extracellular domain of TfR2-α. Similar to TfR1, TfR2-α interacts with holo-Tf but not with apo-Tf at neutral pH.8 Expression of TfR2 mRNA almost exclusively occurs in the liver and erythroid precursor cells.9 Homozygosity for one of several mutations in the TfR2 gene, including the truncation mutation Y250X, has been associated with hereditary hemochromatosis in humans.6 In addition, we recently reported that homozygous TfR2(Y245X) mutant mice, which correspond to the TfR2(Y250X) mutation in humans, showed hepatocellular iron deposition with elevated serum Tf saturations.10 However, the mechanism by which mutations of TfR2 cause hemochromatosis is poorly understood. In this study, we further analyzed the phenotype as well as iron-related gene expression in the TfR2 mutant mice to clarify the role of TfR2 in both iron homeostasis and hematopoiesis.
Materials and methods
Animal experiments
TfR2Y245 mutant mice were generated as previously described (the genetic background of these mice is a mixture of 129/SV and C57 BL/6J).10 To minimize the effect of heterogeneity of genetic background and environmental factors, we mated TfR2 (m/+) mice to generate all 3 genotypes (TfR2 +/+, m/+, and m/m) and used the same gender sibling mice at the same ages for each experiment. Offspring were genotyped by polymerase chain reaction (PCR) analysis of tail genomic DNA.10 The litters were weaned at 3 weeks and fed with a standard diet (Purina Mouse Chow, product code no. 5015, iron content, 223.4 ppm; Purina LabDiet, St. Louis, MO). For iron-loading experiments, 4-month-old either wild-type or homozygous TfR2 Y245X female sibling mice were injected with either 100 μL phosphate-buffered saline (PBS) or 5 mg iron-dextran in 100 μL PBS intraperitoneally twice a week for 3 weeks. One week after the last injection, the mice were killed and the liver, spleen, and duodenum were removed from each mouse for histologic as well as gene expression analyses. To verify the results, 6-month-old male sibling mice (wild-type, heterozygous or homozygous TfR2 Y245X) also were treated with the same procedure. For colony assay, bone marrow cells were collected from the femurs of 2-month-old sibling mice that were either wild-type or homozygous TfR2 Y245X. Animal experiments were performed in accordance with the protocol approved by Cedars-Sinai Medical Center Institutional Animal Care and Use Committee.
Isolation and culture of hepatocytes
The livers of either the wild-type or homozygous TfR2 Y245X female mice (2 months old) were perfused and digested in situ with collagenase, and the dissociated cells were collected in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. Then the hepatocytes were separated from Kupffer cells by centrifugation at 50 g (400 rpm) for 2 minutes followed by 2 washes with the same medium (in situ 2-step liver perfusion method of Seglen).11 The isolated hepatocytes were cultured for 48 hours in the same media supplemented with 5% fetal bovine serum in either the presence or absence of either 15 μM desferrioxamine (DFO), 200 μg/mL human holo-Tf (Sigma-Aldrich, St Louis, MO), 100 ng/mL lipopolysaccharide (LPS), or 20 ng/mL recombinant human interleukin-6 (interleukin-6 [IL-6], Calbiochem, San Diego, CA).
Serum Tf saturations
Serum iron and total iron-binding capacity were measured by using a commercial kit (Sigma-Aldrich). Tf saturation was calculated as (serum iron/total iron-binding capacity) × 100%.
Histologic examination
Tissues were fixed in 10% neutral buffered formalin for 18 hours, subjected to routine histologic processing, and the sections were stained with hematoxylin and eosin. The sections also were stained by the Perl Prussian blue method for the detection of iron storage.
Colorimetric quantitation of cellular iron
Cellular iron was quantified colorimetrically by the method previously described with some modification.10 Briefly, cell lysate was digested overnight in 200 μL 3 M HCl/10% trichloroacetic acid at 65°C, followed by the addition of 400 μL chromagen (0.01% bathophenanthroline sulfonate, 0.1% thioglycolic acid, and 4 M sodium acetate). For a standard curve, serial dilutions of a ferric iron standard (Sigma-Aldrich) were used. Color was allowed to develop for 15 minutes and measured as absorbance at 535 nm. Protein concentrations were determined by using a modified Bradford reagent (Bio-Rad, Hercules, CA).
Northern blot analysis
Total RNA was isolated from the liver and duodenum of mice using Trizol Reagent (Invitrogen). The RNA from each source was denatured in formaldehyde-containing buffer and electrophoresed in 1% agarose/2.2% formaldehyde gel. The RNA was transferred to nylon membranes and immobilized by UV cross-linking. The blots were hybridized with 32P-labeled cDNA probes (murine hepcidin and β-actin) using UrtraHyb buffer (Ambion, Austin, TX) and autoradiographed. The intensity of each band was measured by AlphaImager 2000 Imaging System (Alpha Innotech, San Leandro, CA) with a standard curve made from an autoradiograph of serially diluted 32P-dATP dot blots.
Anti-TfR2 antibody
Maltose binding protein-TfR2-β fusion protein was made using pMAL Protein Fusion and Purification System (New England BioLabs, Beverly, MA). Polyclonal antibody against TfR2 was developed by immunizing a rabbit with the maltose binding protein-TfR2-β fusion protein, followed by purification with a protein-A column (Takara Bio, Otsu, Japan). The specificity of this antibody was confirmed by immunohistochemistry and immunoblotting using CHO-TRVb-Neo, CHO-TRVb-TfR1, and CHO-TRVb-TfR2-α cells7 (only CHO-TRVb-TfR2-α cells were strongly positive, while the others were negative, data not shown).
Western blot analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed under reducing conditions, and the proteins were transferred onto poly(vinylidene difluoride) membranes by electroblotting. These membranes were reacted with 1:1000 dilution of primary antibodies (rabbit anti-human TfR2 and rabbit anti-human ferritin [DAKO, Glostrup, Denmark]), followed by a 1:20 000 dilution of peroxidase-conjugated anti-rabbit IgG (Amersham, Piscataway, NJ). Immunoreactive proteins were detected by using Super-Signal West Dura reagent (Pierce, Rockford, IL), followed by autoradiography.
Real-time polymerase chain reaction
Reverse transcriptase reaction was performed as described.12 Real-time PCR was conducted using HotMaster Taq DNA polymerase (Eppendorf, Hamburg, Germany) and iCycler iQ Multi-Color Real Time PCR Detection System (Bio-Rad). The following primer pairs were used: murine DMT1-exon 1A (5′-AAGCCAAACCAGTCCTGCACCATGG-3′ forward and 5′-TCCAACACCATGGTAGATAC-3′ reverse) and murine β-actin (5′-GCCAACACAGTGCTGTCTG-3′ forward and 5′-CTCAGGAGGAGCAATGATCT-3′ reverse). The fluorescence of Cyber green (PE Biosystems, Foster City, CA) was used for quantitation of amplicons. For DMT1-exon 1A, the PCR conditions were 94°C for 2 minutes, followed by 40 cycles of 94°C (20 seconds), 60°C (10 seconds), 65°C (25 seconds), and 87°C (20 seconds) for data collection. For β-actin, the temperature for data collection was changed to 83°C. Complimentary DNA fragments of murine DMT1 and β-actin were used to make standard curves. Experiments were performed in triplicate, and the mean values from each sample were used as their representatives. The ratios of copy numbers of DMT1 exon 1A and β-actin were calculated for each sample.
Clonogenic assay
Bone marrow cells were isolated aseptically from the femurs of wild-type (n = 4) and homozygous TfR2 mutant (n = 3) mice. For granulocyte macrophage colony-forming units (CFU-GMs) and erythroid burst-forming units (BFU-Es), 2 × 104 cells in methylcellulose (MethoCult; Stem Cell Technologies, Vancouver, BC, Canada) were plated into each well of 6-well plates in the presence of 3 U/mL recombinant human erythropoietin, 10 ng/mL interleukin-3, 10 ng/mL IL-6, and 50 ng/mL stem cell factor. After 7 days of culture, the number of myeloid (CFU-GM) and erythroid (BFU-E) colonies were counted under an inverted microscope. These experiments were done in triplicates, and the mean was taken as the representative value of each sample. For CFU-Meg, 2 × 106 cells in collagen gel (MegaCult-C, Stem Cell Technologies) were plated into each chamber-slide in the presence of 50 ng/mL recombinant human thrombopoietin, 10 ng/mL IL-3, 20 ng/mL IL-6, and 50 ng/mL IL-11. After 7 days of culture, the cells on the slides were fixed with acetone, stained for acetylcholinesterase, and examined under a microscope by following the manufacturer's instruction. This experiment was done in duplicate for each sample.
Statistical analysis
The Mann-Whitney test was used for determination of statistical significance when appropriate. A P value of less than .05 was considered statistically significant.
Results
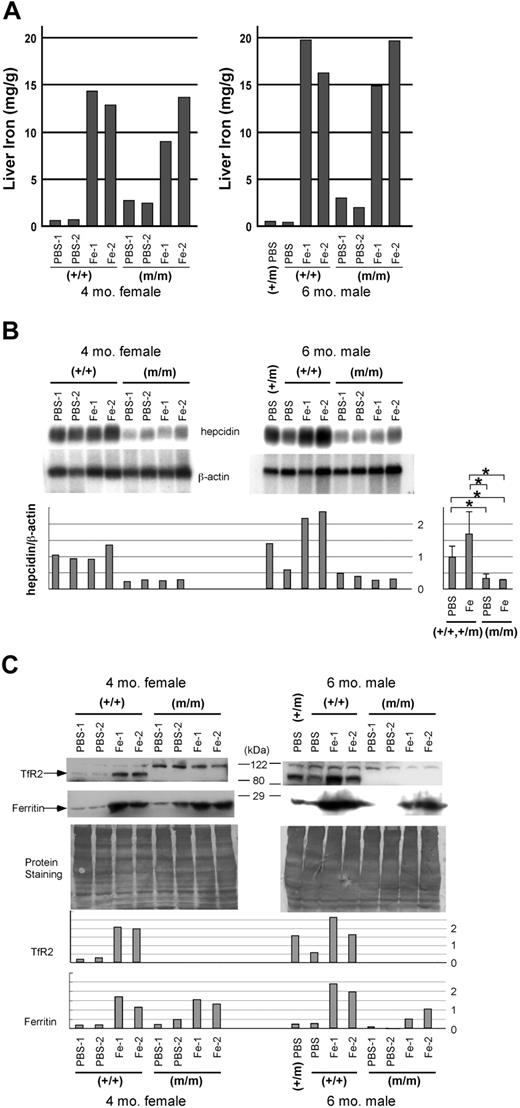
Homozygous TfR2 Y245X mutant and wild-type sibling mice (4-month-old females) were injected intraperitoneally with either 5 μg iron dextran or the same volume of PBS, twice a week for 3 weeks. TfR2 mutant mice showed nearly 100% serum Tf saturation levels irrespective of iron loading, as we previously reported.10 Iron-loaded wild-type mice also had nearly 100% serum Tf saturation levels. Among the mice receiving PBS, predominantly periportal iron deposition was observed in the livers of the TfR2 mutant mice, whereas almost no stainable iron was observed in the livers of wild-type siblings (Figure 1A-B, E-F). After iron loading by intraperitoneal injections, both the TfR2 mutant and wild-type mice showed marked iron deposition in their livers, especially in the Kupffer cells (Figure 1C-D, G-H). These results were consistent with the colorimetric measurement of hepatic iron (Figure 2A).
Histologic examination of the liver. Four-month-old wild-type and homozygous TfR2 Y245X mutant female sibling mice were injected with either PBS or 5 mg iron-dextran intraperitoneally twice a week for 3 weeks. One week after the last injection, the livers were subjected to histologic analysis. (A, E) The liver of a PBS-treated wild-type mouse. (B, F) The liver of a PBS-treated homozygous TfR2 Y245X mutant mouse. (C, G) The liver of an iron-loaded wild-type mouse. (D, H) The liver of an iron-loaded homozygous TfR2 Y245X mutant mouse. (A-D) Hematoxylin eosin staining. (E-H) Perl Prussian blue staining for the detection of iron storage. Original objective magnification of these panels was × 20.
Histologic examination of the liver. Four-month-old wild-type and homozygous TfR2 Y245X mutant female sibling mice were injected with either PBS or 5 mg iron-dextran intraperitoneally twice a week for 3 weeks. One week after the last injection, the livers were subjected to histologic analysis. (A, E) The liver of a PBS-treated wild-type mouse. (B, F) The liver of a PBS-treated homozygous TfR2 Y245X mutant mouse. (C, G) The liver of an iron-loaded wild-type mouse. (D, H) The liver of an iron-loaded homozygous TfR2 Y245X mutant mouse. (A-D) Hematoxylin eosin staining. (E-H) Perl Prussian blue staining for the detection of iron storage. Original objective magnification of these panels was × 20.
Iron content and expression of iron-related genes in the liver. Four-month-old wild-type (+/+) and homozygous TfR2 Y245X mutant (m/m) female sibling mice, as well as 6-month-old wild-type, heterozygous (+/m), and homozygous TfR2 Y245X mutant male murine siblings, were treated with either PBS or iron-dextran for 3 weeks. (A) Iron content in the liver. Iron content in the livers was measured using bathophenanthroline sulfonate as described in “Materials and methods.” (B) Northern blot analysis showing expression of hepcidin in the liver. The intensity of each band was measured by an image analyzer, and the ratio of the intensities of hepcidin and β-actin was calculated. Setting the mean value of the hepcidin/β-actin ratio in PBS-treated wild-type and heterozygous mice as 1, individual values were calculated and shown in a bar graph. The mean values of each group are compared and shown in a bar graph on the right (mean ± SD, *P = .021). (C) Western blot analysis showing expression of TfR2 and ferritin in the liver. Both the antihuman TfR2 and the antihuman ferritin antibodies cross-reacted to their murine counterparts. The bands around 120 kDa seen in the top panels are nonspecific. To show the loading balance, protein staining of the membranes also is shown. The intensity of each band was measured by an image analyzer and shown in bar graphs.
Iron content and expression of iron-related genes in the liver. Four-month-old wild-type (+/+) and homozygous TfR2 Y245X mutant (m/m) female sibling mice, as well as 6-month-old wild-type, heterozygous (+/m), and homozygous TfR2 Y245X mutant male murine siblings, were treated with either PBS or iron-dextran for 3 weeks. (A) Iron content in the liver. Iron content in the livers was measured using bathophenanthroline sulfonate as described in “Materials and methods.” (B) Northern blot analysis showing expression of hepcidin in the liver. The intensity of each band was measured by an image analyzer, and the ratio of the intensities of hepcidin and β-actin was calculated. Setting the mean value of the hepcidin/β-actin ratio in PBS-treated wild-type and heterozygous mice as 1, individual values were calculated and shown in a bar graph. The mean values of each group are compared and shown in a bar graph on the right (mean ± SD, *P = .021). (C) Western blot analysis showing expression of TfR2 and ferritin in the liver. Both the antihuman TfR2 and the antihuman ferritin antibodies cross-reacted to their murine counterparts. The bands around 120 kDa seen in the top panels are nonspecific. To show the loading balance, protein staining of the membranes also is shown. The intensity of each band was measured by an image analyzer and shown in bar graphs.
Next, we analyzed expression of hepcidin in the livers of mice from each of the 4 groups. Northern blot analysis showed that expression of hepcidin mRNA was markedly lower in the TfR2 mutant mice as compared with wild-type littermates. We observed modest up-regulation of hepcidin mRNA in the wild-type mice after iron loading, but no up-regulation was observed in the TfR2 mutant mice even after massive intraperitoneal iron loading (Figure 2B). On Western analysis of the livers, expression of TfR2 was higher in iron-loaded wild-type mice as compared to either PBS-treated wild-type or heterozygous mutant littermates (Figure 2C). No TfR2 protein expression was observed in the TfR2 mutant mice. Up-regulation of ferritin was observed in the iron-treated mice irrespective of their genotypes.
To investigate the mechanism of regulation of hepcidin expression in the liver, we isolated hepatocytes from both TfR2 mutant and wild-type littermates and cultured these cells in the presence of DFO, holo-Tf, IL-6, or LPS for 48 hours. Irrespective of the genotype, IL-6 and LPS up-regulated hepcidin mRNA, while neither DFO nor holo-Tf clearly changed the levels of expression of hepcidin mRNA (Figure 3).
Expression of hepcidin mRNA in isolated hepatocytes. Hepatocytes from either wild-type (+/+) or homozygous TfR2 Y245X mutant (m/m) mice were cultured for 48 hours in culture medium alone (control), DFO, iron-saturated Tf, IL-6, or LPS; their RNA was extracted and Northern blotted using murine hepcidin and β-actin probes.
Expression of hepcidin mRNA in isolated hepatocytes. Hepatocytes from either wild-type (+/+) or homozygous TfR2 Y245X mutant (m/m) mice were cultured for 48 hours in culture medium alone (control), DFO, iron-saturated Tf, IL-6, or LPS; their RNA was extracted and Northern blotted using murine hepcidin and β-actin probes.
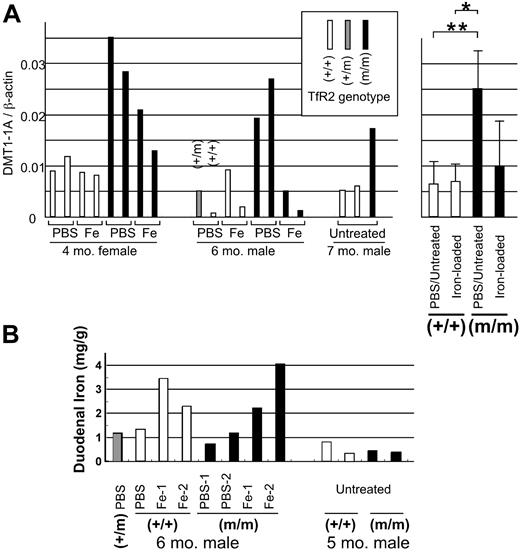
Next, we checked expression of the iron-uptake molecule DMT1 (divalent metal transporter 1; SLC11A2) in the duodenum. At least 4 different transcripts are known for DMT1: one with both exon 1A and an iron-responsive element (IRE), a second with exon 1A without IRE, a third without exon 1A with IRE, and a fourth without exon 1A or IRE. The form most predominantly expressed in the duodenal enterocytes contains both exon 1A and IRE.13 Besides enterocytes, the duodenum contains macrophages, muscles, and other types of cells. We quantified expression of duodenal enterocyte-specific DMT1 transcripts by real-time PCR, using primers specific for the exon 1A isoform. Expression of the DMT1 exon 1A transcript in control (PBS-treated or untreated) TfR2-mutant mice was higher as compared to the levels in the control wild-type mice throughout the 3 sets of sibling mice that we examined (Figure 4A). When combined, these data indicated that this mRNA transcript isoform of DMT1 was increased approximately 3.8-fold in the TfR2 mutant compared with wild-type mice (P = .009). Because the duodenal form of DMT1 was reported to be up-regulated in iron deficiency,14,15 we examined the duodenal iron content in these mice. As seen in Figure 4B, iron-treated mice showed higher iron contents as compared to the mice treated with PBS. In histochemistry, all the stainable iron in the duodenum of these mice was present in macrophages, with no detectable iron in the enterocytes (data not shown). No clear difference was noted between wild-type and homozygous TfR2 Y245X mice in respect to duodenal iron contents or its distribution.
Expression of iron transporter DMT1 and iron content in the duodenum. (A) Four-month-old wild-type (+/+) and homozygous TfR2 Y245X mutant (m/m) female sibling mice were treated with either PBS or iron-dextran for 3 weeks, and expression of the duodenum-specific DMT1 exon 1A transcript was measured by real-time quantitative PCR. Mice used: first group, 4-month-old wild-type and homozygous TfR2 Y245X mutant female sibling mice; second group, 6-month-old wild-type, heterozygous (+/m), and homozygous TfR2 Y245X male siblings. These mice were treated with either PBS or iron-dextran. The third group was composed of 7-month-old wild-type and homozygous TfR2 Y245X mutant mice. (C) Numbers of both DMT1 exon 1A and β-actin transcripts were quantified, and the ratio of these transcripts was calculated for each sample. Open, shaded, and closed rectangles represent wild-type, heterozygous, and homozygous TfR2 Y245X mutant mice, respectively. The mean values of each group are compared and shown in a bar graph on the right (mean ± SD, *P = .014, **P = .009). (B) Iron content in the duodenum. Mice used: first group, 6-month-old wild-type, heterozygous, and homozygous TfR2 Y245X male siblings treated with either PBS or iron-dextran; the second group: 5-month-old wild-type and homozygous TfR2 Y245X mutant mice.
Expression of iron transporter DMT1 and iron content in the duodenum. (A) Four-month-old wild-type (+/+) and homozygous TfR2 Y245X mutant (m/m) female sibling mice were treated with either PBS or iron-dextran for 3 weeks, and expression of the duodenum-specific DMT1 exon 1A transcript was measured by real-time quantitative PCR. Mice used: first group, 4-month-old wild-type and homozygous TfR2 Y245X mutant female sibling mice; second group, 6-month-old wild-type, heterozygous (+/m), and homozygous TfR2 Y245X male siblings. These mice were treated with either PBS or iron-dextran. The third group was composed of 7-month-old wild-type and homozygous TfR2 Y245X mutant mice. (C) Numbers of both DMT1 exon 1A and β-actin transcripts were quantified, and the ratio of these transcripts was calculated for each sample. Open, shaded, and closed rectangles represent wild-type, heterozygous, and homozygous TfR2 Y245X mutant mice, respectively. The mean values of each group are compared and shown in a bar graph on the right (mean ± SD, *P = .014, **P = .009). (B) Iron content in the duodenum. Mice used: first group, 6-month-old wild-type, heterozygous, and homozygous TfR2 Y245X male siblings treated with either PBS or iron-dextran; the second group: 5-month-old wild-type and homozygous TfR2 Y245X mutant mice.
Since expression of TfR2 mRNA has been observed in early erythroid cells, including CD34-positive populations in our previous study,9 we examined hematopoietic parameters of these mice. Consistent with our former report, no significant difference was found in differential cell counts in either the bone marrow or peripheral blood cells between the TfR2 mutant mice and their wild-type littermates.10 Colony assays for bone marrow CFU-GM, BFU-E, and CFU-Meg (megakaryocytic colony forming unit) showed no significant differences between the homozygous TfR2 mutant and wild-type mice (data not shown).
Discussion
Recent evidence indicates that hepcidin, an antimicrobial peptide expressed predominantly in the liver, plays a central role in iron homeostasis in the body.16 Expression of hepcidin in the liver increases in response to iron overload,17 and the expression inversely correlates with duodenal expression of IREG-1 and DMT1 as well as iron absorption from the intestine.18 Transgenic overexpression of hepcidin prevents iron deposition in the liver caused by a homozygous mutation of HFE in mice.19 Furthermore, a hepcidin-producing tumor accompanied by severe iron-refractory microcytic anemia has been reported.20 With removal of this tumor, the hematocrit level reverted to normal. These findings strongly suggest that hepcidin reduces the amount of iron usable by the erythroid cells in the body. Perhaps hepcidin acts in part through down-regulation of either function or expression of genes involved in intestinal iron transport,21 though the mechanism is still unclear.
Inappropriately low expression of hepcidin in the liver was reported in patients with HH type 1 (HFE-mutations) as well as HFE-defective mice,22-24 suggesting that HFE is upstream of hepcidin on the pathway of iron homeostasis of the body. Similarly, in patients with HFE2 (hemojuvelin)-associated juvenile HH, urinary hepcidin levels were decreased, suggesting that hemojuvelin also is upstream of hepcidin.4 In the current study, we demonstrated that homozygous TfR2 mutant mice showed inappropriately low levels of expression of hepcidin mRNA in the liver, indicating that TfR2 also is located upstream of hepcidin in the pathway. The possibility that the regulatory pathways, which include HFE, TfR2, and hemojuvelin, converge on hepcidin may explain why mutations of these genes manifest a similar hemochromatosis phenotype.1,4,6,25
In HH with either HFE or TfR2 mutations, hepatic iron overload is generally mild, and most individuals with these mutations show no clinical symptoms. On the other hand, clinically significant iron overload can occur iatrogenically by repeated blood transfusion or inappropriate intravenous iron administration, even without any genetic background of HH. In this study, to simulate these clinical situations and to present a large excess of iron, we injected iron intraperitoneally into either wild-type or TfR2 mutant mice and analyzed the pattern of iron deposition as well as expression of hepcidin in the livers. We observed periportal iron deposition in the PBS-treated TfR2 mutant mice, whereas we observed massive iron deposition predominantly in the Kupffer cells in the iron-injected wild-type and TfR2 mutant mice. Conceivably, our intraperitoneal iron loading was so large that excess iron, which hepatocytes did not have the capacity to keep, accumulated in Kupffer cells. Pigeon et al17 reported a 4- to 6-fold increase of hepcidin mRNAin the liver with oral iron loading, but our results (Figure 2A) showed only modest up-regulation of hepcidin mRNA after intraperitoneal administration of iron. Perhaps expression of hepcidin in our wild-type mice was already high, nearly saturated, with the regular diet. A similar finding was mentioned in a recent report by Nemeth et al.26 Probably, hepatic expression of hepcidin can be saturated easily with mild oral loading of iron. In contrast, levels of expression of hepcidin mRNA remained low in the TfR2 mutant mice even after massive iron loading (Figure 2), indicating an essential role of TfR2 for the up-regulation of hepcidin induced by iron.
Studies on the distribution of hepcidin and TfR2 in mice and in humans have demonstrated that both genes are predominantly expressed in the liver.7,17,27-29 Gehrke et al30 found a strong correlation between expression of hepcidin and TfR2 in the liver from biopsy samples, irrespective of the underlying disease or the iron status. Serum Tf saturation is closely correlated with expression of hepcidin in the liver in vivo.30 Possibly, the interaction of holoTf with TfR2 in the hepatocyte serves to mediate this relationship. The results shown on Figure 2 demonstrated that expression of both hepcidin mRNA and TfR2 protein was up-regulated by iron loading in wild-type mice; the latter finding is consistent with the observation by Deaglio et al31 that iron-saturated Tf up-regulated TfR2 protein in a hepatoma cell line in vitro. Similarly, ligand-associated up-regulation of their own receptors has been reported for some growth factors, such as epidermal growth factor,32 erythropoietin,33 and granulocyte colony-stimulating factor.34 These findings suggest that TfR2 may function as a sensor of iron-saturated Tf in the liver. However, when isolated hepatocytes or hepatoma cells were used in vitro, iron-saturated Tf did not up-regulate expression of hepcidin mRNA, as shown in Figure 3 and by others.22,30 Up-regulation of hepcidin by iron-saturated Tf may require an additional factor, other than TfR2, that is absent in these in vitro systems.
Expression of hepcidin also increases by inflammatory stimuli including IL-6 and LPS.17 Microcytic anemia, sometimes indistinguishable from that observed in iron deficiency, is a common feature of Castleman disease, multiple myeloma, and chronic inflammations such as rheumatoid arthritis. Among patients with these diseases, elevated levels of serum IL-6 are frequently observed. Studies have reported that IL-6 up-regulates expression of hepcidin,22,30 and our results shown in Figure 3 are consistent with these results. The increased hepcidin may reduce both iron absorption through the duodenum and iron release from macrophages, causing microcytic anemia similar to iron-deficiency anemia (for a review, see Ganz16 ). Isolated hepatocytes from homozygous TfR2 Y245X mutant mice also responded to both IL-6 and LPS (Figure 3), indicating that up-regulation of hepcidin by these inflammatory stimuli does not require expression of TfR2, which is consistent with studies by Lee et al.35 Another recent report by Nemeth et al demonstrated that induction of hepcidin by LPS could be blocked by anti-IL-6 antibody, suggesting that the effect of LPS on expression of hepcidin was dependent on IL-6.26
Liver is one of the candidate organs that may sense iron in the body. In the liver, TfR2, together with HFE or hemojuvelin, may sense iron-Tf and affect expression or release of hepcidin. Hepcidin may regulate dietary iron absorption from the intestine. Recently, Yamaji et al36 reported that hepcidin significantly decreased apical iron uptake by Caco-2 cells, which was accompanied by a decrease in both DMT1(+IRE) protein and mRNA expression. In this scenario, the mutation of TfR2 down-regulates expression and/or release of hepcidin, which may cause up-regulation of the absorption of dietary iron through up-regulation of duodenal iron transporter DMT1. Actually, we showed in the present study that mRNA expression of the duodenum-specific form of DMT1 that contains exon 1A was higher in TfR2 mutant mice when compared to the wild-type littermates (Figure 4A-B). Alternatively, TfR2 may function as a sensor of iron-Tf in the duodenal crypt cells. Trinder et al37 reported that Tf-bound iron uptake from plasma was impaired in the duodenum of HFE-defective mice, implicating involvement of HFE in iron sensing in the duodenum. Although West et al38 found no detectable interaction between HFE and TfR2 in vitro, Griffiths and Cox39 recently demonstrated colocalization of HFE and TfR2 in human duodenal crypt cells in vivo. So, the possibility exists that HFE interacts with TfR2, either directly or indirectly, to cooperate in the sensing of iron in the duodenum.
The underlying mechanisms of maintaining body iron homeostasis are still largely unclear. Which is the main iron-sensing organ, the liver, duodenum, reticuloendothelial system, bone marrow erythroid cells, or others? How does TfR2, together with HFE and hemojuvelin, regulate expression and/or release of hepcidin? How does hepcidin affect the expression and/or function of the duodenal iron transporter DMT1? The precise roles of TfR2 in regulation of iron homeostasis remain to be clarified.
Prepublished online as Blood First Edition Paper, September 2, 2004; DOI 10.1182/blood-2004-04-1416.
Supported in part by grants from the National Institutes of Health, C. and H. Koeffler Fund, Parker Hughes Trust, and a grant-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
H. P. K. holds the Mark Goodson endowed Chair of Oncology and is a member of the Jonsson Cancer Center and the Molecular Biology Institute of UCLA.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal